Abstract
GRACILE (growth retardation, aminoaciduria, cholestasis, iron overload, lactacidosis, and early death) syndrome is a recessively inherited lethal disease characterized by fetal growth retardation, lactic acidosis, aminoaciduria, cholestasis, and abnormalities in iron metabolism. We previously localized the causative gene to a 1.5-cM region on chromosome 2q33-37. In the present study, we report the molecular defect causing this metabolic disorder, by identifying a homozygous missense mutation that results in an S78G amino acid change in the BCS1L gene in Finnish patients with GRACILE syndrome, as well as five different mutations in three British infants. BCS1L, a mitochondrial inner-membrane protein, is a chaperone necessary for the assembly of mitochondrial respiratory chain complex III. Pulse-chase experiments performed in COS-1 cells indicated that the S78G amino acid change results in instability of the polypeptide, and yeast complementation studies revealed a functional defect in the mutated BCS1L protein. Four different mutations in the BCS1L gene have been reported elsewhere, in Turkish patients with a distinctly different phenotype. Interestingly, the British and Turkish patients had complex III deficiency, whereas in the Finnish patients with GRACILE syndrome complex III activity was within the normal range, implying that BCS1L has another cellular function that is uncharacterized but essential and is putatively involved in iron metabolism.
Introduction
GRACILE (growth retardation, aminoaciduria, cholestasis, iron overload, lactacidosis, and early death) syndrome (MIM 603358) is a metabolic disorder with an autosomal recessive mode of inheritance (Fellman et al. 1998; Rapola et al. 2002). Affected infants are severely growth retarded, the average birth weight being only 1,700 g at term. Patients develop fulminant lactic acidosis during the first day of life, with an average arterial blood pH of 7.0. They also have nonspecific aminoaciduria, cholestasis, and iron overload, including hemosiderosis of the liver, increased serum ferritin concentration, hypotransferrinemia with increased transferrin saturation, and free plasma iron. No deficiency in respiratory chain oxygen consumption or enzyme activity has been demonstrated in patients, and they do not have neurological abnormalities or dysmorphic features. Despite intensive care and alkali therapy, about half of the infants die during the first days of life, and the remainder within 4 mo of life.
GRACILE syndrome belongs to the Finnish disease heritage (Norio et al. 1973; Peltonen et al. 1999). On the basis of 13 cases diagnosed in 1991–2000, the incidence is ⩾1/47,000 in Finland. We previously mapped the GRACILE locus to a 1–1.5-cM region between markers D2S2179 and D2S2244 on chromosome 2q33-37 (fig. 1A). The disease locus was initially identified via a genome scan, and the critical chromosomal region was restricted through use of linkage disequilibrium and haplotype analyses in 10 Finnish families (Visapää et al. 1998). All families carried the same ancestral haplotype, indicating one founder mutation in Finland. The physical interval of the critical GRACILE region is 1.4 Mb in the latest NCBI human genome assembly (MapViewer, May 2002, at the Human Genome at NCBI Web site), including as many as 28 known genes and several predicted hypothetical genes. We recently excluded ABCB6, a gene encoding a mitochondrial protein involved in iron homeostasis, mitochondrial respiratory function, and maintenance of the stability of mtDNA (Mitsuhashi et al. 2000), as the causative gene for this disease (Visapää et al. 2002). Another positionally and functionally promising candidate gene, BCS1L, encodes a mitochondrial protein that functions as a chaperone in the assembly of complex III (cytochrome bc1 complex) of the mitochondrial respiratory chain.
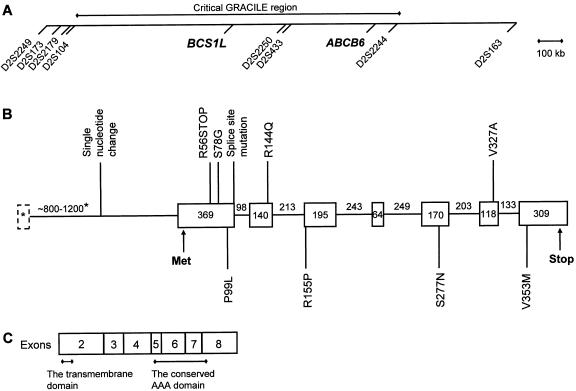
Figure 1.
A, Critical GRACILE region. The locations of the two functionally and positionally promising candidate genes, BCS1L and ABCB6, are shown in relation to the markers demonstrating linkage disequilibrium in GRACILE chromosomes. The marker order and distances are presented as in the NCBI MapViewer, May 2002. B, Genomic structure of the BCS1L gene. The sizes of the exons and introns are indicated in base pairs. The BCS1L mutations reported here (in Finnish and British patients) are indicated above the figure, and the BCS1L mutations described elsewhere (de Lonlay et al. 2001) are marked below. The sizes of the first exon and the first intron (marked with asterisks) vary because of alternative splicing. C, The BCS1L polypeptide of 419 amino acid residues. The numbers indicate the polypeptide regions encoded by distinct exons of the BCS1L gene. Residues 9–32 correspond to the transmembrane region of the yeast BCS1p protein (Fölsch et al. 1996). Residues 224–344 represent the conserved AAA domain, according to CD-Search (Marchler-Bauer et al. 2002) at the NCBI BLAST service.
