Abstract
Recent progress in the field of cellular reprogramming has opened up the doors to a new era of disease modelling, as pluripotent stem cells representing a myriad of genetic diseases can now be produced from patient tissue. These cells can be expanded and differentiated to produce a potentially limitless supply of the affected cell type, which can then be used as a tool to improve understanding of disease mechanisms and test therapeutic interventions. This process requires high levels of scrutiny and validation at every stage, but international standards for the characterisation of pluripotent cells and their progeny have yet to be established. Here we discuss the current state of the art with regard to modelling diseases affecting the ectodermal, mesodermal and endodermal lineages, focussing on studies which have demonstrated a disease phenotype in the tissue of interest. We also discuss the utility of pluripotent cell technology for the modelling of cancer and infectious disease. Finally, we spell out the technical and scientific challenges which must be addressed if the field is to deliver on its potential and produce improved patient outcomes in the clinic.
Keywords: induced pluripotent stem cells, human embryonic stem cells, neurodevelopmental disorders, endodermal disorders, mesodermal disorders, reprogramming, disease modelling
Introduction
The field of cellular reprogramming has advanced in leaps and bounds since the early experiments of Briggs and King in the 1950s which demonstrated that cloning was possible using an embryonic frog model [1]. This approach was refined by John Gurdon in the 1960s and 1970s using nuclear transplantation with differentiated frog cells to generate clones [2–4]. Another advance was made in the 1980s with Harold Weintraub demonstrating that cellular fate could be reassigned via the ectopic expression of the transcription factor and master regulator MyoD [5]. Since then the field of cellular reprogramming has progressed rapidly with the cloning of the first mammal “Dolly the sheep” in the 1990s [6], and more recently the reprogramming of somatic fibroblasts to a pluripotent state by the introduction of four key transcription factors to generate induce pluripotent stem cells (iPSCs) [7–10]. This progress was recently acknowledged with John Gurdon and Shinya Yamanaka being awarded the Nobel Prize for their contributions to the field of reprogramming. Since the inception of induced pluripotent stem cells, the floodgates have opened with respect to refinement of the technology and its application in the generation of patient-specific models of disease [11–13]. For the first time it is now possible to generate a virtually limitless supply of highly characterised, patient-specific cells representing disease-relevant tissues [12]. The power of iPSC technology lies in its ability to model both monogenic and polygenic disorders, as well as disease states where the genetic component has yet to be identified.
To date there are a number of papers that describe the generation of both human embryonic stem cell (hESC) and iPSC lines from a myriad of disease states, however many do not convey the disease phenotype in vitro. In this review we will discuss only the hESC and iPSC lines which present the appropriate phenotype; in addition we will discuss the hurdles that remain before their widespread use in disease modelling and drug discovery can be adopted. Table 1 lists all the diseases which have been successfully modelled in iPSCs to date, and from these we have selected notable studies for in-depth discussion in the text. Our intention is to give as complete a picture of the technology as possible, and represent the wide range of experimental approaches, disease states and translational goals currently attracting interest in this rapidly expanding field.
Table 1.
Human iPSC lines exhibiting disease phenotypes.
| Affected Germ Layer | Disease | Tissue/Cell type affected | Cause/Mutation | Somatic Cell Source | Reprograming Method | Phenotype present in differentiated cells | Drug tests | Genetic Rescue | Ref |
|---|---|---|---|---|---|---|---|---|---|
| Ectodermal | Alzheimer's | Neurons | Presenelin 1(A246E), (L166P), PS2 (N131I), neuron specific APP (695) splice variant BACEl, APP duplication, sporadic Alzheimer's. | Fibroblasts | Retroviral OSKM, Retroviral OSKLN | Diseased neurons have increased amyloid β42 secretion. | γ-secretase inhibitors and modulators have profound effects. | No | [68,91–93] |
| Ectodermal | Amyotrophic lateral sclerosis/Frontotemporal lobar degeneration (ALS/FTLD). | Neurons, Motor neurons | TDP-43 M337V mutation. | Fibroblasts | Retroviral OSKM | Motor neuronscarrying mutation were more susceptible to cell death and to antagonism of PI3K signaling. | No | No | [39] |
| Ectodermal | Familial dysautonomia | Neurons | Skipping of exon 20 of IKBKAP and reduced IKAP protein. | Fibroblasts | Lentiviral OSKM | Higher mutant:normal ration of IKBKAP transcript in both iPS and neurons Reduced expression of genes associated with neurogenesis and differentiation. |
Yes—Kinetin reduced mutant splice form and increased neuronal differentiation. | No | [34] |
| Ectodermal | Huntington Disease (HD) | Neurons | 42/44 CAG, 39/43 CAG, 17/45 CAG. | Fibroblasts, | Lentiviral OSKM | Increase in lysosomal activity in HD-derived neurons. | No | No | [94] |
| Ectodermal | Huntington Disease (HD) | Neurons | 72 CAG. | Fibroblasts | Retroviral OSKM | Increased susceptibility to cell death and changes in mitochondrial activity in NSC. | No | Yes—correction reversed the disease phenotype. | [95] |
| Ectodermal | Spinal muscular atrophy (SMA) | Neurons | Decreased SMN1 expression. | Fibroblasts | Lentiviral OSLN | Differentiated neurons showed reduction in size and overall number as well as defects in synapse formation. | Yes—VPA and tobramycin increased SMN1 protein. | No | [1] |
| Ectodermal | Parkinson's disease (PD) | Dopaminergic neurons | Idiopathic, G2019S mutation in Leucine-Rich Repeat Kinase 2 (LRRK2). | Keratinocyte, Fibroblasts | Retroviral OSK | Reduced neuritis and arborization, accumulation of autophagic vacuoles. | No | No | [96] |
| Ectodermal | Parkinson's disease (PD) | Dopaminergic neurons | PARK2 (Exon 2–4 homozygous deletion and Exon 6–7 homozygous deletion). | Dermal Fibroblasts | Retroviral OSKM | Increased oxidative, reduced levels of GSH, eleveated NRF2. Abnormal mitochondrial morphology observed in neurons. Elevated levels of α-synuclein levels in neurons. | No | No | [97] |
| Ectodermal | Rett Syndrome | Neurons | Heterozygous mutation in MECP2: C916T, 1155del32,C730T, C473T, CDKL5. | Fibroblasts | Lentiviral OSKM, Retroviral OSKM | Reduced spines and density of glutamatergic synapse formation, smaller soma size, altered calcium signaling, electrophysiological defects, maturation defects. | IGF1, Gentamicin—increased glutamatergic synapses and decreased frequency and intensity of spontaneous currents. | No | [37, 98–100] |
| Ectodermal | Timothy syndrome | Neurons | CACNA1C. | Dermal Fibroblasts | Retroviral OSKM | Defects associated with calcium signaling and calcium associated gene expression. Abnormal tyrosine hydrolase expression and increase levels of norepinephrine and dopamine. Neuronal differentiation abnormalities. | Roscovitine treatment reversed these abnormalities. | No | [101] |
