Abstract
We report the identification and characterization of 2,000 human diallelic insertion/deletion polymorphisms (indels) distributed throughout the human genome. Candidate indels were identified by comparison of overlapping genomic or cDNA sequences. Average confirmation rate for indels with a ⩾2-nt allele-length difference was 58%, but the confirmation rate for indels with a 1-nt length difference was only 14%. The vast majority of the human diallelic indels were monomorphic in chimpanzees and gorillas. The ratio of deletion:insertion mutations was 4.1. Allele frequencies for the indels were measured in Europeans, Africans, Japanese, and Native Americans. New alleles were generally lower in frequency than old alleles. This tendency was most pronounced for the Africans, who are likely to be closest among the four groups to the original modern human population. Diallelic indels comprise ∼8% of all human polymorphisms. Their abundance and ease of analysis make them useful for many applications.
Introduction
Nearly all genetics research makes use of DNA sequence variants. Despite this, we know surprisingly little about the numbers and types of variants within human populations. DNA polymorphisms are usually defined as naturally occurring variants for which the most common allele has a frequency of no more than 99% (Gelehrter and Collins 1990).
The vast majority of human DNA polymorphisms can be split into two groups: those based on nucleotide substitutions (commonly called “SNPs”) and those based on insertion or deletion of one or more nucleotides (indels). Indels can in turn be divided into those with multiple alleles (multiallelic) and those with only two alleles (diallelic). Nearly all of the multiallelic indels are based on tandem repeats, mostly STRs. STRPs (also called “microsatellites”) have been the predominant type of polymorphism used in human genetic studies since about 1990. More recently, millions of candidate SNPs have been identified and are beginning to be applied (International SNP Map Working Group 2001).
In contrast, diallelic indels have received very little attention. Diallelic indels vary greatly in length difference between alleles. In rare cases, the length difference between alleles can be tens or even hundreds of kilobase pairs (Lupski et al. 1996). Some diallelic indels differ by the insertion of a retroposon, such as an Alu or L1 element (Watkins et al. 2001). However, by far the largest group of diallelic indels are those with allele-length differences of relatively few nucleotides. The most recently published broad surveys of short indels (covering 80 polymorphisms) were by Krawczak and Cooper (Cooper and Krawczak 1991; Krawczak and Cooper 1991). In the present article, we report basic properties of human diallelic indels determined by the analysis of 2,000 polymorphisms.
Material and Methods
For the Unigene clusters and the SNP Consortium cliques (see the “Results” section), sequences were aligned using the Fragment Assembly System (Genetics Computer Group). Because the single-pass Unigene cDNA sequences were of relatively low quality, candidate indels were considered only if the cluster contained ⩾4 reads and if the minor allele appeared in at least two reads. All BAC-end/BAC overlaps and 14% of the BAC/BAC overlaps were aligned using a customized version of BLAST that distributed jobs nightly to idle laboratory computers. Candidates were considered from alignments of ⩾90% overall identity. Short BAC-end sequences (Zhao et al. 2000) were paired with full BAC sequences from the Sanger, MIT, and Baylor sequencing centers. Sanger Institute public Acedb files were parsed to identify pairs of overlapping BACs (see the Sanger Institute Web site).
The remaining 86% of BAC/BAC overlap candidates were obtained by collection of large insert clone (predominantly BAC) sequences plus associated PHRAP nucleotide-quality values from the public genome sequencing centers (International Human Genome Sequencing Consortium 2001). Overlaps between pairs of clones (∼1.1 Gb of sequence; G. Marth, G. Schuler, R. Yeh, R. Davenport, R. Agarwala, D. Church, S. Wheelan, J. Baker, M. Ward, M. Kholodov, L. Phan, H. Harpending, A. Chakravarti, P.-Y. Kwok, and S. Sherry, unpublished data) were detected primarily with a BLAST similarity search. Putative overlaps were filtered using stringent criteria, to avoid overlaps that represent duplicated segments of the genome. For each overlapping clone pair, a precise, nucleotide-wise alignment was produced using the CROSS_MATCH banded Smith-Waterman dynamic-programming alignment algorithm. These alignments were analyzed with a modified version of POLYBAYES SNP-discovery software (Marth et al. 1999). For substitutions, POLYBAYES computes an SNP confidence value by using the PHRED or PHRAP nucleotide-quality values of the sequences aligned at the candidate polymorphic site. Because nucleotide-quality values do not directly provide information on the likelihood of deleted nucleotides, a similar confidence value cannot be computed for candidate indels. Instead, an experimental, heuristic algorithm was used on the basis of the logic that high-quality, well-resolved nucleotides are unlikely to represent artifactual insertions or deletions due to sequencing error. Accordingly, a high confidence value was assigned to a candidate if (1) insertion nucleotides were of high sequence quality and (2) nucleotides flanking the site of polymorphism in both the long and short alleles were of high sequence quality.