Complex III consists of 11 subunits: core proteins I and II, six small subunits, and three polypeptides involved in electron transport: cytochrome b, cytochrome c1, and the Rieske FeS protein. Cytochrome b is encoded by mtDNA, whereas the other subunits are encoded by nuclear genes. Several sporadic heteroplasmic cytochrome b mutations have been described as causing exercise intolerance, proximal limb weakness, and elevated lactate levels (Andreu et al. 1999; Legros et al. 2001). More-severe multisystem manifestations, including symptoms in the CNS, have also been reported (Keightley et al. 2000; Wibrand et al. 2001). So far, no mutations have been reported in the nuclear-encoded subunits of complex III.
The Bcs1p protein, the Saccharomyces cerevisiae homolog for human BCS1L (BCS1-like), is an assembly factor of complex III. It is a mitochondrial inner-membrane protein with a single transmembrane domain. A short N-terminus is located in the mitochondrial intermembrane space, and the bulk of the protein lies in the matrix. It belongs to the conserved AAA protein family (ATPases associated with various cellular activities) and acts as a chaperone necessary for the assembly of Rieske FeS and Qcr10p subunits into complex III (Nobrega et al. 1992; Fölsch et al. 1996; Cruciat et al. 1999). The human BCS1L mRNA sequence has been identified by searching the EST database with the yeast Bcs1p protein sequence, and it has been proven that human BCS1L is a mitochondrial inner-membrane protein, like yeast Bcs1p (Petruzzella et al. 1998). Here we report that a missense mutation in BCS1L results in GRACILE syndrome and provide data suggesting a new role for BCS1L in human iron metabolism.
Subjects and Methods
Study Subjects
DNA samples from 17 Finnish patients with GRACILE syndrome, 22 parents, 13 siblings, and 3 half-siblings, from 11 families, as well as DNA from 3 British infants, were used for sequence analyses in the present study. The symptoms in the British patients (Morris et al. 1995) resembled those of the Finnish patients with GRACILE syndrome, but there were some remarkable differences (table 1). The British infants had complex III deficiency, whereas, in the Finnish patients, complex III activity did not seem to be affected (Fellman et al. 1998). Moreover, the British patients had neurological symptoms, whereas the Finnish patients with GRACILE syndrome had no neurological or neuropathological abnormalities. We included 494 Finnish individuals as control individuals in mutation screening, 70% of whom originated from eastern and central Finland, where the ancestors of the affected infants had lived (Fellman et al. 1998); we also used a control panel of 50 white individuals (HD50CAU; Coriell Cell Repositories). This control panel consisted of 50 Americans with ancestry in different parts of Europe. Autopsy samples, muscle biopsy specimens, or fibroblast cell lines of nine Finnish patients with GRACILE syndrome were used for RNA extraction and mitochondrial enzyme–complex measurements. This study has been approved by the ethical committee of the Hospital for Children and Adolescents, Helsinki University Central Hospital, Finland, and parental informed consent was obtained.
Table 1.