| Ectodermal | Machado-Joseph disease (Spinocerebeller ataxia type 3) | Neurons | ATXN3. | Dermal Fibroblasts | Retroviral OSKM | Demonstrated l-glutamate induced excitation causes a Calcium dependend proteolysis of ATXN3 aggregates. | Calpain treatment abolishes the phenotype. | No | [102] |
| Mesodermal | LQTS (Type 1 & 2) | Cardiomyocytes | LQT1 (mutation in KCNQ1 - 1893delC), Long QT syndrome (LQTS) type 2 (R176W, A624V in KCNH2 (HERG) gene), G1681A in KCNH2. | Dermal fibroblasts | Retroviral OSKM, Lentiviral OSLN, Lentiviral Slc7a1 + Retroviral OSKM, | Type 1:Increased APD, Increased susceptibility to catecholamine-induced tachyarrhythmia Type 2:Longer action potential duration of the mutant cells compared to controls. Increased inverse correlation between beating rate and repolarization time. |
Type 1:None tested. Type 2:Increased sensitivity to arrhythmogenic drugs (sotalol), IRK blockers (E-4031), cisapride, nifepidine, pinacidil, ranolazine,, Nicorandil, isoprenaline,na-dolol, PD-118057.. |
No | [41, 103, 104] |
| Mesodermal | CPVT1 | Cardiomyocytes | Heterozygous autosomal dominant mutation p.F2483I mutation in RYR2 gene; (S406L mutation in RYR2). | Dermal fibroblasts and Hair Keratinocytes | STEMCCA Lentivirus OSKM | Exhibited arrhythmias and delayed depolarisation after catecholaminergic stimulation. Higher amplitude and duration of calcium induced calcium released events, elevated diastolic calcium as compared to control. EM analysis showed immature myofibrils, enlarged sarcoplasmic reticulum cisternae and reduced caveolae. | Forskolin used to increase cytosolic c-AMP levels and abolish increase of calcium release events; Dantrolene—restored normal Ca(2+) levels and activity. | No | [105] |
| Mesodermal | Familial dilated cardiomyopathy | Cardiomyocytes | (R173W in gene encoding sarcomeric protein cardiac troponin T). | Adipose stem cells | Lentiviral OSKM | Altered calcium regulation, decrease in contractility, abnormal sarcomeric α-actin distribution. | β-adrenergic agonists—cellular stress signs (reduced beating rate, reduced contraction, increase in abnormal α-actin distribution). β-adrenergic blockers rescued the phenotype. | Overexpression of sarcoplasmic reticulum Ca(2+) adenosine triphosphatase (SERCA2a) improved function in disease derived cardiomyocytes. | [106] |
| Mesodermal | Chronic granulomatous disease | Macrophages | p47(phox) or gp91(pho) (x) mutations. | Fibroblasts | Lentiviral OSKM or OSLN | Disease iPSC-derived macrophages have normal phagocytic activity but no reactive oxygen species production, which mimics the clinical pathology. | No | No | [107] |
| Mesodermal | Fanconi Anemia | HSC | FANCA gene, FANCC gene. | Fibroblasts | Retroviral OSKM/polycistronic lentivirus OSKM | Mutation in DNA damage repair pathways causes cells to be refractory to reprogramming. | No | Gene correction leads to reprogramming and mutation free HSC differentiation. | [43–45] |
| Mesodermal | Trisomy 21 (Down Syndrome) | Myeloid Haematopoiesis (other tissues affected) | Trisomy 21. | hES, Fibroblasts and stromal cells | Retroviral OSKM | A developmental stage specific haematopoietic phenotype specifically reduced myelopoiesis and elevated erythropoiesis. | No | Yes, used isogenic controls. | [49, 50] |
| Mesodermal | LEOPARD syndrome | Cardiomyocytes (other tissues and organs) | PTPN11 (SHP2 phosphatase). | Dermal Fibroblasts | Retroviral OSKM | Cardiomyocytes demonstrated elevated sarcomeric organisation, nuclear localisation of NFATc4, elevated levels of phosphor-ERK. | No | No | [108] |
| Mesodermal | Gauchers disease | Macrophage (neurons) |
GBA Type 1 N370S/N370S, Type2 L44P/RecNcil, Type3 L444P/L444P. |
Fibroblasts | STEMCCA Lentivirus OSKM | Low glucocerebrosidase activity and accumulation of sphinolipids and compromised lysosomal function. Defective RBC clearance. | Isofagomine partially restored RBC clearance. | Incubation with recombinant glucocerebrosidase rescued RBC clearance. | [46] |
| Mesodermal | Hutchinson-Gilford progeria syndrome | Mesenchymal lineages: skeletal system, dermis and vascular smooth muscle cells | LMNA gene | Fibroblasts | Retroviral OSKM | Progerin was expressed in differentiated tissues from disease iPSC but not in the pluripotent cells. Misshapen nuclei. Loss of heterochromatin marker H3K9me3. Premature senescence. Reduced telomere lengths. Compromised cell proliferative capacity. Sensitivity to hypoxia. Inability of differentiated MSC to restore circulation in ischemic mouse model. Mislocalization of LAP2. Nuclear lobulation. DNA damage. |
No | Progerin shRNA corrected cells showed improvement in proliferative ability. | [110, 111] |
| Mesodermal | Congenital erythropoietic porphyria | Hematopoietic stem cells | Deficiency in enzymatic activity of uroporphyrinogen III synthase (UROS) | Epidermal keratinocytes | Lentiviral OSKM-shp53 | Disease iPSC derived erythroid cells have low UROS activity and high percentage of fluorocytes compared to control derived cells. | No | Lentiviral gene correction of UROS rescued UROS activity levels. | 112 |
| Endodermal | Diabetes | Pancreatic cells | Mitochondrial DNA A3243G mutation, Type 1 diabetics exhibiting either polyuria and polydypsia or ketoacidiosis. | Fibroblasts | Lentiviral Slc7a1 + Retroviral OSKM, Retroviral OSK | Some iPSC clones showed increase in frequency of mutation while others showed decrease. | No | No | [109] |
| Endodermal | α1-antitrypsin deficiency | Hepatocytes | SERPINA E342K (Z mutation) | Fibroblasts | Retroviral OSKM | Aggregation of misfolded α1-antitrypsin. | No | No | [52] |
| Endodermal | familial hypercholesterolemia (FH) | Hepatocytes | Autosomal dominant mutation in LDLR. | Fibroblasts | Retroviral OSKM Lentiviral OSLN |
Deficient LDL receptor-mediated cholesterol uptake, elevated secretion of APOB-100. | Lovaststatin, increased LDL uptake. | No | [52, 54] |
| Endodermal | glycogen storage disease type 1a | Hepatocytes | Absent hepatic G6Pase enzyme. | Fibroblasts | Retroviral OSKM | Elevated lipid and glycogen accumulation | No | No | [52] |
| Endodermal | Wilson's Disease | Hepatocytes (neurons) | ATP7B (R118L) | Fibroblasts | Retroviral OSKM | Abnormal ATP7B localization and defective copper transport. | Curcumin treatment rescues defect. | Lentiviral delivered ATP7B rescues phenotype. | [55, 56] |
| Endodermal | Cystic Fibrosis (CF) | Airway epithelia Lung progenitors | CF - F508del, G551D. | Skin fibroblasts | Retroviral OSKM mRNA | Rapid degradation of mutant CFTR protein. | VX-809 treatment results in surface localisation of mutant CFTR protein. | No | [57, 58] |
| Endodermal | Hepatitis C infection | Hepatocytes | N/A | Human embryonic lung Fibroblasts, Fibroblast | Retroviral OSKM, Lentiviral OSLN | Hepatocyte-like cell, but not pluripotent cells were able to support hepatitis C infection and proliferation. | Anti-CD81 dose dependently attenuated HCV entry. | No | [66–68] |
| Endodermal | Rotavirus | Intestine like tissue | N/A | Skin keratinocytes | Retroviral OSKM | Supported both the infection and replication of rotavirus. | No | No | [70] |
This table lists the diseases which have been successfully modelled in iPSCs and states the affected gene, the mutation if known, and the lineage in which the disease manifests. The source of the somatic cells and the method of reprogramming are also stated, along with details of the phenotype observed, and if applicable any pharmaceutical or genetic interventions employed.