Candidate indels were further screened by manual inspection. To avoid multiallelic polymorphisms, we excluded sequences if, at the site of polymorphism, the long allele contained more than five uninterrupted, tandem mononucleotide repeats (e.g., (A)6) or more than three uninterrupted, tandem repeats with 2–6-nt repeat lengths (e.g., (AC)4 or (AAAG)5). Candidates were also rejected if they contained >10 unknown nucleotides within the PCR product, if the PCR product fell entirely within an interspersed repetitive element, or if the PCR product contained >10 uninterrupted STRs outside the site of polymorphism (this last criterion was instituted after the first 751 indels). Some putative indels from the Unigene clusters were also manually rejected because of a relatively high level of mismatch among the aligned sequences, indicating low sequence quality. PCR primers were selected using Primer3 software. All PCR primers were outside the putative polymorphic regions.
PCR amplifications were performed in 96-well microtiter plates in 4-μl volumes with the following final concentrations: 10 mM Tris (pH 8.3); 50 mM KCl; 1.5 mM MgCl2; 0.001% gelatin; 0.12 U Taq DNA polymerase (Sigma D1806); 100 μM each dCTP, dGTP, and dTTP; 1.25 μM dATP; 0.28 μCi of α33P-dATP (>2,500 Ci/mmol, 10 μCi/μl; Amersham); 0.6 pmol each primer; and 32 ng of genomic DNA template. Samples were cycled 27 times through steps of 30 s at 94°C, 75 s at 55°C, and 30 s at 72°C, followed by a final 6 min at 72°C. An equal volume of loading buffer that contained 0.3% xylene cyanol, 0.3% bromophenol blue, 10 mM EDTA (pH 8.0), and 90% (v/v) formamide was added to each amplified product. Samples were denatured at 95°C for 10 min and were resolved on 6.5% polyacrylamide gels that contained 7.7 M urea. After electrophoresis, gels were transferred to Whatman 3 MM chromatography paper and were dried. Amplified products were visualized on autoradiographs after exposure for 6 h–30 d.
DNA templates for testing of candidate indels included several individual human DNA samples, pools of human DNA, and chimpanzee and/or gorilla DNA. Individual DNA samples included CEPH family DNA and Polymorphism Discovery Resource (PDR) samples 1–8. PDR samples are from a mix of American donors of European (42%), African (24%), Asian (24%), and Native American (10%) ancestries (see the Coriell Cell Repositories DNA Polymorphism Discovery Resource Web site) (Collins et al. 1998). Five DNA pools were prepared using equal amounts of DNA from 21 Africans (12 Mbuti and 9 Biaka Pygmies), 25 Japanese, 25 Native Americans (14 Karitiana and 11 Rondonian Surui, both from the Amazon), 100 Europeans, and 44 PDR samples (1–44). The African, Japanese, and Native American samples were kindly provided by Ken Kidd (see the ALFRED Web site). The European samples were obtained from unidentified blood samples from 100 consecutive Marshfield Clinic patients. On the basis of a recent Marshfield-area population genetics survey, ∼99% of Marshfield Clinic patients are of European ancestry, and nearly all of these are of northern or central European ancestry. The PDR pool was not used for the first 333 indels.
Allele frequencies were estimated from the DNA pools by scanning of exposed phosphorscreens with a Storm 860 Imaging System (Molecular Dynamics). Frequencies were averaged from two independent PCR amplifications of each pool. Frequencies were not obtained for ∼10% of the indels because of weak bands or interfering nonspecific PCR products. To gauge the accuracy of our method for measurement of allele frequencies, we amplified three of the indels by using DNA from 25 individuals separately and also using a pool that contained equal amounts of DNA from those 25 individuals. The three indels had frequencies for the most common allele (in these 25 individuals) of 0.50, 0.74, and 0.98. Measured differences in allele frequencies between the individual genotypes and the pool ranged from 0.004 to 0.037 and averaged 0.018. From this test, we conclude that the great majority of our allele-frequency estimates made using the DNA pools are within 0.05 of the true allele frequencies.
Results
Identification and Confirmation
We identified candidate diallelic indels by comparing overlapping human genomic or cDNA sequences. The majority of the 2,000 confirmed indels were derived from BAC/BAC overlaps, although substantial numbers also came from Unigene cDNA sequence assemblies, BAC-end/BAC overlaps, and SNP Consortium sequence cliques (table 1). We confirmed candidates by PCR amplification of short (70–220 bp) DNA fragments that encompassed the putative polymorphism, followed by electrophoresis on denaturing polyacrylamide gels. PCR templates for each indel included at least nine individual DNA samples, pools of DNA from different human populations, and at least one great ape sample (for most indels, a single chimpanzee sample). Altogether, we screened a minimum of 360 human chromosomes for each candidate. Criteria for confirmation of the polymorphisms were the presence of no more than two alleles of the expected PCR-product length and the presence of at least one homozygote among the individual DNA samples and/or substantial variation in allele frequency among the different population pools.
