Abstract
In the majority of patients with Pelizaeus-Merzbacher disease, duplication of the proteolipid protein gene PLP1 is responsible, whereas deletion of PLP1 is infrequent. Genomic mechanisms for these submicroscopic chromosomal rearrangements remain unknown. We identified three families with PLP1 deletions (including one family described elsewhere) that arose by three distinct processes. In one family, PLP1 deletion resulted from a maternal balanced submicroscopic insertional translocation of the entire PLP1 gene to the telomere of chromosome 19. PLP1 on the 19qtel is probably inactive by virtue of a position effect, because a healthy male sibling carries the same der(19) chromosome along with a normal X chromosome. Genomic mapping of the deleted segments revealed that the deletions are smaller than most of the PLP1 duplications and involve only two other genes. We hypothesize that the deletion is infrequent, because only the smaller deletions can avoid causing either infertility or lethality. Analyses of the DNA sequence flanking the deletion breakpoints revealed Alu-Alu recombination in the family with translocation. In the other two families, no homologous sequence flanking the breakpoints was found, but the distal breakpoints were embedded in novel low-copy repeats, suggesting the potential involvement of genome architecture in stimulating these rearrangements. In one family, junction sequences revealed a complex recombination event. Our data suggest that PLP1 deletions are likely caused by nonhomologous end joining.
Introduction
The proteolipid protein gene (PLP1) is a dosage-sensitive gene located on chromosome Xq22.2. An extra copy of the PLP1 gene, resulting from large genomic duplications containing the entire gene, affects development of the oligodendrocytes in the CNS and results in a dysmyelinating disease, Pelizaeus-Merzbacher disease (PMD [MIM 312080]; Ellis and Malcolm 1994; Inoue et al. 1996, 1999). PMD is characterized by arrest of oligodendrocyte differentiation and failure to produce myelin in the CNS, resulting in developmental delay recognized from the first year of life in most patients (Hudson 2001). Patients with PMD also present with additional neurological features, including nystagmus, pyramidal and extrapyramidal signs, and cerebellar symptoms. Although PLP1 duplications account for the majority of PMD (60%–70% of patients), sequence alterations in the coding regions and splice site junctions of PLP1 also cause PMD (∼20%) (Garbern et al. 1999). Deletion of PLP1 can also result in PMD, although only one family has been described to date (Raskind et al. 1991).
Molecular characterization of PLP1 duplication revealed that unequal sister-chromatid exchange is probably the mechanism for genomic rearrangement (Woodward et al. 1998; Inoue et al. 1999). During such recombination events in germ cells, reciprocal deletion of the same genomic segments may be generated. However, in a clinical setting, PLP1 deletion has been observed infrequently compared with duplication. Also, in PMD, the sizes of the duplicated genomic segments show extensive variability, distinct from other genomic disorders involving duplication, such as Charcot-Marie-Tooth disease type 1A and dup(17)(p11.2p11.2) (Stankiewicz and Lupski 2002a, 2002b). These observations suggest that the mechanisms for PLP1 genomic rearrangements may differ from nonallelic homologous recombination (NAHR) at paralogous low-copy repeats (LCRs) as shown for recurrent genomic disorders (Inoue and Lupski 2002; Stankiewicz and Lupski 2002a, 2002b).
We studied three independent families with PLP1 deletions, including one family described elsewhere (Raskind et al. 1991). Analyses in family members demonstrated that the deletions arose by three distinct processes: (i) unbalanced inheritance of an insertional translocation, (ii) sister-chromatid exchange in male meiosis, and (iii) complex rearrangement, but each appeared to occur by nonhomologous end joining (NHEJ). Genomic characterization of the deleted segments and recombination products revealed the molecular mechanisms for the genomic rearrangements resulting in the deletions and provided a potential explanation for the infrequent observation of PLP1 deletion.
Patients and Methods
Family HOU542
Patient BAB1379 is a 10-year-old Japanese son of nonconsanguineous parents. There were no problems during the pregnancy and delivery. Nystagmus was noted at the age of 1 mo but later disappeared. Delay in his motor development was noted at 6 mo of age. He rolled over at 12 mo, held his head at 15 mo, and sat at 24 mo of age. Spasticity became evident, particularly in his lower extremities, at 18 mo. He could walk with support at age 3 years, but subsequently his motor function gradually declined. T2-weighted brain MRIs revealed symmetric, diffuse high intensity in the cerebral white matter, suggesting dysmyelination. Brainstem auditory evoked potentials were abnormal. He had normal results in laboratory investigations, including normal 46,XY G-banded chromosomes. He has mildly reduced nerve-conduction velocities (36 m/s in median nerve), with no apparent features of peripheral neuropathies, although this could be masked by severe spasticity and muscle atrophy secondary to disuse. At present, he is wheelchair-bound because of the prominent spasticity. He feeds himself with assistance but requires full support for most of his daily care. He has a healthy male sibling (BAB1380).
His mother (BAB1381) presented with progressive difficulty in walking from her third decade. Subsequently, spasticity and changes in her personality were noted. Mental deterioration was gradually apparent. Because of the worsening spasticity in her lower extremities, she is wheelchair-bound. Her head MRI showed abnormal changes in the cerebral white matter. Results of her laboratory investigations and G-banded chromosome analysis were normal.
Family HOU669
Patient BAB1684, a 10-year-old son of unrelated white parents, was born at term after an uneventful pregnancy and delivery. By 12 mo of age, he showed global developmental delay. At 18 mo, he was still unable to sit unsupported or roll over and had no intelligible speech; however, he was sociable and enjoyed good general health. He showed generalized spasticity, particularly in the lower extremities. No history of abnormal eye movements was noted. Over the next few years he became able to speak in sentences, although he was dysarthric. At age 5 years, brain MRI showed delayed myelination throughout the cerebrum; results of nerve-conduction studies and electroencephalography were normal. He was never able to walk unsupported, and by age 7 years, he was confined to a wheelchair because of severe spasticity. His speech showed marked dysarthria and slowing. He could feed himself without difficulty in swallowing. No involuntary movement or tremor was noted. Follow-up MRI revealed progressive abnormalities in cerebral white matter, particularly marked in the internal capsules and periventricular and subcortical regions. At age 10 years, brainstem auditory and somatosensory evoked potentials were abnormal, and peripheral nerve-conduction velocities were at the lower limit of normal (46.9 m/s). By this age, he was totally dependent on others for feeding and personal hygiene. An ophthalmoscopic examination revealed marked optic atrophy. He has never experienced any seizures.
His 32-year-old mother (BAB1699) denied any significant medical problems. However, neurological examination revealed bilateral pes cavus deformities of the feet, increased deep tendon reflexes, and mildly increased muscle tone in the lower extremities, although Babinski reflexes were negative. She was unable to perform tandem gait. The remainder of the examination was unremarkable. Patient BAB1684 has a healthy female sibling (BAB1701).
Family PMD1
This family was originally described by Raskind et al. (1991). We obtained lymphoblastoid cell lines from two patients (H142 and H152) and a carrier female (H150), which were used for molecular studies.
Interphase and Metaphase FISH
Informed consent was obtained from each patient and family member. Harvested lymphoblastoid cells established from patients and family members were dropped on a glass slide for interphase and metaphase FISH analyses, as described elsewhere (Shaffer et al. 1997). Probes used in the interphase FISH include PLP1 (cosmid c125A1), DXS8096 (RP1-34H10), DXS8075 (RP1-81E11), and the BTK intrachromosomal control probe (RP1-39B6), as described elsewhere (Inoue et al. 1999). For metaphase FISH probes, we used a chromosome 19ptel–specific probe (Vysis) and PLP1 (cosmid c125A1).
Southern Hybridization
We digested 5 μg of genomic DNA from family members of HOU542 with each of the following restriction endonucleases: BamHI, PstI, HindIII, and XbaI, electrophoresed on 1% agarose gel and transferred to a nylon membrane after denaturation. Probe u35G3.20K was obtained by PCR from genomic sequences of the proximal boundary of the deleted region (primer u35G3.20K-U, 5′-GGCTGGGTCTCTTTTTCTAC-3′; and u35G3.20K-L, 5′-GGGGACAATGATGCTTACGA-3′) and used for hybridization.