Abbreviations: Reprogramming factor abbreviations: O, OCT3/4; S, SOX2; K, KLF4; L, LIN28; N, NANOG; M, c-MYC. Disease and gene abbreviations: ALS, amyotrophic lateral sclerosis; APOB-100, apolipoprotein B; APP, amyloid precursor protein; ATP7B, ATPase, Cu2+ transporting, beta polypeptide; ATXN3, ataxin 3; BACE1, beta-secretase 1; CD81, cluster of differentiation 81; CACNA1C, calcium channel, voltage-dependent, L type, alpha 1C subunit; CDKL5, cyclin-dependent kinase-like 5; CF, cystic fibrosis; CFTR, cystic fibrosis transmembrane conductance regulator; CPVT1, catecholaminergic polymorphic ventricular tachycardia; ERK, extracellular-signal-regulated kinases; FANCA, fanconi anemia, complementation group A; FANCC, fanconi anemia, complementation group C; FD, familial dysautonomia; FH, familial hypercholesterolemia; FTLD frontotemporal lobar degeneration; G6Pase, glucose-6-phosphatase; GBA, β-glucocerebrosidase; GSH, glutathione; HD, Huntington disease; HERG, human Ether-à-go-go-Related Gene; IGF1, insulin-like growth factor 1; IKBKAP, inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein; KCNH2, potassium voltage-gated channel, subfamily H (eagrelated), member 2; KNCQ1, potassium voltage-gated channel; LDLR, low-density lipoprotein receptor; LEOPARD, lentigines, electrocardiographic abnormalities, ocular hypertelorism, pulmonary valve stenosis, abnormal genitalia, retardation of growth and deafness; LQTS, long QT syndrome; LRRK2, leucine rich repeat kinase; MECP2, methyl CpG binding protein 2; NFATc4, nuclear factor of activated T-cells, cytoplasmic 4; NRF2, nuclear factor (erythroid-derived 2)-like 2; PARK2, parkinson protein 2 or E3 ubiquitin protein ligase; PD, Parkinson's disease; PTPN11 (SHP2), tyrosine-protein phosphatase non-receptor type 11; PI3K, phosphatidylinositol 3-kinase; RYR2, ryanodine receptor 2; SERCA2a, sarcoplasmic reticulum Ca2+ ATPase; SERPINA, serine protease inhibitor A (alpha-1-antitrypsin); SMA, spinal muscular atrophy; SMN1, survival of motor neuron 1; TDP-43, TAR DNA-binding protein 43. Other Abbreviations: c-AMP, cyclic adenosine monophosphate; APD, action potential duration; CD81, cluster of differentiation 81; DNA, deoxyribonucleic acid; EM, electron microscopy; HCV, hepatitis C virus; hESC, human embryonic stem cell; HSC, haematopoietic stem cell; iPSC, induced pluripotent stem cell; LDL, low density lipoprotein; NSC, neuronal stem cell; RBC, red blood cell; VPA, valproic acid.
Drawbacks of current approaches in modelling diseases
Up to this point, the majority of disease modelling has been conducted in animal models, patient fibroblast cultures / primary tissue, or by the overexpression of affected genes in previously characterised cell lines. Though these approaches are informative and have contributed to our overall understanding, each has its limitations. For example, due to differences in both gene expression and physiology, animal models do not always fully recapitulate disease states as they present in the human system, and this has led in part to high attrition rates on translation from animal to clinical studies [14–17]. The use of primary cells from affected individuals is limited both by access to donors and the lifespan of the tissue in culture, as well as the difficulty in accessing particular cell types from living patients such as neural and cardiac material. Finally, the overexpression of proteins in cell lines may not accurately reflect the pathophysiology of disease states, for example due to variability in transgene integration points and copy number states. With iPSC technology it is possible to isolate a patient biopsy, culture the cells, induce pluripotency, and differentiate the resulting iPSCs into the specific cell type afflicted with the disease. This opens up the possibility of studying the disease in situ in the relevant cell type under the correct genetic background, allowing the penetrance of the disease to be considered.
Human embryonic stem cells for modelling disease
Embryonic stem cell research has laid the groundwork for the development and use of iPSC technology. Following their initial derivation in 1998 by Thomson and colleagues [18], hESCs were predicted to provide a powerful platform for the scientific community to interrogate disease, as well as a limitless supply of somatic cells for therapy and translation. However, their widespread adoption has been slowed by the ethical concerns which still surround the hESC derivation process. In addition to leading the way to iPSC technology, hESCs have also provided insight into disease in their own right through several different approaches.
One method involves manipulating the genome of the hESC line, as exemplified by the modelling of Lesh-Nyhan Syndrome. This model was generated via gene targeting to introduce a mutated form of the disease gene HPRT1. The generated lines exhibited some of the attributes of the disease, both lack of HPRT1 activity and elevated levels of uric acid [19]. This approach is both difficult and laborious due to technical limitations, but recent advances in genome engineering with zinc finger nucleases (ZFNs) and transcription activator like effector nucleases (TALENs) which can modify the genome with precision will potentially allow the modification of hESC genomes more routinely [20–23]. By modifying target sequences in the genome, ZFN and TALEN-based gene editing may be able to introduce or correct disease-causing mutations in iPSCs. This will allow the rapid and precise generation of genetically well-defined and homogeneous iPSCs for disease modelling. This approach will be applicable to defined monogenic disease states where penetrance of the disease is not an issue.
Another approach that has been exploited in hESCs is pre-implantation genetic diagnosis (PGD) [24]. PGD is carried out when one or both parents harbour either a known mutation such as those seen in X-linked monogenic diseases or a known chromosomal abnormality; testing is performed to determine if these are present in the embryo. To date a number of studies have described PGD derived disease models including Huntington, Charcot Marie Tooth type 1A, Fragile-X and cystic fibrosis [19, 25–31]. Although many lines have been derived following PGD, there are very few publications that describe further characterisation in terms of phenotype recapitulation. A few notable studies have delved deeper, including one describing an hESC line containing Fragile-X mutation, in which mutant FMR1 gene was shown to be actively transcribed in the pluripotent state, and silenced upon differentiation. There are again a number of drawbacks to this approach in that it is dependent on access to PGD embryos carrying the required disease state and suffers from a lack of patient information, which is especially important in determining the penetrance of many disorders. This coupled with ethical issues surrounding the use of hESCs has led researchers to embrace alternative strategies, chiefly those based on human iPSC technology.
Human induced pluripotent stem cells for modelling disease
iPSC technology has moved at great speed with respect to both the refinement of derivation techniques, and the diversity of tissue types that can be reprogrammed [11]. In addition we have seen the translation of this technology to other arenas including the derivation of iPSCs from endangered species and farm animals [32]. Disease specific lines were first reported in 2008 by Park and colleagues, who describe 10 disease specific iPSC lines [12]. Since this publication the number of disease specific lines has exploded, though many studies fail to demonstrate a disease phenotype. In the following passages we discuss only the hiPSC models that present a phenotype; for a comprehensive list, see Table 1.
Disease modelling - Ectodermal lineages
In addition to the difficulties in using animal models to study human diseases, as discussed above, one of the major limitations hampering research into diseases which affect the neural lineages has been the difficulty in obtaining representative human tissue samples. Traditionally these samples have been obtained post-mortem. While these post-mortem samples have contributed immensely to our understanding, they are representative of the end point of the disease, making it difficult to unravel early events, progression and the overall etiology. With the advent of induced pluripotent stem cell technology and downstream procedures to produce regionalised neuronal subtypes, patients specific lines representative of many neurological and neurodegenerative disorders can now be studied.
The use of pluripotent cells for researching the neural lineage is well established, both in neurological and neurodegenerative disease. One of the earliest models to be described was spinal muscular atrophy (SMA), an autosomal recessive disorder caused by mutations in the survival of the motor neuron 1 gene (SMN1), which leads to death in infants. Ebert and colleagues derived iPSCs from a SMA patient and demonstrated that these could be differentiated to both neurons and astrocytes [33]. The striking observation was that the mutant line recapitulated the disease state in terms of lack of SMN1, and also exhibited selective death of motor neurons. Additionally, the authors demonstrated that SMN1 levels could be modulated by treatment with compounds known to induce SMN1, thus providing a “proof of concept” that iPSC models can be used as a platform for screening therapeutic interventions.