Table 1.
Sequence Sources and Confirmation Rates
| Source | No. (%) | Confirmation Ratea(%) |
| Unigene clusters | 176 (8.8) | 40.1 |
| BAC end/BAC overlaps | 254 (12.7) | 40.5 |
| BAC/BAC overlaps | 1,477 (73.8) | 65.6 |
| SNP Consortium cliques | 93 (4.7) | 69.4 |
Of the PCR primer pairs that supported successful amplification, the fraction that led to confirmed polymorphisms. Data in this table cover only the 2,000 indels with a ⩾2-nt length difference between alleles.
Of the total 3,721 primer pairs tested, 92.7% supported amplification of DNA of the expected length. Of the primer pairs with successful PCR, 58.0% led to confirmed polymorphisms. The Unigene and BAC-end/BAC overlap sequence sources had the lowest confirmation rates; the BAC/BAC and SNP Consortium sources had the highest rates (table 1). The great majority of unconfirmed candidates gave a PCR product of only a single length in all individuals and pools. Approximately 3% of the primer pairs that supported amplification yielded both long and short alleles in approximately equal amounts in all individuals and pools. We believe that most of the aligned sequences in these cases are paralogs—nearly identical sequences from two or more distinct genomic locations with long alleles at one locus and short alleles at a second.
As shown in table 2, confirmation rates increased dramatically as the length difference between alleles increased from 1 to 4 nt. Above 4 nt, confirmation rates slowly drifted downward. All of the 2,000 indels described in the present article have ⩾2 nt between alleles. The confirmation rate for 1-nt length differences was so low that we abandoned efforts on this group early in the project. Most (85%) of the 1-nt-length-difference candidates had mononucleotide tandem repeats of ⩾2 nt—for example, (A)3—in the long allele. These candidates had a confirmation rate of only 11%. For the remaining 15% of candidates without mononucleotide runs, the confirmation rate was 31%.
Table 2.
Confirmation Rates by Allele-Length Difference
| Allele-LengthDifference (in nt) | Primer PairsTesteda | Confirmation Rate(%) |
| 1 | 343 | 14.3 |
| 2 | 1,037 | 46.9 |
| 3 | 719 | 60.8 |
| 4 | 710 | 69.6 |
| 5 | 273 | 66.7 |
| 6 | 154 | 61.7 |
| ⩾7 | 558 | 54.8 |
Numbers include only those primer pairs that supported successful PCR amplification.
Evolution
To study evolution of the indels and to determine ancestral state, we amplified DNA from chimpanzees and gorillas. We attempted to amplify the first 100 indels in a set of six individual gorilla samples and/or in pools of two to five chimpanzee DNAs. Of the 87 indels that were amplified successfully, only three showed any evidence of length polymorphism in the ape DNA, and in none of these cases did both ape alleles match both human alleles in length. For a second group of 100 indels, we amplified DNA from four unrelated chimpanzees. Only one of these indels displayed length variation in the chimpanzees, and the alleles in that case were also different than they were in humans. For the remaining 1,800 indels, the single chimpanzee DNA sample tested appeared to carry both human alleles in only two cases. Therefore, only very rarely are the human length variations shared with chimpanzees or gorillas. Our data indicate that nearly all of the 2,000 indels arose since the divergence of the human/chimpanzee/gorilla common ancestors. The monomorphic alleles in chimpanzees and gorillas very likely represent the ancestral states of these sequences.
Typing of the ape DNA therefore allowed us to split the 2,000 indels into four mutation groups (table 3). Deletion or insertion mutations were indicated when the chimpanzee allele matched exactly in length, respectively, the long or short human allele. We also observed a significant number of cases (∼8%) in which the chimpanzee allele was different in length from either human allele (see “Other” in table 3). The ratio of deletions:insertions was 4.1. Classification of the polymorphisms as either deletions or insertions offered a convenient way to compare and contrast these two types of mutations.
Table 3.
Mutation Events Leading to Diallelic Indels
| Event | No. (%) |
| Deletion | 1,348 (67.4) |
| Insertion | 331 (16.6) |
| Othera | 161 (8.0) |
| No amplificationb | 160 (8.0) |
The amplified chimpanzee/gorilla “allele” had a different length than either human allele.
When chimpanzee or gorilla DNA was used as template, the human PCR primers did not amplify any DNA fragments close in length to the human PCR products.