PCR Amplification of the PLP1 Exons and STS Markers
Genomic DNA was extracted from peripheral blood cells and/or lymphoblastoid cells. Each exon of the PLP1 gene was amplified, with these genomic DNAs as the template (Osaka et al. 1999). We used STSs adjacent to the PLP1 locus, including DXS8096, SG45649, SG45650, DXS1191, and DXS8075, for STS-content mapping of the deletion. Additional primers for STSs were identified from genome mapping and sequence databases as well as PAC end-sequencing analyses, to further narrow down the recombination breakpoints by STS-content mapping (Tp-A, 5′-CCACTCCCTTTCTGCTTCACTGCTC-3′ and 5′-GGTCCTGGCAAACCCTTTCATCAGC-3′; Tp-B, 5′-CCAATGCAAAGACCAACACT-3′ and 5′-GGAGCAGAAAGAAACTATCA-3′; Td-A, 5′-TGTTGACAAGGCTTCAGTAT-3′ and 5′-AGGCACTTTTTAGTTAGGAG-3′; Td-B, 5′-GTCCTCAATGCTGTAATCCC-3′ and 5′-GAAATCCAATTAAGTTCTGTATTT-3′; Gp-A, 5′-GAGATTAAGCCATTTTCCAT-3′ and 5′-GCTTTTACATGACCAGACTA-3′; Gp-B, 5′-GCTCTGTAAGGCTAAATGTT-3′ and 5′-TGAACTTGGGCTGGTGGTAT-3′; Gd-A, 5′-CCAACATCACTTATTCACCA-3′ and 5′-CCACTTCTCACCCATCTCAG-3′; Gd-B, 5′-CTGGAACTTGGGAGGTGACC-3′ and 5′-GGCAAGAAAGGGACTGACTG-3′; Wp-A, 5′-TTAGTTGCCTGCCCTGATGA-3′ and 5′-TCCTTCTGCCCTCTGTGTGG-3′; Wp-B, 5′-CCAGAAAAGGGTCAGAGAGG-3′ and 5′-TGGAGCAAGCAGAACAAATG-3′; Wd-A, 5′-CTGGAACTTGGGAGGTGACC-3′ and 5′-GCTGTGACCGTTTCTTCATT-3′; Wd-B, 5′-TAATGCAGCTCAAAGGAAAG-3′ and 5′-CAGGGACATAAATCTCAATC-3′). The PCR products were electrophoresed on 2% agarose gels, and ethidium bromide staining was performed for visualization.
X-Inactivation Studies
We tested for random X inactivation, using a PCR-based assay with slight modifications (Allen et al. 1992). Genomic DNA was digested with the methylation-sensitive restriction endonuclease HpaII and was amplified with a set of fluorescently labeled primers from the androgen-receptor gene (AR) (Allen et al. 1992). PCR products were analyzed by an ABI 377 automated sequencer, with Genescan software (Applied Biosystems) for haplotyping and signal quantification.
Haplotype Analysis
We used four fluorescence-labeled primers for STR markers—DXS8096, CA-PLP (Mimault et al. 1995), DXS1191, and DXS8075—to determine the haplotype of family members from HOU669. To examine recombinations within an interval between AR and PLP1, we used seven STR markers that span these two genes (DXS991-AR-DXS986-DXS990-DXS8077-DXS8020-DXS1106-PLP1-DXS8075). PCR products were separated by electrophoresis, using an ABI 377 (Applied Biosystems), and were analyzed by the appropriate software, Genescan and Genotyper (Applied Biosystems).
Degenerate Oligonucleotide Primer (DOP) PCR for Junction Fragment Cloning
We used DOP-PCR to span the recombination breakpoint and to obtain DNA sequence of the junction fragment for each deletion. A pair of oligonucleotide primers, remote and nested, was designed from the proximal side of the breakpoint for each family (u36G3.20.1K-U, 5′-CCACTCCCTTTCTGCTTCACTGCTC-3′, and u36G3.20.1K-N, 5′-TGCTGATGAAAGGGTTTGCCAGGAC-3′ for family HOU542; GproxDOP.R182, 5′-GCAGGAAGAGAAGCACAGGCAAAGGGAGTA-3′, and GproxDOP.N289, 5′-CTAATGACAGAGGACACCAGGGAGCAGAAT-3′ for family HOU699; and WproxDOP.R49, 5′-GACTCCTATTAGTTGCCTGCCCTGATGAGG-3′, and WproxDOP.N195, 5′-AGTGCTGCTTGTGCTGGCTCCAATGCTGTG-3′ for family PMD1). We used a degenerate primer, 6MW (5′-CCGACTCGAGNNNNNNATGTGG-3′), in the primer extension and after PCR amplification, as described elsewhere (Wu et al. 1996), with slight modification. After the sequential PCR using the remote/6MW and nested/6MW primer sets, we separated PCR products by agarose gel electrophoresis. A second round of PCR products was purified and sequenced directly. On the basis of the sequence data, we designed primers flanking the breakpoints to amplify deletion-specific junction fragments, to confirm the results from the DOP-PCR analyses (Tjct, 5′-CCACTCCCTTTCTGCTTCACTGCTC-3′ and 5′-GGAGCAGAAAGAAACTATCA-3′; Gjct, 5′-CACAGACTTCACTTGGAATG-3′ and 5′-CCATTTGAAAACATAAGCAA-3′; Wjct, 5′-AGTGCTGCTTGTGCTGGCTCCAATGCTGTG-3′ and 5′-TAAGTCGTTTCTATTTTGAGTTCCTTCTTG-3′). The following primer set was used to investigate the possibility of an inversion rearrangement in family PMD1 (breakpoint PCR, 5′-TCCAAAGGAGAAAGCAACCACAGAT-3′ and 5′-CCCAGAATATTTACCAACAGAGGAG-3′).
Genome Sequence Analyses of the PLP1 Region
A 1.5-Mb genomic segment flanking PLP1 was obtained from the University of California, Santa Cruz Human Genome Project Working Draft. Genes and predicted genes in the region were identified with an integrated Web-based homology search and annotation tool, BLAT (Kent 2002), and a genome sequence–annotation database, Ensembl (Hubbard et al. 2002). Genome sequence was locally assembled with Sequencher (Genecode) sequence analysis software. Each predicted gene in the Ensembl database was evaluated individually for possible coding sequence, existence of ESTs with perfect sequence match, and similarity to other genes, using various Web-based sequence analysis software including RepeatMasker, BLAST, Fgenesh, Grail, and MZEF, as described elsewhere (Inoue et al. 2001a). PipMaker with Dotplot analyses was used to identify potential LCRs in this region (Schwartz et al. 2000). The sequences of potential LCRs were analyzed for identity with each other by BLAST2.
Results
Deletion of the Entire PLP1 Gene
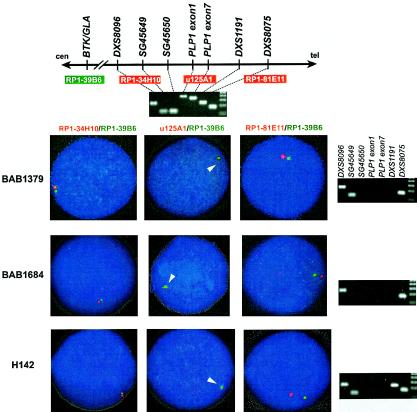
Interphase FISH using a PLP1 probe revealed that the patients from all three families (BAB1379, BAB1684, and H142) carried deletion of the PLP1 gene (fig. 1). However, probes for two markers that flank PLP1 (RP1-34H10, distal to DXS8096, and RP1-81E11, for DXS8075) exhibited a normal pattern of hybridization signals (fig. 1), indicating that these deletion rearrangements are smaller than those usually observed with PMD-associated duplications (Inoue et al. 1999).