Clinical Characteristics of the Finnish Patients with GRACILE Syndrome, Compared with Three British Infants with Different BSC1L Mutations
|
Patients with GRACILE Syndromea |
British Patientsb |
|||||||
| Clinical Characteristic | NormalRange | n | Median | IQ25 | IQ75 | Patient 1 | Patient 2 | Patient 3 |
| Male/female | 5/12 | M | F | F | ||||
| Gestational age (wk) | 17 | 37.6 | 35.3 | 38.4 | 38 | 39 | 38 | |
| Birth weight (g) | 17 | 1,560 | 1,310 | 1,790 | 2,020 | 1,830 | 1,840 | |
| Birth weight (SD score) | 17 | −4.0 | −4.2 | −3.6 | −3.3 | −3.8 | −3.5 | |
| Birth length (cm) | 17 | 43 | 40 | 45 | 47 | 45 | 48 | |
| Head circumference (cm) | 17 | 31 | 30 | 32 | 31 | 31 | 31 | |
| 1-min Apgar score | 17 | 8 | 7 | 9 | 9 | ND | ND | |
| Umbilical arterial pH | >7.20 | 17 | 7.30 | 7.26 | 7.30 | ND | ND | ND |
| pH at admission to intensive care | 7.35 to 7.43 | 17 | 7.00 | 6.95 | 7.09 | 7.13 | 7.09 | 7.09 |
| Base excess (mmol/liter) | −2.5 to 2.5 | 17 | −21 | −25 | −20 | −20 | −20 | −13 |
| Blood lactate (mmol/liter) | .7 to 1.8 | 17 | 11.8 | 6.5 | 19.9 | 23.5 | 10.9 | 13.8 |
| Blood pyruvate (mmol/liter) | .04 to .07 | 17 | .12 | .08 | .16 | .31 | .27 | .33 |
| Lactate:pyruvate ratio | <25 | 17 | 81 | 52 | 130 | 76 | 40 | 42 |
| Age at death (d) | 17 | 7 | 2 | 68 | 2 | 42 | 105 | |
| Aminoaciduria | 17 | All | Yes | Yes | Yes | |||
| Cholestasis | 17 | All | No | Yes | Yes | |||
| Neurological symptoms: | ||||||||
| Hypotonia | 17 | None | Yes | Yes | Yes | |||
| Seizures | 17 | None | Yes | No | No | |||
| Iron metabolism: | ||||||||
| Serum ferritin (μg/liter) | <150 | 6 | 1,314 | 1,193 | 2,148 | ND | ND | ND |
| Serum iron (μmol/liter) | 19 to 48 | 6 | 14.0 | 11.6 | 16.4 | ND | ND | ND |
| Serum transferrin (g/liter) | 1.1 to 1.8 | 5 | .72 | .72 | .76 | ND | ND | ND |
| Transferrin saturation (%) | 30 to 60 | 5 | 81 | 61 | 91 | ND | ND | ND |
| Hemosiderosis of liver (score 0–4) | 0 | 13 | 3 | 1 | 4 | ND | ND | ND |
| Liver iron content (μg/mg dry weight) | 990 to 2,800 | 5 | 5,470 | 3,990 | 5,700 | ND | ND | ND |
IQ = interquartile range.
ND = not determined. British patients 1–3 have been described by Morris et al. (1995).
Determining the Localization and Structure of the BCS1L Gene
BCS1L has been localized to the critical GRACILE region (Visapää et al. 1998) in both the NCBI MapViewer (Human Genome at NCBI Web site) and the UCSC Human Genome Project Working Draft (UCSC Genome Bioinformatics Web site). Localization of BCS1L was also confirmed by radiation hybrid (RH) mapping using Stanford medium-resolution RH panel G3 (Research Genetics).
Both the mRNA sequence and the genomic structure of the BCS1L gene have been published elsewhere (Petruzzella et al. 1998; de Lonlay et al. 2001). However, the genomic sequence published by de Lonlay et al. (2001) was partially incomplete for the noncoding part of the first exons, which we determined using biocomputing tools. The genomic clone (RP11 1077K22) containing BCS1L was identified by a BLAST search with BCS1L mRNA sequence. Because we had noticed variations in the 5′ UTRs in the various BCS1L mRNA sequences, we searched the human EST database and the nucleotide sequence database with only the coding part of the BCS1L mRNA sequence, using the BLASTN algorithm at the NCBI BLAST service, in order to find all the different splice variants of this gene. The EST and mRNA sequences were aligned with the genomic clone through use of the Sequencher computer program and were divided into exons so that all the exon-intron boundaries followed the GT-AG rule (Mount 1982).
Sequencing and Mutation Screening of the BCS1L Gene
Sequencing was performed using a BigDye terminator kit and ABI 377 and ABI 3700 automated sequencers (Applied Biosystems). The genomic clone RP11 1077K22 was used for primer planning. The whole BCS1L genomic sequence was amplified by PCR from the DNA samples from two Finnish patients with GRACILE syndrome, three British patients with similar symptoms (Morris et al. 1995), and two control individuals, in 13 overlapping pieces, and was sequenced using the same primers. The sequence covered 5.5 kb, starting 2,020 bp upstream from the start codon and ending 1,040 bp downstream from the stop codon of BCS1L.
The coding region of BCS1L was also sequenced using RT-PCR products. For RT-PCR sequencing, total RNA was extracted from the liver tissue of one patient with GRACILE syndrome and two control individuals, through use of the RNeasy Mini Kit (Qiagen). The GenBank AF038195 mRNA sequence was used as a reference.
Screening for the 232A→G (S78G) point mutation in the Finnish patients with GRACILE syndrome and their parents was performed by direct sequencing (ABI; Applied Biosystems). Mutation screening in siblings and control samples was performed using solid-phase minisequencing (Syvänen et al. 1993). The PCR primer sequences for minisequencing were CTATGCCTGGTTGCTTAGCTG (5′ end biotinylated) and CTTCGTTCTACCCGAATCCATT, and the detection primer sequence was AAGGTACGAAGTCTCGACAC. For other mutations, the control individuals were screened by direct sequencing.