We also compared allele lengths for indels that were amplified using both gorilla and chimpanzee DNA. Of the 65 indels for which we had data from both apes and for which the gorilla allele matched either the long or short human allele, chimpanzee alleles were the same length as the gorilla allele in 61 cases. This provides further support that the ape DNA represents the ancestral state. We also examined a group of 70 indels for which either the chimpanzee (most cases) or gorilla DNA was different in length from either human allele (taken from the “Other” category in table 3). In 30 cases (43%), the allele from the second ape species matched one of the human alleles in length. These cases can easily be explained by an independent deletion or insertion event within the PCR product in the chimpanzee or gorilla ancestral line after divergence from the common human/chimpanzee/gorilla ancestor. In 32 cases (46%), both chimpanzee and gorilla alleles were the same length but differed in length from either human allele. These cases can most easily be explained by two separate indel mutations in the human ancestral line after divergence from the common ancestor. The first mutation became fixed in the human line, and the second led to the current, observed polymorphism. This interpretation is consistent with the lower nucleotide diversity observed in humans compared to chimpanzees or gorillas (Kaessmann et al. 2001). In eight cases (11%), the chimpanzee and gorilla alleles differed in length from each other, as well as from both human alleles. These cases can be explained by the occurrence of independent indel mutations in both chimpanzee and gorilla lines after divergence. In six of these last eight cases, a relatively highly mutable mononucleotide run of 6–15 nt was present within the PCR product (but not at the site of the human polymorphism).
Distribution of length differences between long and short alleles for the 2,000 indels is shown in table 4. There were approximately equal numbers of indels with 2-, 3-, or 4-nt length differences, and these three groups comprised 71% of the total. Beyond 4-nt length differences, the numbers of indels dropped off with increasing length difference. There were 10 indels with a ⩾30-nt length difference; the greatest length difference was 55 nt. Indels with greater length differences are increasingly more difficult to detect, because the length difference adversely affects sequence-alignment algorithms. Insertions had a modest dearth of 2-nt length differences and an excess of 4-nt length differences compared to deletions, but, overall, the two groups did not differ greatly.
Table 4.
Allele-Length–Difference Distributions
| Allele-LengthDifference (in nt) | No. (%)All Indels | No. (%)Deletions | No. (%)Insertions |
| 2 | 486 (24.3) | 350 (26.0) | 61 (18.4) |
| 3 | 437 (21.8) | 301 (22.3) | 73 (22.1) |
| 4 | 494 (24.7) | 310 (23.0) | 87 (26.3) |
| 5 | 182 (9.1) | 123 (9.1) | 29 (8.8) |
| 6 | 95 (4.8) | 66 (4.9) | 19 (5.7) |
| 7 | 51 (2.6) | 36 (2.7) | 10 (3.0) |
| 8 | 48 (2.4) | 26 (1.9) | 16 (4.8) |
| 9 | 24 (1.2) | 14 (1.0) | 6 (1.8) |
| 10 | 30 (1.5) | 21 (1.6) | 4 (1.2) |
| 11 | 27 (1.4) | 17 (1.3) | 5 (1.5) |
| 12 | 15 (.8) | 10 (.7) | 2 (.6) |
| 13 | 15 (.8) | 14 (1.0) | 0 (.0) |
| 14 | 10 (.5) | 6 (.4) | 2 (.6) |
| 15 | 6 (.3) | 6 (.4) | 0 (.0) |
| ⩾16 | 80 (4.0) | 48 (3.6) | 17 (5.1) |
The 2,000 diallelic indels mapped to all 24 chromosomes. The distribution of indels among the chromosomes, however, was biased. For example, 45% of the indels mapped to chromosomes 5, 6, 7, and 22, whereas only 2.8% of the indels mapped to chromosomes 4 and 8. We believe that this bias can be largely or entirely explained by differential availability among the chromosomes of overlapping sequences at the time of indel development.