Figure 1.
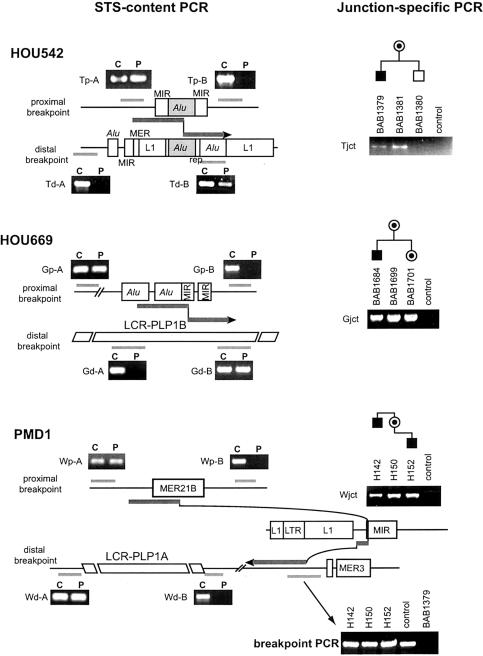
Detection of PLP1 deletion and breakpoint mapping by STS-content mapping and interphase FISH analyses. Map (top) shows selected STS markers surrounding the PLP1 locus that were used in this study. Results of agarose gel electrophoresis of PCR products for each STS-PCR of a normal control are shown below. Probes used in the interphase FISH studies were assigned with corresponding color in the photograph. The green probe, RP1-39B6, was used as an intrachromosomal control. Results for one proband from each family are shown. Interphase FISH using u125A1 that contains the entire PLP1 gene (middle column) revealed no red signal in each patient, whereas interphase FISH using RP1-34H10 and RP1-81E11 (left and right columns) reveal signals for markers proximal and distal to the breakpoints for the deletion, respectively. Accordingly, agarose gel electrophoresis analyses (right column) of STS-content PCR products for each patient revealed no amplification from STSs in the deleted segment, including PLP1 exonic sequences. STS-content-mapping analyses revealed distinct locations for the deletion breakpoints in each family.
PCR analyses did not amplify PLP1 exonic sequences in any of the three male probands. Subsequent STS-content mapping revealed that all deletion segments are flanked by DXS8096 and DXS8075, confirming the results from interphase FISH analyses. STS mapping with SG45649, SG45650, and DXS1191 showed that the deleted segments in these three families are of different sizes. Together with a normal karyotype for the three male patients (data not shown), our data suggest that each patient has a submicroscopic deletion of PLP1, but distinct genomic segments are involved.
Interstitial Translocation of the PLP1 Gene in Family HOU542
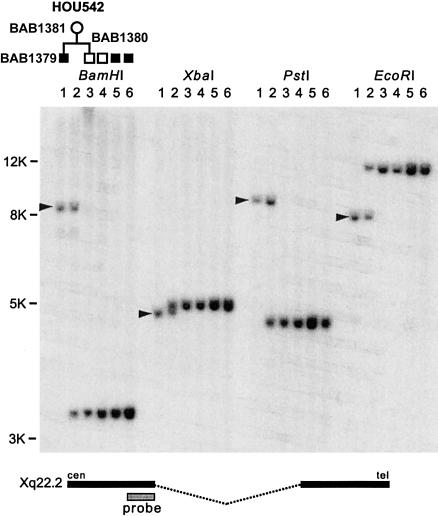
Genomic Southern blotting analysis of family HOU542, using a probe from the X chromosome, revealed deletion-specific junction fragments in genomic DNA from the proband (BAB1379) and his mother (BAB1381) (fig. 2). These data indicate that the deleted X chromosome was inherited from the mother. The proband’s healthy brother (BAB1380) does not have this junction fragment, which indicates that he does not carry the deleted X chromosome.
Figure 2.
Southern analysis identified a recombination-specific junction fragment in family HOU542. Genomic DNA from proband (BAB1379; lane 1), mother (BAB1381; lane 2), healthy male sibling (BAB1380; lane 3), normal control (lane 5), and two patients with PLP1 duplication (lanes 5 and 6) were used for genomic Southern hybridization analysis using four different endonucleases. We observed deletion-specific junction fragments (arrowheads) in the patient and mother, but not in other individuals. Bottom, Location of the hybridization probe, u35G3.20K, is shown.
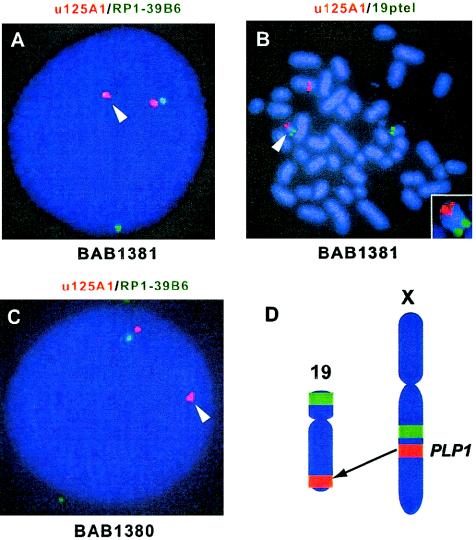
Interphase FISH analyses of BAB1381 showed two PLP1 signals (red) and two intrachromosomal control signals (green), but one PLP1 signal appeared to be distant from the control signal (fig. 3A). Metaphase FISH showed one PLP1 signal on chromosome 19qtel (fig. 3B). These observations suggest that the submicroscopic PLP1 deletion in BAB1379 resulted from inheritance of an unbalanced translocation. The patient BAB1379 did not inherit the derivative chromosome 19 (no PLP1 signal was observed in either FISH or PCR analysis; fig. 1). However, to our surprise, his healthy male sibling, BAB1380, carries this derivative chromosome 19, containing PLP1 (fig. 3C). Thus, he carries two copies of PLP1: one on his X chromosome and the other on one chromosome 19 [46,XY.ish der(19)ins(19;X)(q13.4;q22.2q22.2)(PLP1+)].
Figure 3.
Interphase and metaphase FISH of family HOU542 revealed transposition of PLP1 to chromosome 19. A, Interphase FISH using probes for PLP1 (u125A1; red) and an intrachromosomal control (RP1-39B6; green) revealed one PLP1 signal (arrowhead) appearing distant from the control signal in the mother (BAB1381). B, In the same individual, metaphase FISH using chromosome 19-specific probe (green) localized one copy of PLP1 (red) on 19qtel (arrowhead). The small box shows an enlarged image of derivative chromosome 19. C, Interphase FISH of the healthy brother (BAB1380) revealed that he also has the derivative chromosome 19 with PLP1 signal (arrowhead). D, An ideogram representing insertional translocation of PLP1 from Xq22.2 to 19qtel (red). Green signals indicate intrachromosomal controls that were used in FISH analyses.
Meiotic Recombination in the Maternal Grandfather Generated the Deletion in Family HOU669
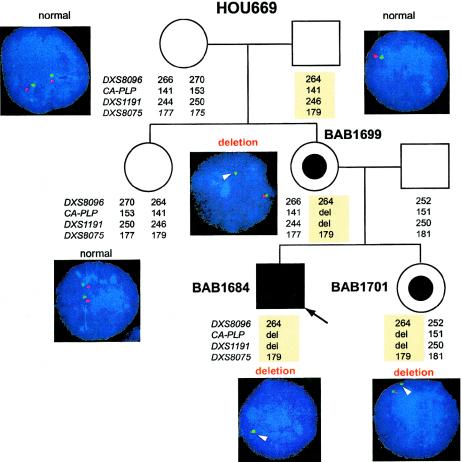
Interphase FISH analyses in family HOU669 revealed that the patient’s healthy sister and mother are carriers for the deletion. Both maternal grandparents had normal FISH results from their blood cells and have no neurological phenotype, indicating de novo rearrangement in the mother. Haplotype analyses of the family members, using four STR markers flanking PLP1 (DXS9096, CA-PLP, DXS1191 and DXS8075), showed that the deleted X chromosome in the patient was derived from his maternal grandfather’s X chromosome. Because the grandfather’s somatic cells do not have a deletion, our findings suggest that the deletion event occurred in the grandfather’s germ cells (fig. 4).