Another early study published in 2009 by the Studer laboratory describes patient-specific iPSCs representing familial dysautonomia (FD), a rare but fatal neuronal disease [34]. FD is caused by a point mutation in the IKBKAP gene, and its disruption leads to a tissue specific splicing defect causing loss of autonomic and sensory neurons [35]. Owing to a lack of suitable model systems the role of the FD defect in the aetiology of the disease remained elusive. To this end, Studer and colleagues derived iPSCs from 3 FD patients and non-affected control patients, and confirmed the presence of the FD causing mutation in IKBKAP. The iPSCs were differentiated into cells deriving from all three lineages, further confirming pluripotency. On differentiation to neural precursors, no notable difference in differentiation efficiency was detected. However, markedly lower levels of the normal IKBKAP transcript were observed in the mutant line-derived neural crest precursors compared to controls. The mutant lines also demonstrated a decreased rate of neurogenesis and migration potential when subjected to a wound healing assay. These observations correlated well with the in vivo pathophysiology of the disease, demonstrating an observable relevant phenotype. As a “proof of concept” various compounds were used to assess rescue of phenotype with respect to splicing defects, rate of neurogenesis and migration potential. Of the compounds tested, kinetin, a plant hormone which had previously shown to rescue splicing defect in FD-lymphoblast lines [35], rescued this phenotype in neural crest precursors. Interestingly, a short term treatment was only able to rescue the splicing defect, with no effect observed against neurogenesis or migration. Extended treatment with kinetin produced a significant rescue of neurogenesis, but no improvement of migration. In a recent review, Studer and colleagues have gone on to outline the future of research into FD and indeed other diseases with iPSC technology [36]. They identify three key areas of future work based on FD-iPSCs: genetic correction of the disease mutation, mechanistic studies of the disease pathology, and the use of FD-iPSC derived neural precursors as a platform for drug screening.
The FD study was successful because there is a clearly defined mutation which presents a definite phenotype in vitro that correlates to the clinical presentation of the disease in human patients; this is true now of many monogenic disorders (see Table 1). For more complex neuronal diseases such as the autism spectrum disorder (ASD), new genomic sequencing and genome wide association (GWA) studies are highlighting novel ASD-relevant genes to study using iPSC technology [37]. Using iPSCs derived from patients harbouring more highly characterised genetic aberrations as a model for autism, it will be possible to create a platform for drug discovery which could be effective in alleviating the symptoms of patients with a more complex cause of the disease.
An important factor that needs to be taken into consideration is the time of onset of the disease. For example amyotrophic lateral sclerosis (ALS) is a progressive disorder that usually manifests around the age of 50 [38]. Other neurodegenerative disease such as Parkinson's and Alzheimer's also develop over time. All of these have been modelled in iPSCs and all exhibit some of the phenotypic signatures of the disease state. For example, Bilican and colleagues recently demonstrated that an iPSC model of ALS (mutation in the gene TARDBP) exhibited the pathological hallmarks of the disease, in that mutant motor neurons (MNs) contained detergent resistant TDP-43 protein [39]. In addition, the functionality of MNs was not compromised, but a reduced survival rate was observed. The above studies give credence to the use of iPSC technology for the interrogation of late onset disease types.
Disease modelling - Mesodermal lineages
The generation of mesodermal tissue from pluripotent stem cells has been the focus of the pharmaceutical industry, namely the production of cardiomyocytes to investigate cardiotoxicity to improve drug safety and potentially accelerate drug discovery. The rationale for this is to provide a supply of cells that can stop gap the shortage of primary material. Additionally, researchers have targeted therapeutically relevant cell types for use in cellular therapy. Now researchers armed with protocols to generate the afflicted cell types, combined with access to patients and iPSC technology will provide a powerful platform to study disease.
There are now a number of studies that describe the modelling of diseases that affect mesodermal tissues. These include the cardiac diseases, and to date there have been a number of reports that describe iPSC models of the cardiac arrhythmias, including the congenital long QT syndrome (LQTS) which affects cardiomyocytes [40]. In a study by Itzhaki and colleagues, type-2 LQTS patient specific iPSCs were derived containing an A614V missense mutation in the KCNH2 gene [41]. On analysis of differentiated cardiomyocytes from these cells by patch clamping, a prolonged action potential was revealed in comparison to control. Upon further investigation, it was revealed that the defect was caused by a reduction in the cardiac potassium current (Ikr), which is crucial for cardiomyocyte function. Itzhaki and colleagues then tested various pharmacological compounds to assess their influence on the observed phenotype [41]. The rationale behind this was to demonstrate the utility of iPSC models in emulating disease states and producing platforms for drug screening, and also to investigate drug interactions that could potentiate the problem. This was highlighted when Itzhaki and colleagues observed increased arrhythmogenicity in LQTS-iPSC derived cardiomyocytes following exposure to cisapride, a drug that was removed from market due to pro-arrthythmic mortality [41]. Developing this technology further will create a platform to help reduce drug attrition and provide personalised screening for adverse drug reactions.
There are now a number of studies which describe the modelling of disease states affecting hematopoietic tissues. One such disease is Fanconi anaemia (FA), which is a bone marrow failure disease caused by mutations in a large number of genes in the FA pathway, but whose mechanism remains elusive [42]. A study by Raya and colleagues described the unresponsiveness of FA cells to reprogramming, and reported that iPSCs from FA patients could only be derived after correction of the affected gene [43]. This raised the question of whether this was due to the refractory nature of FA cells to reprogramming, or if the limitations of current technology were a contributing factor. In 2012 Muller and colleagues demonstrated the latter, using physiological normoxia (5% O2) and a polycistronic lentivirus to produce FA-iPSCs, albeit at a reduced efficiency [44, 45]. Now that both corrected and uncorrected FA-iPSC lines have been derived it will be possible to investigate the disease in an isogenic background.
A number of recent papers describe the generation of iPSCs from patients with diseases affecting several different tissue types. Panicker and colleagues [46] derived iPSCs from patients with Gaucher's disease (GD), an inherited lipid storage disease with a high prevalence in the Ashkenazi Jewish population [47, 48]. Patients with GD present with many symptoms including haematological abnormalities, enlarged liver and spleen, and bone disease, with varying severity depending on the type of GD. Types 1 to 3 were modelled in this study [46]. The GD-iPSCs were differentiated to macrophages, the cell type most influenced by this disease, and to neurons which are also affected. Both the macrophages and neurons presented with the expected reduced levels of glucoceroroside activity, along with accumulation of sphingolipids and loss of lysosomal function. In the macrophages this was further reinforced by their inability to clear red blood cells, a phenotype representative of the severity of the mutation (type 1, 2 or 3). Interestingly, this phenotype could be rescued with recombinant glucoceroroside, and partially rescued with the chaperone isofagomine, in agreement to clinical observations.
Two other recent publications describe the derivation of trisomy 21 iPSCs for the modelling of the haematological abnormalities associated with Down's syndrome, which previously had been impossible [49, 50]. Both groups produced iPSC lines, but MacLean and colleagues derived isogenic iPSCs that were di- or trisomic with respect to chromosome 21. In addition they utilised a hESC line containing trisomic 21 as a control. Both groups observed differences in differentiation potential, with a reduction of myeloid potential and increased erythroid potential. The added value of using isogenic controls not only here but in other model systems is that this will allow further dissection of the molecular mechanism of the disease state. In an independent study by Li and colleagues [51], they describe the derivation of Down's syndrome-iPSCs and the successful removal of one copy of chromosome 21, thus reverting it back to a normal chromosomal complement. This was achieved by targeting of one copy of chromosome 21 with thymidine kinase (TK) neomycin cassette in the presence of G418 selection, and then subsequent counter-selection for TK with ganciclovir. This approach has potential implications for modelling Down's syndrome with direct isolation of isogenic control iPSC line. However, as reported above by MacLean and colleagues [50], disomics for chromosome 21 can arise spontaneously, and cells could be generated in this way rather than through laborious genetic manipulation.