Allele Frequencies
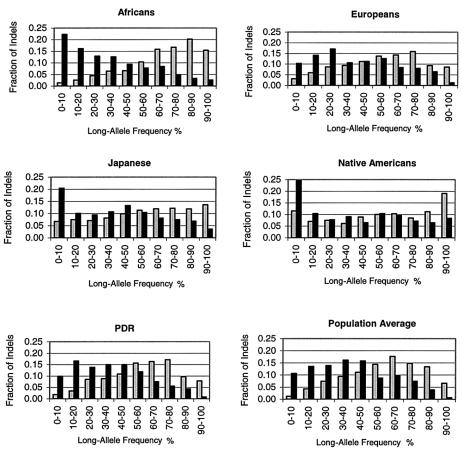
Using DNA pools from different human populations, we measured allele frequencies for ∼90% of the diallelic indels. Distributions of long-allele frequencies in five populations plus an average of the populations are displayed in figure 1. As expected from comparison (in most cases) of only two overlapping sequences, the indels generally had high informativeness. Fifty-one percent of the indels had population-average frequencies of the minor allele that were in the range of 30%–50%, and only 8% had population-average minor-allele frequencies of 0–10%. Note that, for indels that arose by DNA insertion, long-allele frequencies were shifted toward low values. For indels that arose by DNA deletion, the shift was toward high values (see also table 5). This trend was most pronounced for the Africans and was least pronounced for the Native Americans. The European and “mixed” population (PDR and population average) distributions were hump-shaped, with relatively few indels at extreme frequencies, whereas the Native American distributions were bowl- or U-shaped, with the greatest number of indels at frequency extremes. Among the “unmixed” populations (Africans, Europeans, Japanese, and Native Americans) for both deletions and insertions, Africans had the greatest mean long-allele–frequency deviations from 0.50, and Native Americans had the highest SDs (table 5). The average long-allele frequencies of all indels combined are >0.50 because of the predominance of deletions.
Figure 1.
Long-allele–frequency distributions. Distributions are shown for the five indicated populations plus a population average. Gray bars indicate the deletions, and black bars indicate the insertions.
Table 5.
Long-Allele Frequencies
|
Mean ± SD Frequency (No. of Indels) in |
||||||
| Indel Set | Africans | Europeans | Japanese | Native Americans | PDR | Population Average |
| All indels | .60 ± .27 (1,802) | .53 ± .25 (1,805) | .53 ± .29 (1,795) | .53 ± .32 (1,804) | .54 ± .24 (1,519) | .55 ± .24 (1,806) |
| Deletions | .67 ± .23 (1,212) | .56 ± .24 (1,213) | .57 ± .28 (1,207) | .56 ± .31 (1,213) | .58 ± .22 (1,023) | .59 ± .22 (1,214) |
| Insertions | .34 ± .25 (310) | .40 ± .25 (311) | .40 ± .28 (309) | .40 ± .31 (310) | .38 ± .23 (254) | .39 ± .23 (311) |
We next considered indels that were informative or uninformative in only one or two populations (table 6). Europeans, Africans, and Europeans/Africans combined had by far the largest number of indels informative in only those populations. Africans, Native Americans, and Japanese/Native Americans combined had the largest numbers of uninformative indels.
Table 6.
Numbers of Indels Informative or Uninformative in Single Populations or Pairs of Populations[Note]
| Alonea | Africans | Europeans | Japanese | NativeAmericans | |
| Aloneb | 24 | 7 | 1 | 0 | |
| Africans | 36 | 19 | 2 | 1 | |
| Europeans | 2 | 1 | 5 | 1 | |
| Japanese | 3 | 1 | 1 | 1 | |
| Native Americans | 38 | 4 | 2 | 20 |
Note.— Numbers of indels that are informative only in each population or each pair of populations are listed in the upper right half of the array (italic); numbers of indels that are uninformative only in each population or each pair of populations are listed in the lower left half of the array (boldface). In this table, informative indels are defined as having allele (long or short allele) frequencies ⩾20.0% in the population or pair of populations under consideration and frequencies of the same allele ⩽5.0% in the other two or three populations. Uninformative indels are defined as having allele frequencies ⩽5.0% in the population or pair of populations under consideration and frequencies ⩾20.0% in the other populations.
Numbers of uninformative indels in the individual populations.
Numbers of informative indels in the individual populations.
As a final comparison of the populations, we plotted long-allele frequencies from one population against the others in pairs. Linear correlation coefficients for these plots are shown in table 7. Not unexpectedly, Africans were the most divergent population. Among the unmixed populations, Europeans/Japanese and Japanese/Native Americans had the highest correlations. Correlations with the PDR pool were generally high, with the European/PDR value being highest of all.
Table 7.
Correlation Coefficients among Populations Studied[Note]
| Europeans | Japanese | Native Americans | PDR | |
| Africans | .32 | .30 | .22 | .48 |
| Europeans | .58 | .49 | .85 | |
| Japanese | .58 | .75 | ||
| Native Americans | .64 |
Note.— Linear correlation coefficients are given for plots of long-allele frequencies between the population pairs.
Complete information for all 2,000 indels is available at the dbSNP (Sherry et al. 2000) and Marshfield Web sites. Tables with indel data, including population allele frequencies, can be downloaded from the Marshfield Web site.
Discussion
The overall confirmation rate for the 2,000 diallelic indels was 58%. When the highest-quality candidate polymorphisms were utilized (accounting for PHRED or PHRAP nucleotide-quality values), this rate climbed to ∼70% (table 1). Even so, the rate for the indels was lower than published confirmation rates for SNPs of ∼83% (International SNP Map Working Group 2001; Marth et al. 2001). Possible reasons for this difference include the lack of sequence-quality values for missing nucleotides (see the “Material and Methods” section), increased rates of indel-sequencing errors compared to substitutions, and increased artifacts that occurred during Escherichia coli subcloning.