Figure 4.
Results of interphase FISH and haplotype analyses of family HOU669. A pedigree of the family HOU669 is shown, with haplotypes at DXS8096, CA-PLP, DXS1191, and DXS8075, as well as interphase FISH images using both PLP1 (red) and control (green) probes. The healthy sister and mother carry a deleted chromosome shown by both FISH and haplotype analyses. Neither of the maternal grandparents revealed deletion by FISH. The haplotype analysis shows that the deletion chromosome (light yellow boxes) was derived from the grandfather.
Slightly Skewed X Inactivation in HOU542 and HOU669
In unaffected females, the X-inactivation pattern is represented by a “bell-shaped” distribution with a 50:50 average ratio for the active versus inactive X chromosome, respectively. Therefore, a skewed inactivation pattern is not uncommon in unaffected females. In fact, a ratio of ⩾80:20 can be observed in 5%–10% of unaffected females (Willard 2001). This skewing may result in a disease phenotype when one has a deleterious mutation in a gene that is usually subjected to X inactivation.
In family HOU542, the mother (BAB1381) revealed slightly skewed X inactivation (active vs. inactive X chromosome 81:19) in the white blood cells (data not shown). Haplotype analysis of family members, using STR markers between Xq13.1 (AR) and Xq22.2 (PLP1), showed that the 81% represents the chromosome bearing the deleted allele (data not shown). Similarly, in family HOU669, the mother (BAB1699) showed slight skewing (82:18), and the carrier sister (BAB1701) revealed essentially random inactivation (66:34; data not shown). Haplotype analysis demonstrated a recombination between DXS8077 and DXS8020 in the patient, and thus the deletion allele is represented by 82% and 66% in the mother and sister, respectively (data not shown). In family PMD1, a carrier female, H150, revealed random inactivation (41:59), with the former number representing the deletion allele based on haplotype analysis (data not shown).
Mapping of the Recombination Breakpoints and Cloning of the Junction Fragments
Using STS markers that map to the PLP1 region, we characterized the size of the deleted genomic segments and recombination breakpoints in each family by STS-content mapping. In family HOU542, the proximal breakpoint occurred 47 Kb centromeric to PLP1, within cosmid u35G3 (GenBank accession number Z93848). STS mapping localized the proximal breakpoint within a 1-Kb genomic interval (fig. 5). With draft genomic sequence information about the PLP1 proximal region used as a guide, a recombination-specific junction fragment was cloned, using DOP-PCR. DNA sequencing of this junction fragment allowed us to identify the recombination breakpoint and localize the distal end at ∼700 Kb telomeric to PLP1, within PAC RP3-513M9 (AL049631) (figs. 6 and 7). Deletion-specific junction fragments were identified in the patient and his mother with primers spanning the recombination breakpoints, both confirming the recombination breakpoint and its segregation in the family (fig. 5). As anticipated, no deletion junction fragment was identified in the unaffected sibling. The deletion in HOU542 spans ⩾750 Kb (fig. 7).
Figure 5.
STS-content PCR and junction-specific PCR analyses. Flanking genomic regions of the proximal and distal breakpoints are shown for each family. Interspersed repeats are marked. In each region, the centromere is to the left and the telomere to the right. Recombination breakpoints and flanking sequence are shown as thick arrows. Light gray bars represent the position of the target regions for STS-content PCR, and the corresponding agarose gel photographs are presented. In each experiment, results for a proband (P) and a normal control (C) are shown. Note that STS within the deleted genomic region resulted in no amplification in probands. Results of the agarose gel electrophoresis for junction-specific PCR analyses (right) revealed amplification from patients and carriers, but not from noncarriers or normal control. Breakpoint PCR for family PMD1 showed PCR amplification in all three individuals (two patients and one carrier mother) as well as in a normal control, using primers flanking the recombination breakpoint, suggesting the existence of contiguous wild-type sequence of this region. We used BAB1379 as a negative control to determine the specificity of this PCR assay.
Figure 6.
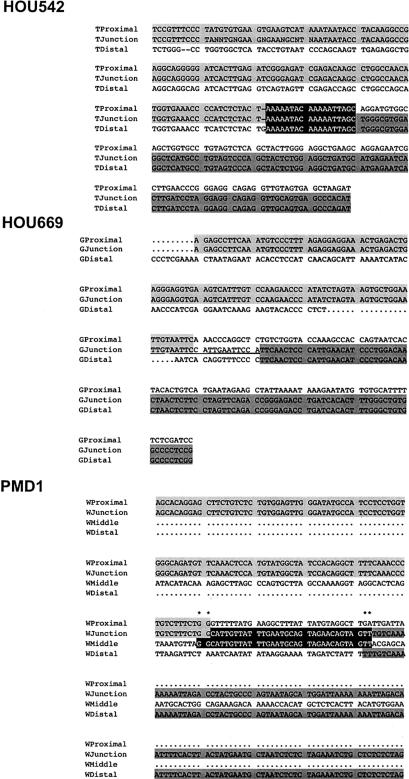
DNA sequence of recombination junction fragments. DNA sequence for each deletion-specific junction fragment obtained by DOP-PCR was perfectly aligned to the wild-type flanking finished genomic sequences for both proximal and distal breakpoints. Alignments with the proximal boundary were shaded in light gray, and those with the distal boundary were shaded in dark gray. Top, Sequence alignment in family HOU542. The recombination breakpoint was embedded in an 18-bp stretch of perfect sequence alignment (shaded in black with white letters). Middle, Sequence alignment in family HOU669. The recombination breakpoint was located within a 12-bp segment that has partial sequence identity to proximal boundary sequence (underlined). The origin of this 12-bp segment is unknown. Bottom, Sequence alignment in family PMD1. The sequences of the junction fragment consist of three segments (see fig. 5). In addition to the proximal and distal boundaries, a 34-bp middle fragment is shown in black shading. This middle segment has 2-bp overlaps with proximal and distal segments, respectively (asterisks above the alignment).
Figure 7.
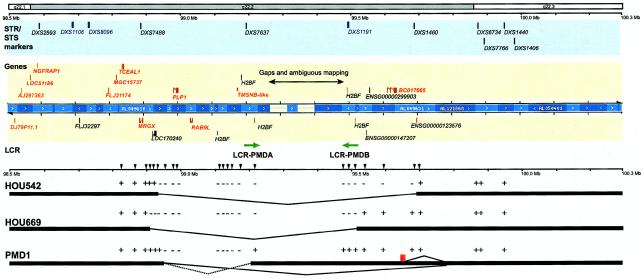
STS-content mapping of the deletion intervals in the genomic physical map of the Xq22.2 region. The top rectangles represent chromosomal bands, and the solid horizontal line below shows physical distances from the telomeric end of the short arm (left). The light blue rectangle below displays STR (blue) and STS (black) markers in this region. The light yellow rectangle represents genes (red) and predicted genes (black) in this region. Blue bars indicate a contig of large-insert genomic bacterial clones (BACs, PACs, and cosmids) that were sequenced and annotated by the draft Human Genome Sequencing project. The contig was obtained from the Ensembl genome browser. Annotation of each gene was determined on the basis of the Ensembl genome database project and additional computational analyses of each gene. Two LCRs, LCR-PMDA and LCR-PMDB, are shown (green arrows). Thick horizontal bars and intermediate thin lines indicate the deleted genomic segment in each family, determined by the STS-content mapping of deletion segments using STS markers in this region (arrowheads). The dotted line in family PMD1 represents the deleted genomic region estimated by STS-content mapping, whereas solid lines show the actual genomic rearrangements identified by junction-fragment sequence analysis. The result of each PCR experiment is shown as a plus (+) or a minus (−) symbol. A red rectangle in family PMD1 indicates the location of the 5-Kb probe used in the interphase FISH analysis.