Disease modelling - Endodermal lineages
The generation of endoderm is of particular interest to the pharmaceutical industry, as a large proportion of drug toxicity relates to the liver. The main goal of the field has been to ameliorate the dearth of hepatocytes for toxicology, drug metabolism and drug efficacy, and to reduce drug attrition. In addition researchers have focussed heavily on the production of therapeutically relevant endodermal cell types for use in cellular therapy, in particular hepatocytes and pancreatic β-cells. The knowledge acquired from developmental studies has informed a number of publications describing the production of functional hepatic endoderm and pancreatic β-cells from human pluripotent stem cells. Now that the full potential of iPSCs is realised as discussed above, efforts are underway to utilise this technology to generate endodermal tissues for disease modelling.
One class of diseases being intensely investigated are the inherited metabolic disorders, which manifest due to mutations that perturb hepatocyte function. Rashid and colleagues [52] describe the production, characterisation and differentiation of 14 iPSC lines representing 5 different metabolic diseases. They then further derived three α1-antitrypsin deficiency (A1ATD) lines from different patients, a familial hypercholesterolemia (FH) line, and a glycogen storage disease type 1A (GSD1A) line, a model also generated by Ghodsizadeh and colleagues who did not describe a phenotype [53]. These 3 models all presented a phenotype following differentiation to hepatocytes that reflected some of the pathology of each disease. This highlights the utility of these models for potential platforms to screen therapeutic interventions. One salient point to be taken from this report was the phenotypic variation observed in the A1ATD disease lines which contained the same mutation (E342K), put down to the recalcitrant nature of one of the lines to the hepatocyte differentiation procedure. This highlights the potential pitfalls of the technology and reinforces the requirement for multiple independent clones and subjects. Cayo and colleagues have taken the preliminary study of Rashid and colleagues further to address key pathophysiological question of FH [54]; they investigated whether patient-derived iPSCs would reflect key defects in this metabolic disorder. Importantly, Cayo et al tailored the experimental approach to take into account the variability associated with reprogramming by employing multiple clones. Hepatocytes derived from these lines demonstrated a reduced ability to take up low-density lipoprotein (LDL), as well as increased apoB-100/VLDL secretion, in concurrence with clinical observations. They investigated the effect of lovastatin, an inhibitor of HMG-CoA reductase which reduces LDL, and on treatment they demonstrated increased LDL uptake. They also highlighted the issue of differential response to the statins; with a large number of individuals being poor responders due to the polymorphic variation inherent in the population. One possible solution to this would be the construction of a library from individuals representative of these polymorphisms who exhibit elevated cholesterol.
Two independent studies describe the generation of iPSCs from patients with Wilson's disease, a metabolic disorder caused by mutation of the ATP7B gene which leads to accumulations of copper in the liver and central nervous system [55, 56]. Both studies generated hepatocytes and demonstrated the presence of ATP7B at the transcription and protein level. In addition, Zhang and colleagues [55] demonstrated copper transport dysfunction, which was rescued through overexpression of ATB7B via lentivirus or treatment with the drug curcumin. The mechanisms behind copper accumulation in Wilson's disease remain unclear, and a tool such as this is likely to aid understanding and facilitate the development of new drugs and treatments. The number of iPSC models that affect the hepatic lineages is growing and now includes tyrosinemia type 1, progressive familial hereditary cholestasis, and Crigler-Najjar syndrome [52, 53].
Aside from the liver, other potential uses of endodermal cells in disease modelling relate to airway diseases and Type 1 diabetes. Two independent groups describe the derivation of patient specific cystic fibrosis (CF) iPSC lines [57, 58]. Mou and colleagues [58] describe the generation of lung and airway progenitors from the derived mutant iPSCs but were unable to produce mature lung epithelium ex vivo; the authors speculate this may be due to a lack of the correct epigenetic patterning. Wong and colleagues, appear to overcome some issues of maturation and describe the production of mature airway epithelia, but acknowledge the issue of variability and heterogeneity of differentiation [57]. The resulting epithelia presented CF protein in the expected polarity, on the apical plasma membrane. CF transport function was demonstrated but due to the aforementioned heterogeneity was variable. In CF mutants in this case (F508del) it has been shown that the mutant protein is not incorporated into the plasma membrane but targeted for degradation. This was recapitulated in the mutant lines with the F508 deletion, and could be partially rescued with VX-809, a therapeutic known to promote re-localisation currently in phase 2 trails. Overall function was however not recovered as ascertained by efflux potential. These studies are very promising and highlight the power of iPSC technology but also illustrate the limitations that can be encountered in terms of being able to generate homogeneous, mature cell types representative of the in vivo situation.
Type 1 and Type 2 diabetes has had been the focus of intense research, and as stated above the main area that has been addressed is the generation of mature β-cells ultimately for cell replacement therapy. However, researchers have seen the utility of iPSC technology to facilitate the dissection of monogenic, and some more complex diseases. A handful of papers describing the production of patient specific iPSC lines from both Type 1 and 2 diabetes are now published, but as of yet none describes a phenotype [59, 60]. For an insight into the direction of the use of pluripotent stem cells in diabetes research, see [61, 62].
Modelling Infectious Disease
An emerging area that is beginning to exploit iPSC technology is the study of infectious disease. To date the availability of models to explore infectious agents, pathogens and parasites, and their interactions with the host remain sparse. Some reports now describe the use of iPSCs to investigate a number of viral systems including the human immunodeficiency virus (HIV) and hepatitis C [63–68]. The recent publication by Spence and colleagues [69] describing the directed differentiation of human pluripotent stem cells to intestinal tissue has many implications in this area [70]. Very recently a publication by Finkbeiner and colleagues described the use of this system to model rotavirus infection in so called “induced human intestinal organoids” [70]. A potential implication of this study is the reduction of animals (African green monkeys) used to produce primary material. With this area rapidly developing, researchers outside the stem cell field are now becoming aware of the implications and versatility of this emerging technology [71–73].
Modelling Cancer
Another area gaining momentum is the use of iPSC technology to understand the molecular events involved in cancer and tumourigenicity. This is not a new area of investigation as early work describing the nuclear transfer of mouse embryonal carcinoma lines was performed in 2004, to investigate tumourigenic and developmental effects and the contribution of epigenetic and genetic components to the process [74].
The potential of pluripotent stem cells to give rise to all three lineages is a cornerstone of their potential in regenerative medicine. There are however concerns about their tumourigenic nature, exemplified by the formation of teratomas. These cells in turn are now being used to dissect the mechanism of their oncogenic potential [75]. The reprogramming process itself shares a lot of features with tumourigenesis, and indeed there are a number of studies that describe changes to the genome after this process [76, 77]. Insights gained from these early events may provide the tools to further understand cancer [78].