Confirmation rates for candidate indels with 1- and, to a lesser degree, 2-nt allele-length differences were especially low (table 2). This is important because indels with 1-nt allele-length differences are most abundant of all (Antonarakis et al. 2000; Berger et al. 2001; Halangoda et al. 2001; Wicks et al. 2001; also see below). The confirmation rate improved somewhat (to 31%) for indels with 1-nt allele-length differences that did not contain runs of mononucleotides at the site of polymorphism.
We observed a ratio of deletion:insertion mutation events leading to the indels of 4.1 (table 3). This value agrees reasonably well with the ratio of 2.7 taken from the Human Gene Mutation Database and with somatic mutation studies of the lacI (ratio 3.7) and p53 (ratio 3.4) genes (Halangoda et al. 2001). Support for our categorization of the indels as either insertions or deletions is justified by large differences in allele-frequency distributions between the two groups (fig. 1) and by large differences between the two groups in mechanism of mutation (J. L. Weber, R. Boudreau, and D. David, unpublished data).
The finding that nearly all human diallelic indels are apparently monomorphic in chimpanzees and gorillas closely matches results reported previously for SNPs (Hacia et al. 1999). The average lifetimes for both types of polymorphisms appear to be significantly shorter than the ∼6 million years since the common human/chimpanzee ancestor (Clark 1997; Miller et al. 2001).
When considering allele frequencies for the indels, it is important to recognize the ascertainment bias in informativeness due to the nature of the overlapping sequences used to identify the indels. The great majority of the sequence overlaps that we used contained only two sequences. With only two sequences, the probability of detecting the polymorphism is equal to the heterozygosity. In addition, there is a likely population bias in the identification of the polymorphisms. The ancestries of the DNA donors for the bulk of the public human genome sequencing were not reported (International Human Genome Sequencing Consortium 2001). Our data indicate, however, that, of the four populations that we studied, Europeans are closest to the major DNA donors for sequencing. Support for this conclusion comes from the relatively large numbers of indels informative in only the Europeans (or Europeans/Africans) and from the relatively small number uninformative in only the Europeans (table 6). Europeans also had the fewest number of indels with minor-allele frequency <10% or <2%, the highest average heterozygosity at 37% (Japanese and Africans each had 33%, and Native Americans had 30%), and hump-shaped distributions for long-allele frequencies (fig. 1).
Even with the limitations described above, some population genetics and evolutionary conclusions can be drawn from our data. Of the four populations studied, African Pygmy new-allele–frequency distributions are clearly closest to the shape expected for neutral alleles in a population of constant size (Fu 1995; Subrahmanyan et al. 2001). The Africans have the greatest bias toward low frequencies for the new alleles (fig. 1 and table 5). Watkins et al. (2001) obtained very similar results for polymorphisms based on insertion of Alu elements and for a relatively small group of SNPs. The Africans appear to be most similar to the original modern human population and to have undergone no severe population bottlenecks (Tishkoff et al. 2000; Jorde et al. 2001).
In contrast, Europeans, Japanese, and Native Americans all appear to have undergone at least one relatively severe population bottleneck. These populations have less bias toward low frequencies for new alleles and have higher SDs for average allele frequencies than the Africans. The Native Americans probably passed through more than one severe bottleneck, since they show bowl-shaped long-allele–frequency distributions (fig. 1). As a population passes through a bottleneck, allele frequencies tend to change rapidly; rare neutral alleles usually drop in frequency, but some increase dramatically. The 36 indels uninformative in only the Africans (table 6) may be examples of the latter case.
Our results are also consistent with many evolutionary trees that have been drawn for modern human populations. Africans are clearly the most distant group compared to the other three (table 7). Of the three “out of Africa” populations, the Japanese and Native Americans are clearly the most closely related pair, as demonstrated by their relatively high correlation coefficient (table 7) and also by the relatively large numbers of indels that are uninformative in only the Native Americans/Japanese (table 6).
Nearly all diallelic polymorphisms, even those with high average informativeness, will have low informativeness in some human populations. As an example, of the 909 indels with population-average long-allele frequencies between 30% and 70%, 176 (19%) had a minor-allele frequency of ⩽10% in at least one of the four populations that we studied. Care will have to be taken to ensure that diallelic polymorphisms chosen for generic screening sets have reasonable informativeness in all major world populations.