Sequence comparison between the recombinant junction fragment and wild-type genomic sequence revealed that the translocation event occurred between two Alu repetitive sequences (subgroup Alu-Sq for the proximal and Alu-Sx for the distal copies, respectively), in which the overall sequence identity was 85% for 160 bp between proximal and distal copies (figs. 5 and 6). Perfect sequence alignment was observed at the recombination breakpoint for 18 bp (fig. 6). Other than these Alu sequences, there was no apparent homology between the genomic regions around the two breakpoints.
In the family HOU669, STS mapping localized the proximal breakpoint to ∼84 Kb centromeric to PLP1, within PAC RP5-1055C14 (AL049610). Similarly, we used DOP-PCR to span the recombinant breakpoint to the distal end. DNA sequence of the DOP-PCR fragments revealed that the distal breakpoint is located ∼500 Kb telomeric to PLP1 within cosmid u240C2 (Z73497) (figs. 5 and 6). This finding was independently confirmed by STS-PCR mapping (fig. 7). Thus, this deletion spans ∼600 Kb. Junction-specific PCR amplification confirmed the genomic interval for the deletion and segregation in the family members (fig. 5). Sequence analysis of the junction fragment revealed that there is no homologous sequence between proximal and distal boundaries (fig. 6). The proximal breakpoint is embedded in a short stretch of MIR (mammalian-wide interspersed repeats) sequence adjacent to two contiguous Alu sequences. The distal junction–flanking sequence contains no interspersed repetitive elements for >1 Kb surrounding the breakpoint (fig. 5). No sequence overlap was found at the breakpoint, suggesting that the recombination was mediated by NHEJ (fig. 6). There is an unrecognized 12-bp sequence between proximal and distal breakpoints, which contains an incomplete 9-bp direct repeat.
In family PMD1, STS mapping identified the proximal breakpoint ∼30 Kb centromeric to PLP1, within cosmid u35G3 (Z93848). The distal breakpoint mapped ∼200 Kb telomeric to PLP1 (figs. 5 and 7). DOP-PCR and sequence analyses, however, indicated that junction sequences are inconsistent with the STS mapping results. We obtained 30 bp of sequence adjacent to the proximal breakpoint that completely matched a unique sequence ∼760 Kb telomeric to PLP1, within BAC RP11-541I12 (AL121868) in an inverted orientation (figs. 5 and 6). There was no homology between the flanking sequences before and after the breakpoint, other than a 2-bp overlap. Sequences of the junction fragment after this 30-bp segment aligned to a region ∼640 Kb telomeric to PLP1, within PAC RP3-513M9 (AL049631), also in an inverted orientation (figs. 5 and 7). PCR using primers spanning these multiple breakpoints amplified the recombination specific-junction fragments that extend ⩾5 Kb further in the centromeric direction (data not shown). No homologous sequence was identified in the region flanking the deletion breakpoints, other than a 2-bp overlap.
The complicated and multidirectional recombination events—as well as the discrepancy between STS mapping and junction fragment analyses—indicate that the chromosome rearrangement in this family is complex. Experimental evidence suggests that it is not a simple deletion but rather is accompanied by inversion or inverted duplication events. To examine these complex rearrangements, we investigated further the genomic region ∼640 Kb telomeric to PLP1. Two possibilities were investigated. First, a large inversion event between regions ∼200 Kb and ∼640 Kb telomeric from PLP1, adjacent to the deletion, may be associated with the deletion. Second, an inverted duplication of part or the entire length of the same 200–640-Kb region may be involved in the complex rearrangement.
The first possibility was examined by PCR using a set of primers spanning the potential inversion breakpoint at the 640 Kb region. The inversion should split the interval of this PCR amplification; thus, no amplification would be predicted in the case of inversion. However, we identified amplification in two patients, H142 and H152, as well as in the carrier female and controls (fig. 5, PMD1 breakpoint PCR). Therefore, we concluded that inversion is unlikely.
To test the second possibility, we used interphase FISH analysis with a 5-Kb genomic probe adjacent to the 640 Kb breakpoint. This should allow the highest resolution to detect a relatively small inverted duplication. The interphase FISH for patient H142, however, failed to detect a duplicated signal (data not shown). We therefore did not find evidence for duplication at this resolution.
Genes Contained within the Genomic Deletions
The genomic sequence of the deleted regions, including one gap of unknown length at ∼260 Kb telomeric to PLP1, were analyzed. Deletion breakpoints were placed on the physical map (fig. 7). Proximal breakpoints are all located within an ∼100-Kb interval between PLP1 and MRGX. There is only one predicted gene, LOC170240, within this interval that is deleted in family HOU669. The distance from PLP1 to each distal breakpoint revealed larger variation in length, but each involved two genes, RAB9L and TMSNB, in addition to PLP1.
RAB9L was identified by a similarity search, using RAB9 sequences (Seki et al. 2000). RAB9L has three exons spanning 7 Kb. It contains an ATP/GTP-binding site motif A, which defines RAB9L as a member of the RAS superfamily, but its function has not been determined. TMSNB is a member of the thymosin β family. It spans 3.5 Kb and encodes a small (5-KDa) polypeptide that likely binds to a monomer actin and prevents actin polymerization (Huff et al. 2001). Deletion of TMSNB may not be deleterious, because another copy of TMSNB exists ∼1.2 Mb proximal to PLP1, based on the genome sequence annotation (data not shown). Although the sequence identity for the entire mRNA between these two copies of TMSNB is only 91%, the coding sequences are completely identical (99.7% sequence identity), except for one silent substitution at the fourth amino acid residue (K4K G→A12). Probably both copies are expressed, because multiple corresponding mRNA for either gene was identified in the EST database (e.g., BI438503 and BC000183 for the copies on Xq22.2 and Xq22.1, respectively).
Some predicted genes were identified within the deletion intervals (fig. 7). Five copies of H2B-like genes were identified, which are likely contained within an LCR, LCR-PMD, described below. No matching ESTs have been identified for these H2B-like genes. LOC1170240 was computationally predicted to encode seven exons and 337 amino acids, with significant homology to the glycine receptor aZ2 subunit. However, no EST that aligns to this predicted gene is identified in the GenBank database. RP1-233G16.1 spans six exons and likely encodes a 388–amino acid protein with unknown function. Three predicted genes assigned by the Ensembl database (ENSG00000147207, ENSG00000299903, and ENSG00000123576) had no matching ESTs.
It appears that the genomic region distal to PLP1 contains fewer genes than the proximal region. There are only two known genes (RAB9L and TMSNB) in the ∼1-Mb interval distal to PLP1, whereas at least eight known genes are located within a 0.5-Mb proximal genomic interval (fig. 7). Therefore, it is likely that deletions involving large genomic segments in the proximal region of PLP1 result in deletion of several genes, which may be deleterious. However, each gene and the predicted genes need to be elucidated with regard to the phenotypic consequence of deletion.
LCR and Its Potential Involvement in the Deletions
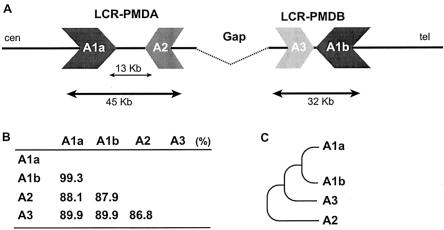
An analysis of the genome sequence at the telomeric end of two deletions revealed complex genomic architecture. A large-scale genome comparison revealed a pair of LCRs (designated LCR-PMDA and LCR-PMDB, respectively) flanking the gap of the draft genome sequence (fig. 8A). LCR-PMDA spans ∼45 Kb and contains a 13-Kb internal segment of mostly interspersed repeat elements (>94% of total length), with two inverted homologous segments (88.1% identity), designated A1a and A2. LCR-PMDB spans ∼32 Kb with no interruption and also contains two inverted segments, A1b and A3, with 89.9% identity. The segments A1a and A1b share high sequence identity (99.3%) for >20 Kb, whereas segments A2 and A3 show 86.8% sequence identity (fig. 8B). Segment A2 has the lowest homology to other segments (87%–88%) and reveals fragmentation by insertion/deletion events. These findings suggest that the division between segments A2 and A3 is the more ancient event and that between A1a and A1b is the more recent (fig. 8C).