A number of studies have now described the reprogramming of cancer cells to a pluripotent state [79–83]. A striking observation is that upon differentiation of the cancer iPSCs, their cell progeny appear to lose or at least reduce their tumourigenic potential. It is speculated that this is driven by epigenetic remodelling which reverses the observed dysfunction [82]. In another study Lin and colleagues [80] compared the microRNA profile of the original cancer tissue to the cancer-derived iPSCs, and identified the hESC specific microRNA Mir-302. This was capable of reprogramming cancer cell lines, including Colo and PC3, to iPSC like cells with respect to self-renewal and pluripotency. Interestingly, they suggested this could be utilised to produce transgene free iPSCs from somatic cell types, which has now been described by Anokye-Danso et al [84]. More recently, the potential therapeutic application of this technology has been highlighted by the findings of Zhang et al [85]. This study demonstrated that sarcomas with complex karyotypes could be reprogrammed to an iPSC status, and in turn differentiated into cell types including connective tissue and red blood cells. Zhang et al. found that the differentiated tissues lost their proliferative capacity as well as their tumourigenicity. By analysing the transcriptome and the epigenome of the original cancer cells and comparing them to the reprogrammed cells, they found that extensive epigenetic remodelling of tumour suppressor genes and oncogenes arose during the reprogramming process [85]. The critical finding of this study was that it is possible to reprogram cancer cells, differentiate them to a tissue type of interest and essentially erase the characteristic hallmarks of the cancer. Through further study of the transcriptome and epigenome of cancer cells and their reprogrammed progeny it may be possible to understand the precise changes which lead to cancer being so effective in colonising the human body.
Challenges
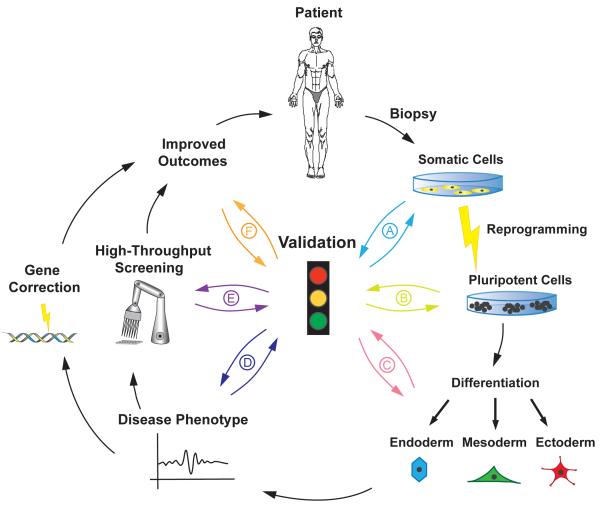
There are several key challenges which must be addressed if the power of iPSC technology is to be fully utilised in the modelling of disease. Firstly, bona fide and well characterised iPSC lines must be generated from disease patients as well as age and sex matched control lines, and if possible isogenic controls should be used for comparative studies. Secondly, there must be robust and efficient in vitro directed differentiation protocols available in order to generate the cell types of interest. These should be extensively characterised and possess the same properties as their in vivo counterparts. Importantly, the derived progeny should express the allele affected and manifest an observable phenotype (see Figure 1).
Figure 1. The process of creating improved patient care using reprogramming and differentiation of donor tissue.
A biopsy is taken from a patient, reprogrammed to a state of pluripotency before being differentiated to a cell type of interest. Disease-specific tissues can then be used to improve current understanding of disease states and aid the drug development process. The induction of pluripotency allows for limitless expandability of the cell population. Validation is critical to the success of the process; here we identify six key steps which must be addressed. (A). Tissue from the patient must present the genetic traits of the disease state. (B). Reprogrammed cells must demonstrate pluripotency as assessed through a rigorous, standardised validation process. (C). Differentiated cells must demonstrate the key characteristics of the mature cell type as assessed by marker expression and functionality. (D). The differentiated cells should present the disease phenotype. (E). Genetic and drug interventions should be able to correct the phenotype. The cell model should predict the response of current therapies. This will lead to increased knowledge of the disease mechanism. (F). Patient benefit must be assessed through clinical trials.
Another challenge relates to the types of diseases which can be modelled, as the majority of diseases described to date are monogenic, and many present with a phenotype (see Table 1). This may become more problematic with respect to complex diseases, as in many cases we are uncertain about the contribution of genetic and epigenetic components. This could prove to be an issue particularly with respect to iPSC derived cancer models as discussed above.
Next, the way in which iPSCs are derived has an influence on their epigenetic landscape, which is extremely pertinent when modelling X linked diseases [86]. Taken further it has been demonstrated that the sex of cell lines needs to be taken into account as Anguera and colleagues have shown that female iPSCs may be epigenetically less stable [87].
Assuming that these criteria are met, one key issue remains: the maturity of pluripotent stem cell derived progeny. It has recently been shown that differentiated products of hESCs and iPSCs retain an immature phenotype even when terminally differentiated [88]. This group performed comprehensive transcriptomic analysis on pluripotent cell derived progeny and compared them to their tissue derived equivalents. In doing so they found that although both hESCs and iPSCs made equivalent progeny when differentiated to all three lineages, the pluripotent stem cell derived tissue retained expression of genes which are associated with very early mammalian development in the order of 6 weeks of gestation, regardless of the tissue type. While they found that expression of pluripotency genes was quickly extinguished upon initiation of differentiation, other genes associated with embryonic tissue such as LIN28A, LIN28B, and DPPA4 were not silenced. This data raises serious concerns about disease modelling using pluripotent stem cells, as well as their future clinical application, and thus represents one of the key challenges to be tackled in the future, as it is critical that stem cell derived progeny are functionally mature in order to provide accurate information about the diseases being modelled. There are preliminary indications that culture conditions and microenvironments have a critical impact on their resulting maturity and function [89]. Finding the best possible combination of media composition, oxygen levels, growth factors and matrices, as well as considering 2D versus 3D culture, will very likely lead to dramatic improvements in the maturity and functionality of pluripotent stem cell derived progeny. iPSC technology is still a young, rapidly evolving field and the majority of these shortcomings are technical limitations which are likely to be overcome in the near future.
Conclusions
While tremendous progress has been made in recent years modelling diseases using patient derived iPSCs, much work remains to be done. It is clear that iPSC technology represents a powerful resource which can provide great insights into disease pathology, but it is essential that a number of key steps in the process be standardised (see Figure 1). The research community will need to agree on what defines a high quality iPSC, standardise methods of confirming pluripotency, and establish the minimum number of clones, subjects and controls deemed necessary to cover genetic variance. A number of groups are now working to address some of these issues, for example Muller et al. suggest a bioinformatical approach to define pluripotency and potentially the quality of cells [90]. In addition, through greater definition one may be able to predict the differentiation potential of lines thus allowing the researcher to choose lines fit for purpose. The most immediate benefit with regard to disease modelling and therapy is the use of these cells as a platform to discover novel compounds, which can ameliorate disease phenotypes as described above. While gaps remain in our understanding of the very nature of iPSCs and their relation to hESCs, it is clear that they have the potential to make a substantial contribution to our understanding of many of the diseases which affect the population.