It is important to keep in mind that, although substitutions (i.e., SNPs) are the most abundant class of human polymorphisms, indels are also quite common. (Although the term “SNP” has occasionally been used to cover indels with a 1-nt allele-length difference, we recommend that this term be restricted to substitutions.) As shown in table 8, indels comprise ∼20% of all human DNA polymorphisms. The numbers in table 8 that are from the Human Gene Mutation Database may be biased toward indels because this catalog contains mostly mutations that severely disrupt gene function. The estimates from the overlapping BACs for the whole genome and specifically for chromosome 22 are less biased and probably are a more accurate reflection of the true situation. The fraction of polymorphisms that are indels in humans is consistent with numbers from three model organisms (table 8). In the many species with more genetic diversity than humans, average spacing between indels will be impressively low.
Table 8.
Breakdown of DNA Polymorphisms by Type
| Species | Indels | Substitutions | Reference |
| Arabidopsis thaliana | 37% | 63% | Arabidopsis Genome Initiative 2000 |
| Caenorhabditis elegans | 25% | 75% | Wicks et al. 2001 |
| Drosophila melanogaster | 16% | 84% | Berger et al. 2001 |
| Homo sapiens: | |||
| Human Gene Mutation Database | 30% | 70% | Antonarakis et al. 2000 |
| Overlapping BACs | 21% | 79% | G. Marth, G. Schuler, R. Yeh, R. Davenport, R. Agarwala, D. Church, S. Wheelan, J. Baker, M. Ward, M. Kholodov, L. Phan, H. Harpending, A. Chakravarti, P.-Y. Kwok, and S. Sherry, unpublished data |
| Chromosome 22 | 18% | 82% | Dawson et al. 2000 |
Human indel candidates from the ∼1.1 Gb of overlapping BAC sequences (see the “Material and Methods” section) can be further divided into an ∼60:40 ratio of multiallelic STRPs and diallelic indels, respectively. Division of the indels into these two groups is based on the rules listed in the “Material and Methods” section and is therefore somewhat arbitrary. Many sequences categorized as multiallelic will likely turn out to be diallelic, and at least a few of the sequences categorized as diallelic will likely have more than two alleles. Repeat lengths for STRP candidates followed expected patterns (Tóth et al. 2000). Mononucleotide repeats were most abundant (73% of total), followed by dinucleotide repeats (18%), tetranucleotide repeats (6%) and trinucleotide repeats (2%). For diallelic candidates, most had 1-nt length differences between alleles (76%). The distribution of candidates with ⩾2-nt allele-length differences was very close to table 4.
Indels can be easily genotyped using just PCR and gel electrophoresis. Diallelic indels can also be genotyped using the various methods developed for SNPs. The significant difference in sequence between many of the diallelic indels allows these polymorphisms to be efficiently analyzed in a highly automated fashion by allele-specific PCR (J. L. Weber, J. Che, A. Yu, N. Ghebranious, and M. Doktycz, unpublished data). We recommend indels for most genetic studies.
Acknowledgments
This work was supported by grant HL62681 and contract HV48141 from the National Heart, Lung, and Blood Institute. We thank Drs. Ken Kidd (Yale), Gay Reinartz (Milwaukee Zoo), and Oliver Ryder (San Diego Zoo) for providing human, bonobo, and gorilla DNA samples, respectively. Jayme Opolka, Ryan Boudreau, Jianhong Che, Patti Franckowiak, Kelly Gebert, Jennifer Imm, Fay Jahr, Jessica Kayhart, Obrad Kokanovic, Melissa Krall, Sarah Merz, Keith Pulvermacher, Bryndon Schank, Ann Solatycki, Dan Tomaszewski, and Maggie Yin provided excellent laboratory technical assistance. We also thank Andrew Clark for helpful comments.