Figure 8.
Genomic structure of LCR-PMDA and LCR-PMDB. A, Flanking the gap in the draft genome sequence, two inverted repeats, LCR-PMDA and LCR-PMDB, span 45 Kb and 32 Kb, respectively. LCR-PMDA has an inset of a 13-Kb genomic segment that is abundant in interspersed repeat elements (>94%). Each LCR consists of two homologous segments, designated A1a and A2 for LCR-PMDA, as well as A1b and A3 for LCR-PMDB. B, Sequence identity analysis among the segments, using BLAST2. Segments A1a and A1b reveal the highest identity, whereas A2 and A3 show the lowest identity. C, Phylogenic tree shows evolution of each segment in LCR-PMDA and LCR-PMDB, based on the sequence identity and genomic structure.
The distal recombination breakpoint for family HOU669 appears to be located within segment A1b of LCR-PMDB. STS-content mapping for family PMD1 placed the distal breakpoint within LCR-PMDA. However, the information from the PCR assay was limited because of the interference of LCR-PMDB and the high interspersed repeat content. Although the cloning and sequence analysis of the junction fragment revealed that the breakpoints are not located within the LCR-PMDA, formation of the complex genomic rearrangement in this family may be associated with the genome architecture involving LCR-PMDA and, possibly, LCR-PMDB.
Discussion
Genomic rearrangements play a major role in the pathogenesis of PMD, which defines PMD as a genomic disorder (Lupski 1998; Inoue and Lupski 2002). Duplications of a genomic fragment, usually >500 Kb and encompassing the entire PLP1 gene, account for PMD in 60%–70% of patients (Sistermans et al. 1998; Inoue et al. 1999). In contrast, complete deletion of PLP1 is observed infrequently. Only one family with PLP1 deletion was described elsewhere (Raskind et al. 1991), and the present study adds two new families. We explored the products of genomic recombination resulting in PLP1 deletion and present a model for their molecular mechanisms and phenotypic consequences.
Genomic Mechanisms for Chromosomal Rearrangements Resulting in the PLP1 Deletions
The deletion of the genomic fragment that contains the entire PLP1 gene in the two novel families described in this study arose by at least two distinct processes: an insertional translocation event and a sister-chromatid exchange in male meiosis. The third family revealed a complex genomic rearrangement; further characterization will be required to elucidate how the recombinant products were derived.
The insertional translocation is probably a rare event; no such recurrent cases with similar PLP1 translocations have been reported. However, because the size of the translocated fragment was not visible by routine G-banding microscopic analysis (i.e., submicroscopic), some cases with similar recombination may have been overlooked. Notably, there are three unrelated families with PMD with interstitial submicroscopic PLP1 insertion within the X chromosome—two in Xp22.1 and the other in Xq26—resulting in PLP1 duplications (Hodes et al. 2000). One of the cases was accompanied by pericentric inversion. Together with the interstitial translocation identified in this study, this propensity for translocation of PLP1 suggests that the genomic region surrounding PLP1 may contain sequence resulting in susceptibility to transposition.
On the other hand, sister-chromatid exchange might be more common, because deletion may be generated as a reciprocal recombination event of the duplication. Despite the high frequency of PLP1 duplications as products of sister-chromatid exchanges, however, family HOU669 is in fact the first reported instance of a sister-chromatid–exchange event resulting in PLP1 deletion. Given our observations, compared with duplications, deleted genomic segments are relatively small and probably involve only three genes (PLP1, RAB9L, and TMSNB), whereas duplication can be more variable in size and can involve more genes (Inoue et al. 1999). Yet PLP1 deletion is relatively infrequent compared with PLP1 duplication. We hypothesize that the viable size for deletion is limited. Larger deletions might, in turn, result in a reduced fertility or embryonic lethality, whereas duplication of the same segment does not. This may explain the infrequent observation of PLP1 deletion; however, we cannot formally exclude the possibility that duplication and deletion arise by separate mechanisms with different frequencies.
Complex Rearrangements and NHEJ as the Mechanism for the DNA Recombination
Molecular dissection of the PLP1 deletion–rearrangement breakpoints at the DNA sequence level revealed mechanisms for the recombination event in each family. In family HOU542, with insertional translocation, the DNA sequence revealed that the breakpoint is located within two overlapping Alu sequences in direct orientation with an 18-bp perfect sequence identity at the breakpoint. Given the observation of insertional translocation in the mother, the chromosome rearrangement likely did not result from a reciprocal recombination between two different chromosomes, but rather resulted from a transposition of the PLP1-containing genomic segment. Excision of the genomic segment was probably mediated by an Alu-Alu recombination on the X chromosome. No other genomic architectural features supporting susceptibility for the transposition event—such as palindromes, LCRs, or low-complexity sequence—were found around the breakpoints. Analyses of the maternal grandparents and characterization of chromosome 19 breakpoints are required for precise elucidation of the interchromosomal transposition of PLP1.
Sequence analyses of junction fragments in the other two families revealed a complex genomic recombination. Each of the deletion breakpoints was mapped to a different location; no common breakpoint was observed. Together with the absence of homologous sequences flanking the deleted genomic interval, it is unlikely that nonallelic homologous recombination mediates the rearrangement. Our findings from the analyses of the products of recombination are consistent with a mechanism of NHEJ. In both families, a small piece of DNA sequence that does not belong to either proximal or distal flanking sequences was identified between the proximal and distal breakpoints. In family HOU669, a 12-bp fragment of unknown origin was found at the junction. Because this 12-bp fragment consists of partial direct repeats, it may be synthesized during the process of double-strand break repair and NHEJ.
Family PMD1 revealed a more complex recombination event. A 30-bp short fragment was inserted at the junction, which originated >100 Kb away from the distal junction. No sequence homology was found at proximal, interstitial, or distal boundary sequences, indicating that NHEJ likely mediated the recombination. Furthermore, the DNA recombination likely involves a complex process, as shown in figures 5, 6, and 7. We investigated two possibilities—inversion or inverted duplication accompanied by the deletion—to explain the genomic rearrangement in this family. Neither a PCR assay for inversion breakpoints nor interphase FISH revealed evidence in support of either of these possibilities. Because these assays do not completely exclude the possibility of variant inversion or duplication, further intensive genomic investigation is required to elucidate the genomic mechanism of this complex rearrangement.
Genome Architecture Analysis of Complex LCRs that May Instigate the Genomic Rearrangements
We identified two novel LCRs, LCR-PMDA and LCR-PMDB, both telomeric to PLP1, flanking a gap in the draft genome sequence. Both LCRs are likely associated with the genomic recombination that resulted in deletions in two families. These LCRs do not serve as substrates for NAHR, at least in the DNA rearrangements resulting in PLP1 deletion, but may be associated with susceptibility to initiate DNA rearrangements, perhaps by stimulating double-strand breaks. Involvement of LCRs in chromosomal translocations was indicated in chromosome 22–associated chromosomal rearrangements (Spiteri et al. 2001). No unique sequence structures or homology segments were recognized at the recombinant breakpoints of DMD duplications (Hu et al. 1991); however, DMD rearrangement breakpoints appear to cluster (Baumbach et al. 1989), and the molecular basis for the nonrandom nature of the breakpoints has not been determined at the DNA sequence level. These rearrangement-breakpoint hot spots may reflect unique genome-architectural features.
Of interest, our preliminary data indicate that at least some of the breakpoints for PLP1 duplications are also located within the intervals that contain the LCR-PMDA and LCR-PMDB (data not shown). As observed in other regions of human genome, this gap may contain additional complex or repeat structures that are difficult to clone and sequence (Eichler 2001). Further investigation of this genomic region may clarify the molecular basis of PLP1 duplication and deletion and a role for these LCRs and genomic architecture in susceptibility to genomic rearrangements.