Acknowledgements
We thank Carina Knudsen at the University of Oslo Photo and Graphics Service for the illustration. We also thank the members of the Norwegian Stem Cell Centre for helpful discussions. Gareth Sullivan was supported by the Research Council of Norway and Helse Sør Øst. Richard Siller and Sebastian Greenhough contributed equally to this work. In-Hyun Park was partly supported by NIH (GM0099130-01), YCCMD, Charles Hood Foundation and by CTSA Grant UL1 RR025750 from the National Center for Advancing Translational Science (NCATS), a component of the National Institutes of Health (NIH), and NIH roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
References
- 1.Briggs R, King TJ. Transplantation of Living Nuclei From Blastula Cells into Enucleated Frogs' Eggs. Proc Natl Acad Sci U S A. 1952;38(5):455–63. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elsdale TR, Gurdon JB, Fischberg M. A description of the technique for nuclear transplantation in Xenopus laevis. J Embryol Exp Morphol. 1960;8:437–44. [PubMed] [Google Scholar]
- 3.Gurdon JB. Factors responsible for the abnormal development of embryos obtained by nuclear transplantation in Xenopus laevis. J Embryol Exp Morphol. 1960;8:327–40. [PubMed] [Google Scholar]
- 4.Gurdon JB, Laskey RA. The transplantation of nuclei from single cultured cells into enucleate frogs' eggs. J Embryol Exp Morphol. 1970;24(2):227–48. [PubMed] [Google Scholar]
- 5.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 6.Wilmut I, et al. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385(6619):810–3. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K, et al. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007;2(12):3081–9. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 11.Mochiduki Y, Okita K. Methods for iPS cell generation for basic research and clinical applications. Biotechnol J. 2012;7(6):789–97. doi: 10.1002/biot.201100356. [DOI] [PubMed] [Google Scholar]
- 12.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134(5):877–86. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilmut I, Sullivan G, Chambers I. The evolving biology of cell reprogramming. Philos Trans R Soc Lond B Biol Sci. 2011;366(1575):2183–97. doi: 10.1098/rstb.2011.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeves RH, et al. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11(2):177–84. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 15.Hackam DG, Redelmeier DA. Translation of research evidence from animals to humans. JAMA. 2006;296(14):1731–2. doi: 10.1001/jama.296.14.1731. [DOI] [PubMed] [Google Scholar]
- 16.van der Worp HB, et al. Can animal models of disease reliably inform human studies? PLoS Med. 2010;7(3):e1000245. doi: 10.1371/journal.pmed.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi Y, et al. Species Differences in Tissue Distribution and Enzyme Activities of Arylacetamide Deacetylase in Human, Rat, and Mouse. Drug Metabolism and Disposition. 2012;40(4):671–679. doi: 10.1124/dmd.111.043067. [DOI] [PubMed] [Google Scholar]
- 18.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 19.Urbach A, et al. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6(5):407–11. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soldner F, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146(2):318–31. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–4. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carroll D. Zinc-finger nucleases: a panoramic view. Curr Gene Ther. 2011;11(1):2–10. doi: 10.2174/156652311794520076. [DOI] [PubMed] [Google Scholar]
- 23.Silva G, et al. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther. 2011;11(1):11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Yosef D, Malcov M, Eiges R. PGD-derived human embryonic stem cell lines as a powerful tool for the study of human genetic disorders. Mol Cell Endocrinol. 2008;282(1–2):153–8. doi: 10.1016/j.mce.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Bradley CK, et al. Derivation of Huntington's disease-affected human embryonic stem cell lines. Stem Cells Dev. 2011;20(3):495–502. doi: 10.1089/scd.2010.0120. [DOI] [PubMed] [Google Scholar]
- 26.Pickering SJ, et al. Generation of a human embryonic stem cell line encoding the cystic fibrosis mutation deltaF508, using preimplantation genetic diagnosis. Reprod Biomed Online. 2005;10(3):390–7. doi: 10.1016/s1472-6483(10)61801-9. [DOI] [PubMed] [Google Scholar]
- 27.Tropel P, et al. High-efficiency derivation of human embryonic stem cell lines following preimplantation genetic diagnosis. In Vitro Cell Dev Biol Anim. 2010;46(3–4):376–85. doi: 10.1007/s11626-010-9300-8. [DOI] [PubMed] [Google Scholar]
- 28.Mateizel I, et al. Derivation of human embryonic stem cell lines from embryos obtained after IVF and after PGD for monogenic disorders. Hum Reprod. 2006;21(2):503–11. doi: 10.1093/humrep/dei345. [DOI] [PubMed] [Google Scholar]
- 29.Mateizel I, et al. Derivation, culture, and characterization of VUB hESC lines. In Vitro Cell Dev Biol Anim. 2010;46(3–4):300–8. doi: 10.1007/s11626-010-9284-4. [DOI] [PubMed] [Google Scholar]
- 30.Eiges R, et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1(5):568–77. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Lannon BM, et al. Derivation of the first human embryonic stem cell line with Charcot Marie Tooth disease. Fertility and sterility. 2009;92(3):S171–S172. [Google Scholar]
- 32.Ben-Nun IF, et al. Induced pluripotent stem cells from highly endangered species. Nat Methods. 2011;8(10):829–31. doi: 10.1038/nmeth.1706. [DOI] [PubMed] [Google Scholar]
- 33.Ebert AD, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457(7227):277–80. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee G, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461(7262):402–6. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slaugenhaupt SA, et al. Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum Mol Genet. 2004;13(4):429–36. doi: 10.1093/hmg/ddh046. [DOI] [PubMed] [Google Scholar]
- 36.Lee G, Studer L. Modelling familial dysautonomia in human induced pluripotent stem cells. Philos Trans R Soc Lond B Biol Sci. 2011;366(1575):2286–96. doi: 10.1098/rstb.2011.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim KY, et al. Cellular reprogramming: a novel tool for investigating autism spectrum disorders. Trends Mol Med. 2012;18(8):463–71. doi: 10.1016/j.molmed.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7(9):710–23. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 39.Bilican B, et al. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc Natl Acad Sci USA. 2012;109(15):5803–8. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoekstra M, et al. Induced pluripotent stem cell derived cardiomyocytes as models for cardiac arrhythmias. Front Physiol. 2012;3:346. doi: 10.3389/fphys.2012.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itzhaki I, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471(7337):225–9. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 42.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8(10):735–48. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 43.Raya A, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460(7251):53–9. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller LU, et al. Induced pluripotent stem cells as a tool for gaining new insights into Fanconi anemia. Cell Cycle. 2012;11(16):2985–90. doi: 10.4161/cc.21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller LU, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119(23):5449–57. doi: 10.1182/blood-2012-02-408674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panicker LM, et al. Induced pluripotent stem cell model recapitulates pathologic hallmarks of Gaucher disease. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1207889109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horowitz M, et al. Prevalence of glucocerebrosidase mutations in the Israeli Ashkenazi Jewish population. Hum Mutat. 1998;12(4):240–4. doi: 10.1002/(SICI)1098-1004(1998)12:4<240::AID-HUMU4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 48.Beutler E, et al. Gaucher disease: gene frequencies in the Ashkenazi Jewish population. Am J Hum Genet. 1993;52(1):85–8. [PMC free article] [PubMed] [Google Scholar]
- 49.Chou ST, et al. Trisomy 21-associated defects in human primitive hematopoiesis revealed through induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109(43):17573–8. doi: 10.1073/pnas.1211175109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maclean GA, et al. Altered hematopoiesis in trisomy 21 as revealed through in vitro differentiation of isogenic human pluripotent cells. Proc Natl Acad Sci USA. 2012;109(43):17567–72. doi: 10.1073/pnas.1215468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li LB, et al. Trisomy Correction in Down Syndrome Induced Pluripotent Stem Cells. Cell Stem Cell. 2012 doi: 10.1016/j.stem.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rashid ST, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120(9):3127–36. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghodsizadeh A, et al. Generation of liver disease-specific induced pluripotent stem cells along with efficient differentiation to functional hepatocyte-like cells. Stem Cell Rev. 2010;6(4):622–32. doi: 10.1007/s12015-010-9189-3. [DOI] [PubMed] [Google Scholar]
- 54.Cayo MA, et al. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology. 2012 doi: 10.1002/hep.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang S, et al. Rescue of ATP7B function in hepatocyte-like cells from Wilson's disease induced pluripotent stem cells using gene therapy or the chaperone drug curcumin. Hum Mol Genet. 2011;20(16):3176–87. doi: 10.1093/hmg/ddr223. [DOI] [PubMed] [Google Scholar]