Electronic-Database Information
URLs for data presented herein are as follows:
- ALFRED, http://alfred.med.yale.edu/alfred/
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- Coriell Cell Repositories DNA Polymorphism Discovery Resource, http://locus.umdnj.edu/nigms/pdr.html
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/
- Human Gene Mutation Database, http://www.hgmd.org/
- Primer3 Software Distribution, http://www-genome.wi.mit.edu/genome_software/other/primer3.html
- Sanger Institute, The, http://www.sanger.ac.uk/HGP/
References
- Antonarakis SE, Krawczak M, Copper DN (2000) Disease-causing mutations in the human genome. Eur J Pediatr 159 Suppl 3:S173–S178 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815 [DOI] [PubMed] [Google Scholar]
- Berger J, Suzuki T, Senti K-A, Stubbs J, Schaffner G, Dickson BJ (2001) Genetic mapping with SNP markers in Drosophila. Nat Genet 29:475–481 [DOI] [PubMed] [Google Scholar]
- Clark AG (1997) Neutral behavior of shared polymorphism. Proc Natl Acad Sci USA 94:7730–7734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Brooks LD, Chakravarti A (1998) A DNA Polymorphism Discovery Resource for research on human genetic variation. Genome Res 8:1229–1231 [DOI] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M (1991) Mechanisms of insertional mutagenesis in human genes causing genetic disease. Hum Genet 87:409–415 [DOI] [PubMed] [Google Scholar]
- Dawson E, Chen Y, Hunt S, Smink LJ, Hunt A, Rice K, Livingston S, Bumpstead S, Bruskiewich R, Sham P, Ganske R, Adams M, Kawasaki K, Shimizu N, Minoshima S, Roe B, Bentley D, Dunham I (2001) A SNP resource for human chromosome 22: extracting dense clusters of SNPs from the genomic sequence. Genome Res 11:170–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y-X (1995) Statistical properties of segregating sites. Theor Popul Biol 48:172–197 [DOI] [PubMed] [Google Scholar]
- Gelehrter TD, Collins FS (1990) Principles of medical genetics. Williams & Wilkins, Baltimore, p 55 [Google Scholar]
- Hacia JG, Fan JB, Ryder O, Jin L, Edgemon K, Ghandour G, Mayer RA, Sun B, Hsie L, Robbins CM, Brody LC, Wang D, Lander ES, Lipshutz R, Fodor SP, Collins FS (1999) Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat Genet 22:164–167 [DOI] [PubMed] [Google Scholar]
- Halangoda A, Still JG, Hill KA, Sommer SS (2001) Spontaneous microdeletions and microinsertions in a transgenic mouse mutation detection system: analysis of age, tissue, and sequence specificity. Environ Mol Mutagen 37:311–313 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- International SNP Map Working Group (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928–933 [DOI] [PubMed] [Google Scholar]
- Jorde LB, Watkins WS, Bamshad MJ (2001) Population genomics: a bridge from evolutionary history to genetic medicine. Hum Mol Genet 10:2199–2207 [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Wiebe V, Weiss G, Pääbo S (2001) Great ape DNA sequences reveal a reduced diversity and an expansion in humans. Nat Genet 27:155–156 [DOI] [PubMed] [Google Scholar]
- Krawczak M, Cooper DN (1991) Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum Genet 86:425–441 [DOI] [PubMed] [Google Scholar]
- Lupski JR, Roth JR, Weinstock GM (1996) Chromosomal duplications in bacteria, fruit flies, and humans. Am J Hum Genet 58:21–27 [PMC free article] [PubMed] [Google Scholar]
- Marth GT, Korf I, Yandell MD, Yeh RT, Gu Z, Zakeri H, Stitziel NO, Hillier L, Kwok P-Y, Gish WR (1999) A general approach to single-nucleotide polymorphism discovery. Nat Genet 23:452–456 [DOI] [PubMed] [Google Scholar]
- Marth G, Yeh R, Minton M, Donaldson R, Li Q, Duan S, Davenport R, Miller RD, Kwok P-Y (2001) Single-nucleotide polymorphisms in the public domain: how useful are they? Nat Genet 27:371–372 [DOI] [PubMed] [Google Scholar]
- Miller RD, Taillon-Miller P, Kwok P-Y (2001) Regions of low single-nucleotide polymorphism incidence in human and orangutan Xq: deserts and recent coalescences. Genomics 71:78–88 [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward M, Sirotkin K (2000) Use of molecular variation in the NCBI dbSNP database. Hum Mutat 15:68–75 [DOI] [PubMed] [Google Scholar]
- Subrahmanyan L, Eberle MA, Clark AG, Kruglyak L, Nickerson DA (2001) Sequence variation and linkage disequilibrium in the human T-cell receptor β (TCRB) locus. Am J Hum Genet 69:381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Pakstis AJ, Stoneking M, Kidd JR, Destro-Bisol G, Sanjantila A, Lu RB, Deinard AS, Sirugo G, Jenkins T, Kidd KK, Clark AG (2000) Short tandem-repeat polymorphism/Alu haplotype variation at the PLAT locus: implications for modern human origins. Am J Hum Genet 67:901–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth G, Gáspári Z, Jurka J (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res 10:967–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins WS, Ricker CE, Bamshad MJ, Carroll ML, Nguyen SV, Batzer MA, Harpending HC, Rogers AR, Jorde LB (2001) Patterns of ancestral human diversity: an analysis of Alu-insertion and restriction-site polymorphisms. Am J Hum Genet 68:738–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RHA (2001) Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet 28:160–164 [DOI] [PubMed] [Google Scholar]
- Zhao S, Malek J, Mahairas G, Fu L, Nierman W, Venter JC, Adams MD (2000) Human BAC ends quality assessment and sequence analyses. Genomics 63:321–332 [DOI] [PubMed] [Google Scholar]