Phenotypic Consequence of PLP1 Deletions in Males and Females
Each male patient with PLP1 deletion had a mild form of PMD or a complicated form of spastic paraplegia type 2. In addition, two families with nonsense mutations in exon 1, which resulted in the termination at the second codon of PLP1 and presumably null alleles, also had a mild form of PMD (Sistermans et al. 1996; Garbern et al. 1997). Of note, null PLP1 results in peripheral myelinopathy in addition to CNS myelinopathy; peripheral neuropathy has not been associated with other defects of the PLP1 gene (Garbern et al. 1997). Furthermore, in mice, Plp1 is not necessary for myelin compaction during development (Klugmann et al. 1997; Griffiths et al. 1998). These observations suggest a complex pathology for gene dosage abnormalities at the PLP1 locus, in which dominant-negative and loss-of-function alleles of the PLP1 gene may result in different pathogenesis with distinct phenotypic consequences.
Female carriers add even more complexity to the mechanism for phenotypic manifestation because of X inactivation. We observed symptomatic carriers with mild late-onset spastic diplegia of varying severity in the families with deletions. Such families with symptomatic carriers are mostly associated with mutations that result in a mild phenotype but not with mutations resulting in a severe phenotype. The cellular mechanism for this paradoxical presentation can be explained by mosaicism of the oligodendrocyte population in the mild mutations (Hudson 2001; Inoue et al. 2001b).
In family HOU542, the mother with an insertional translocation of PLP1 from chromosome X to 19 presented with a more severe phenotype than did carrier females in other families. On the other hand, the male sibling who has the derivative chromosome 19, thus carrying two copies of PLP1, manifests no apparent clinical phenotype, suggesting silencing of the translocated PLP1 copy. This phenotypic presentation in the mother may simply be explained by mildly skewed X inactivation in the unfavorable direction (i.e., the active X chromosome contains the deleted PLP1 allele), although it is unclear whether X inactivation in peripheral blood reflects that in the CNS. In addition, altered expression of PLP1 due to the translocation, which could either increase or decrease expression, may be associated with the phenotypic manifestation. Such a change in gene expression due to chromosomal rearrangements, referred to as a position effect, has been observed in other human disorders (reviewed in Kleinjan and van Heyningen 1998).
Two distinct mechanisms can be associated with a position effect (Kleinjan and van Heyningen 1998). First, the translocation might separate upstream cis-acting regulatory elements from PLP1 exons, resulting in inappropriate expression of PLP1. This includes disruption of a locus-control region, which is well-documented in the β-globin gene (Kioussis et al. 1983). Although the deletion in HOU542 contains 47 Kb of upstream sequence, which is likely enough for specific PLP1 expression (Wight et al. 1993; Ikenaka and Kagawa 1995), it may not contain a potential locus-control region, which is required to overcome a position effect (Milot et al. 1996; Grosveld et al. 1987). However, a locus-control region in PLP1 has not been identified. Alternatively, a portion of the genomic segment could be truncated during the process of translocation. In this model, all of the expressing cells may reveal an equally altered level of expression. Given the equally strong FISH signals from chromosomes X and 19 by the cosmid probe for the PLP1 gene, a gross loss of genomic segment is unlikely.
Second, a mechanism analogous with Drosophila melanogaster position-effect variegation (PEV) may suppress PLP1 transcription if the translocated PLP1 is embedded in the telomeric heterochromatic-repeat region (Wakimoto 1998). When a functional copy of a euchromatic gene is translocated to a heterochromatic region, the heterochromatinized state of DNA may spread into the juxtaposed euchromatic gene, thereby silencing the transcription (Kleinjan and van Heyningen 1998). PEV results in a stable silencing of genes in a clonal subpopulation of cells; thus the expression appears to be mosaic (Festenstein et al. 1996). Such a heterochromatic suppression effect on a gene inserted adjacent to telomeric repeats in human cells was experimentally observed (Baur et al. 2001). Together with the mosaicism due to the X inactivation, the PEV model may result in significant complexity as to the PLP1 expression level in the mother of HOU542. To date, we have obtained no evidence to favor either mechanism. Further genomic characterization of the position of translocation and the cis-regulatory sequence of translocated PLP1 gene is required to address this question.
In summary, analysis of the products of recombination in patients with PMD who harbor PLP1 deletion suggests NHEJ as a predominant mechanism. Breakpoint mapping reveals genome architecture consisting of a complex LCR, which may result in susceptibility to rearrangements. Phenotypic analyses indicate that genomic rearrangement may have complex consequences for genes that manifest dosage effects, particularly if they are located on the X chromosome. Although many genomic disorders are mediated through homologous recombination mechanisms (Lupski 1998; Emanuel and Shaikh 2001; Inoue and Lupski 2002; Stankiewicz and Lupski 2002a, 2002b), the cases presented here suggest that many, if not most, other genomic and chromosomal rearrangements are mediated through sometimes very complex but certainly distinct mechanisms, such as NHEJ. In addition, cases like this allow for studies of genes outside their normal chromosomal environments and will aid in our understanding of control of gene expression and position effects.
Acknowledgments
We thank the patients and families for their contributions to this study; Drs. Philip J. Hastings, David Nelson, and Pawel Stankievicz and Ms. Jennifer Lee for their critical reviews and helpful advice; and the members of the Kleberg Cytogenetics Laboratory, Baylor College of Medicine, especially Catherine Kashork and Jessica Wu, for their expert FISH on these cases. This study is dedicated to the memory of Dr. M. E. Hodes, who died September 29, 2001. K.I. is supported by a development grant from the Muscular Dystrophy Association. This study was supported in part by grants from the National Institute for Neurological Disorders and Stroke, NIH (R01 NS27042), the National Institute for Child Health and Development (P01 HD38420), and the Muscular Dystrophy Association (to J.R.L.), and by the Baylor College of Medicine Mental Retardation Research Center (NIH P30 HD24064).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ (for sequence identity analysis)
- Fgenesh, http://genomic.sanger.ac.uk/gf/gf.shtml (for sequence analysis)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html
- Grail, http://grail.lsd.ornl.gov/ (for sequence analysis)
- MZEF, http://argon.cshl.org/genefinder/ (for sequence analysis)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PMD [MIM 312080])
- PipMaker, http://bio.cse.psu.edu/pipmaker/ (for DNA alignment computations)
- RepeatMasker, http://ftp.genome.washington.edu/cgi-bin/RepeatMasker (for sequence analysis)
- UCSC Genome Bioinformatics, http://www.genome.ucsc.edu/
- Wellcome Trust Sanger Institute, Ensembl Genome Browser, http://www.ensembl.org/
References
- Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW (1992) Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 51:1229–1239 [PMC free article] [PubMed] [Google Scholar]
- Baumbach LL, Chamberlain JS, Ward PA, Farwell NJ, Caskey CT (1989) Molecular and clinical correlation of deletion leading to Duchenne and Becker muscular dystrophies. Neurology 39:465–474 [DOI] [PubMed] [Google Scholar]
- Baur JA, Zou Y, Shay JW, Wright WE (2001) Telomere position effect in human cells. Science 292:2075–2077 [DOI] [PubMed] [Google Scholar]