- 56.Yi F, et al. Establishment of hepatic and neural differentiation platforms of Wilson's disease specific induced pluripotent stem cells. Protein Cell. 2012 doi: 10.1007/s13238-012-2064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong AP, et al. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat Biotechnol. 2012 doi: 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mou H, et al. Generation of multipotent lung and airway progenitors from mouse ESCs and patient-specific cystic fibrosis iPSCs. Cell Stem Cell. 2012;10(4):385–97. doi: 10.1016/j.stem.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maehr R, et al. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci USA. 2009;106(37):15768–73. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohmine S, et al. Reprogrammed keratinocytes from elderly type 2 diabetes patients suppress senescence genes to acquire induced pluripotency. Aging (Albany NY) 2012;4(1):60–73. doi: 10.18632/aging.100428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maehr R. iPS cells in type 1 diabetes research and treatment. Clin Pharmacol Ther. 2011;89(5):750–3. doi: 10.1038/clpt.2011.1. [DOI] [PubMed] [Google Scholar]
- 62.Kao DI, Chen S. Pluripotent stem cell-derived pancreatic beta-cells: potential for regenerative medicine in diabetes. Regen Med. 2012;7(4):583–93. doi: 10.2217/rme.12.27. [DOI] [PubMed] [Google Scholar]
- 63.Yoshizaki S, et al. Vaccination with Human Induced Pluripotent Stem Cells Creates an Antigen-Specific Immune Response Against HIV-1 gp160. Front Microbiol. 2011;2:27. doi: 10.3389/fmicb.2011.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ni Z, et al. Human pluripotent stem cells produce natural killer cells that mediate anti-HIV-1 activity by utilizing diverse cellular mechanisms. J Virol. 2011;85(1):43–50. doi: 10.1128/JVI.01774-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jerebtsova M, et al. HIV-1 Resistant CDK2-Knockdown Macrophage-Like Cells Generated from 293T Cell-Derived Human Induced Pluripotent Stem Cells. Biology (Basel) 2012;1(2):175–195. doi: 10.3390/biology1020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwartz RE, et al. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109(7):2544–8. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu X, et al. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathog. 2012;8(4):e1002617. doi: 10.1371/journal.ppat.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshida T, et al. Use of human hepatocyte-like cells derived from induced pluripotent stem cells as a model for hepatocytes in hepatitis C virus infection. Biochem Biophys Res Commun. 2011;416(1–2):119–24. doi: 10.1016/j.bbrc.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 69.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470(7332):105–9. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finkbeiner SR, et al. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio. 2012;3(4):e00159–12. doi: 10.1128/mBio.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klotz C, Aebischer T, Seeber F. Stem cell-derived cell cultures and organoids for protozoan parasite propagation and studying host-parasite interaction. Int J Med Microbiol. 2012;302(4–5):203–9. doi: 10.1016/j.ijmm.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 72.Howell JC, Wells JM. Generating intestinal tissue from stem cells: potential for research and therapy. Regen Med. 2011;6(6):743–55. doi: 10.2217/rme.11.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang A, Sander M. Generating cells of the gastrointestinal system: current approaches and applications for the differentiation of human pluripotent stem cells. J Mol Med (Berl) 2012;90(7):763–71. doi: 10.1007/s00109-012-0923-y. [DOI] [PubMed] [Google Scholar]
- 74.Blelloch RH, et al. Nuclear cloning of embryonal carcinoma cells. Proc Natl Acad Sci USA. 2004;101(39):13985–90. doi: 10.1073/pnas.0405015101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ghosh Z, et al. Dissecting the oncogenic and tumorigenic potential of differentiated human induced pluripotent stem cells and human embryonic stem cells. Cancer Res. 2011;71(14):5030–9. doi: 10.1158/0008-5472.CAN-10-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gore A, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471(7336):63–7. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hussein SM, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471(7336):58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 78.Bernhardt M, et al. Mediators of induced pluripotency and their role in cancer cells - current scientific knowledge and future perspectives. Biotechnol J. 2012;7(6):810–21. doi: 10.1002/biot.201100347. [DOI] [PubMed] [Google Scholar]
- 79.Hu K, et al. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117(14):e109–19. doi: 10.1182/blood-2010-07-298331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin SL, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14(10):2115–24. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Utikal J, et al. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci. 2009;122(Pt 19):3502–10. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mahalingam D, et al. Reversal of aberrant cancer methylome and transcriptome upon direct reprogramming of lung cancer cells. Sci Rep. 2012;2:592. doi: 10.1038/srep00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miyoshi N, et al. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci USA. 2010;107(1):40–5. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Anokye-Danso F, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8(4):376–88. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang X, et al. Terminal differentiation and loss of tumorigenicity of human cancers via pluripotency-based reprogramming. Oncogene. 2012 doi: 10.1038/onc.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tomoda K, et al. Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell. 2012;11(1):91–9. doi: 10.1016/j.stem.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anguera MC, et al. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11(1):75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patterson M, et al. Defining the nature of human pluripotent stem cell progeny. Cell Res. 2012;22(1):178–93. doi: 10.1038/cr.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thery M. Micropatterning as a tool to decipher cell morphogenesis and functions. J Cell Sci. 2010;123(Pt 24):4201–13. doi: 10.1242/jcs.075150. [DOI] [PubMed] [Google Scholar]
- 90.Muller FJ, et al. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8(4):315–7. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Israel MA, et al. Probing sporadic and familial Alzheimer's disease using induced pluripotent stem cells. Nature. 2012;482(7384):216–20. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yagi T, et al. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Hum Mol Genet. 2011;20(23):4530–9. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- 93.Yahata N, et al. Anti-Abeta drug screening platform using human iPS cell-derived neurons for the treatment of Alzheimer's disease. PLoS One. 2011;6(9):e25788. doi: 10.1371/journal.pone.0025788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Camnasio S, et al. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington's disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol Dis. 2012;46(1):41–51. doi: 10.1016/j.nbd.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 95.An MC, et al. Genetic correction of Huntington's disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11(2):253–63. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sanchez-Danes A, et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson's disease. EMBO Mol Med. 2012;4(5):380–95. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Imaizumi Y, et al. Mitochondrial dysfunction associated with increased oxidative stress and alpha-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol Brain. 2012;5(1):35. doi: 10.1186/1756-6606-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hotta A, et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat Methods. 2009;6(5):370–6. doi: 10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- 99.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143(4):527–39. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Amenduni M, et al. iPS cells to model CDKL5-related disorders. Eur J Hum Genet. 2011;19(12):1246–55. doi: 10.1038/ejhg.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pasca SP, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17(12):1657–62. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Koch P, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480(7378):543–6. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 103.Moretti A, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363(15):1397–409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 104.Lahti AL, et al. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech. 2012;5(2):220–30. doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Novak A, et al. Cardiomyocytes generated from CPVTD307H patients are arrhythmogenic in response to beta-adrenergic stimulation. J Cell Mol Med. 2012;16(3):468–82. doi: 10.1111/j.1582-4934.2011.01476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun N, et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4(130):130ra47. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jiang Y, et al. Derivation and functional analysis of patient-specific induced pluripotent stem cells as an in vitro model of chronic granulomatous disease. Stem Cells. 2012;30(4):599–611. doi: 10.1002/stem.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Carvajal-Vergara X, et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465(7299):808–12. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fujikura J, et al. Induced pluripotent stem cells generated from diabetic patients with mitochondrial DNA A3243G mutation. Diabetologia. 2012;55(6):1689–98. doi: 10.1007/s00125-012-2508-2. [DOI] [PubMed] [Google Scholar]
- 110.Zhang J, et al. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8(1):31–45. doi: 10.1016/j.stem.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 111.Liu G, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472(7342):221–5. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bedel A, et al. Metabolic correction of congenital erythropoietic porphyria with iPSCs free of reprogramming factors. Am. J. Hum. Genet. 2012;91(1):109–21. doi: 10.1016/j.ajhg.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]