- Eichler EE (2001) Segmental duplications: what's missing, misassigned, and misassembled—and should we care? Genome Res 11:653–656 [DOI] [PubMed] [Google Scholar]
- Ellis D, Malcolm S (1994) Proteolipid protein gene dosage effect in Pelizaeus-Merzbacher disease. Nat Genet 6:333–334 [DOI] [PubMed] [Google Scholar]
- Emanuel BS, Shaikh TH (2001) Segmental duplications: an “expanding” role in genomic instability and disease. Nat Rev Genet 2:791–800 [DOI] [PubMed] [Google Scholar]
- Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kioussis D (1996) Locus control region function and heterochromatin-induced position effect variegation. Science 271:1123–1125 [DOI] [PubMed] [Google Scholar]
- Garbern J, Cambi F, Shy M, Kamholz J (1999) The molecular pathogenesis of Pelizaeus-Merzbacher disease. Arch Neurol 56:1210–1214 [DOI] [PubMed] [Google Scholar]
- Garbern JY, Cambi F, Tang X-M, Sima AA, Vallat JM, Bosch EP, Lewis R, Shy M, Sohi J, Kraft G, Chen KL, Joshi I, Leonard DG, Johnson W, Raskind W, Dlouhy SR, Pratt V, Hodes ME, Bird T, Kamholz J (1997) Proteolipid protein is necessary in peripheral as well as central myelin. Neuron 19:205–218 [DOI] [PubMed] [Google Scholar]
- Griffiths I, Klugmann M, Anderson T, Yool D, Thomson C, Schwab MH, Schneider A, Zimmermann F, McCulloch M, Nadon N, Nave KA (1998) Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science 280:1610–1613 [DOI] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves DR, Kollias G (1987) Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell 51:975–985 [DOI] [PubMed] [Google Scholar]
- Hodes ME, Woodward K, Spinner NB, Emanuel BS, Enrico-Simon A, Kamholz J, Stambolian D, Zackai EH, Pratt VM, Thomas IT, Crandall K, Dlouhy SR, Malcolm S (2000) Additional copies of the proteolipid protein gene causing Pelizaeus-Merzbacher disease arise by separate integration into the X chromosome. Am J Hum Genet 67:14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X-Y; Ray PN, Worton RG (1991) Mechanisms of tandem duplication in the Duchenne muscular dystrophy gene include both homologous and nonhomologous intrachromosomal recombination. EMBO J 10:2471–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, et al (2002) The Ensembl genome database project. Nucleic Acids Res 30:38–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson LD (2001) Pelizaeus-Merzbacher disease and the allelic disorder X-linked spastic paraplegia type 2. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular basis of inherited diseases. McGraw-Hill, New York, pp 5789–5798 [Google Scholar]
- Huff T, Muller CS, Otto AM, Netzker R, Hannappel E (2001) β-Thymosins, small acidic peptides with multiple functions. Int J Biochem Cell Biol 33:205–220 [DOI] [PubMed] [Google Scholar]
- Ikenaka K, Kagawa T (1995) Transgenic systems in studying myelin gene expression. Dev Neurosci 17:127–136 [DOI] [PubMed] [Google Scholar]
- Inoue K, Dewar K, Katsanis N, Reiter LT, Lander ES, Devon KL, Wyman DW, Lupski JR, Birren B (2001a) The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res 11:1018–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Lupski JR (2002) Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet 3:199–242 [DOI] [PubMed] [Google Scholar]
- Inoue K, Osaka H, Imaizumi K, Nezu A, Takanashi J, Arii J, Murayama K, et al (1999) Proteolipid protein gene duplications causing Pelizaeus-Merzbacher disease: molecular mechanism and phenotypic manifestations. Ann Neurol 45:624–632 [PubMed] [Google Scholar]
- Inoue K, Osaka H, Sugiyama N, Kawanishi C, Onishi H, Nezu A, Kimura K, Kimura S, Yamada Y, Kosaka K (1996) A duplicated PLP gene causing Pelizaeus-Merzbacher disease detected by comparative multiplex PCR. Am J Hum Genet 59:32–39 [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Tanaka H, Scaglia F, Araki A, Shaffer LG, Lupski JR (2001b) Compensating for central nervous system dysmyelination: females with a proteolipid protein gene duplication and sustained clinical improvement. Ann Neurol 50:747–754 [DOI] [PubMed] [Google Scholar]
- Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussis D, Vanin E, deLange T, Flavell RA, Grosveld FG (1983) β-Globin gene inactivation by DNA translocation in γβ-thalassaemia. Nature 306:662–666 [DOI] [PubMed] [Google Scholar]
- Kleinjan DJ, van Heyningen V (1998) Position effect in human genetic disease. Hum Mol Genet 7:1611–1618 [DOI] [PubMed] [Google Scholar]
- Klugmann M, Schwab MH, Puhlhofer A, Schneider A, Zimmermann F, Griffiths IR, Nave KA (1997) Assembly of CNS myelin in the absence of proteolipid protein. Neuron 18:59–70 [DOI] [PubMed] [Google Scholar]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 [DOI] [PubMed] [Google Scholar]
- Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P (1996) Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell 87:105–114 [DOI] [PubMed] [Google Scholar]
- Mimault C, Cailloux F, Giraud G, Dastugue B, Boespflug-Tanguy O (1995) Dinucleotide repeat polymorphism in the proteolipoprotein (PLP) gene. Hum Genet 96:236 [DOI] [PubMed] [Google Scholar]
- Osaka H, Kawanishi C, Inoue K, Onishi H, Kobayashi T, Sugiyama N, Kosaka K, Nezu A, Fujii K, Sugita K, Kodama K, Murayama K, Murayama S, Kanazawa I, Kimura S (1999) Pelizaeus-Merzbacher disease: three novel mutations and implication for locus heterogeneity. Ann Neurol 45:59–64 [DOI] [PubMed] [Google Scholar]
- Raskind WH, Williams CA, Hudson LD, Bird TD (1991) Complete deletion of the proteolipid protein gene (PLP) in a family with X-linked Pelizaeus-Merzbacher disease. Am J Hum Genet 49:1355–1360 [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res 10:577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki N, Azuma T, Yoshikawa T, Masuho Y, Muramatsu M, Saito T (2000) cDNA cloning of a new member of the Ras superfamily, RAB9-like, on the human chromosome Xq22.1-q22.3 region. J Hum Genet 45:318–322 [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Kennedy GM, Spikes AS, Lupski JR (1997) Diagnosis of CMT1A duplications and HNPP deletions by interphase FISH: implications for testing in the cytogenetics laboratory. Am J Med Genet 69:325–331 [PubMed] [Google Scholar]
- Sistermans EA, de Coo RFM, De Wijs IJ, Van Oost BA (1998) Duplication of the proteolipid protein gene is the major cause of Pelizaeus-Merzbacher disease. Neurology 50:1749–1754 [DOI] [PubMed] [Google Scholar]
- Sistermans EA, de Wijs IJ, de Coo RFM, Smit LM, Menko FH, van Oost BA (1996) A (G-to-A) mutation in the initiation codon of the proteolipid protein gene causing a relatively mild form of Pelizaeus-Merzbacher disease in a Dutch family. Hum Genet 97:337–339 [DOI] [PubMed] [Google Scholar]
- Spiteri E, Goldberg R, Edelmann L, Gogineni SW, Pulijaal VR, Shanske A, Wakui K, Kashork CD, Shaffer LG, Morrow BE (2001) Molecular and cytogenetic characterization of translocations involving 22q11. Am J Hum Genet 69:327–34011452361 [Google Scholar]
- Stankiewicz P, Lupski JR (2002a) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 [DOI] [PubMed] [Google Scholar]
- ——— (2002b) Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev 12:312–319 [DOI] [PubMed] [Google Scholar]
- Wakimoto BT (1998) Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell 93:321–324 [DOI] [PubMed] [Google Scholar]
- Wight PA, Duchala CS, Readhead C, Macklin WB (1993) A myelin proteolipid protein-LacZ fusion protein is developmentally regulated and targeted to the myelin membrane in transgenic mice. J Cell Biol 123:443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard HF (2001) The sex chromosomes and X chromosome inactivation. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular basis of inherited diseases. McGraw-Hill, New York, pp 1191–1211 [Google Scholar]
- Woodward K, Kendall E, Vetrie D, Malcolm S (1998) Pelizaeus-Merzbacher disease: identification of Xq22 proteolipid-protein duplications and characterization of breakpoints by interphase FISH. Am J Hum Genet 63:207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Zhu S, Simpson S, de Jong PJ (1996) DOP-vector PCR: a method for rapid isolation and sequencing of insert termini from PAC clones. Nucleic Acids Res 24:2614–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]