Northern Blot Analysis
Messenger RNAs for northern blot analysis were extracted from two GRACILE and one control fibroblast cell line, as well as from the liver tissue of one patient with GRACILE syndrome and one control individual, through use of the Oligotex mRNA Direct Mini kit (Qiagen). The control liver tissue was derived from an adult who died in an accident, and the control fibroblast cell line was also adult-derived. Northern blotting to Hybond-XL nylon membrane (Amersham Pharmacia Biotech) was performed using standard procedures. A 493-bp probe was produced by RT-PCR using the primers TTCTGGCTCTGAAGGACAATCC and GCTGTGTACATCACGGTCTTCC and was labeled with 32P, using a random primer DNA labeling system (Life Technologies). Hybridizations were performed in Expresshyb hybridization solution (Clontech), as suggested by the manufacturer.
Expression Plasmid Construction, Cell Culture, and Transfections
BCS1L cDNA was PCR-cloned to a pGEM-T Easy vector (Promega), through use of linker primers introducing the FLAG sequence immediately before the stop codon. IMAGE clone 44576 was used as a template in the PCR. The 232A→G mutagenesis, resulting in an S78G amino acid change, was performed in pGEM-T Easy vector, through use of the QuickChange site-directed mutagenesis kit, according to the manufacturer’s instructions (Stratagene). Wild-type and mutant BCS1L cDNAs with a C-terminal FLAG tag were then subcloned to pCMV5 expression vector (Andersson et al. 1989). The constructs were confirmed by sequencing.
COS-1 cells were obtained from the American Type Culture Collection and were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Cellgro), supplemented with 10% fetal bovine serum (FBS; Cellgro) and antibiotics (Life Technologies) in 5% CO2 at 37°C. One hundred thousand cells per well were plated in six-well plates 1 d prior to transfection. Cells were transiently transfected using Lipofectamine and PLUS reagent (Life Technologies), following the manufacturer’s guidelines.
Immunofluorescence Microscopy
For immunofluorescence microscopy, COS-1 cells were plated on cover slips and were transfected as described above. Forty-eight hours after transfection, cells were incubated in DMEM without FBS, in the presence of 50 μg/ml of cycloheximide (Sigma) to halt protein synthesis, for 1 h, and then for an additional 30 min with cycloheximide and 200 nM MitoTracker Red CMXRos (Molecular Probes), for mitochondrial staining. Thereafter, the cells were fixed with methanol, were blocked with 0.5% BSA/0.2% saponin (Sigma), and were incubated with anti-FLAG M2 antibody (Stratagene; 1/500 dilution). The cells were washed with 0.5% BSA/0.2% saponin and were stained with fluorescein isothiocyanate (FITC)–conjugated anti-mouse secondary antibody (Sigma; 1/200 dilution). After washing with PBS, the cells were mounted in glycerol and were viewed with a Leica DMR immunofluorescence microscope (Leica Microscope and Scientific Instruments Group), using a Quips FISH image capture system (Applied Imaging Corporation).
Western Blot Analysis
For western blot analysis, COS-1 cells were lysed 48 h after transfection with lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% (octylphenoxy)polyethoxyethanol, 0.5% deoxycholic acid, 0.1% SDS) supplemented with protease inhibitors (Complete; Roche Diagnostics), via incubation at 4°C for 20 min with shaking, followed by centrifugation and retention of the supernatant. Samples were denatured at 60°C for 5 min and were electrophoresed on a 10% SDS-PAGE gel with a Low Range protein marker (BioRad). Western blotting was performed according to standard procedures, using the Hybond-P membrane (Amersham Pharmacia Biotech). The membrane was stained with anti-FLAG M2 antibody (Stratagene; 1/1,000 dilution), the secondary antibody being peroxidase-conjugated anti-mouse IgG (Sigma; 1/10,000 dilution). The signals were detected using ECL western-blotting detection reagents (Amersham Pharmacia Biotech).
Pulse-Chase Analysis
The half-lives of the wild-type and S78G mutant BCS1L polypeptides were determined by pulse-chase experiments. Cells were metabolically labeled 36 h after transfection, by starving them in methionine- and cysteine-free medium (Life Technologies) for 1 h and thereafter labeling them with 50 μCi/ml of both [35S]methionine and [35S]cysteine (Amersham Pharmacia Biotech) for 1 h. The cells were then incubated for 0, 12, 24, 36, or 48 h, in chase media (DMEM, 2% FBS, and antibiotics), and were lysed with lysis buffer, as described above. The lysed cells were immunoprecipitated with anti-FLAG M2 antibody (Stratagene; 1/1,000 dilution) and Protein A/G Sepharose (Santa Cruz Biotechnology). Immunocomplexes were separated on 10% SDS-PAGE and were visualized by fluorography, using Amplify reagent (Amersham Pharmacia Biotech). The results were analyzed by densitometry.
Yeast Complementation Studies
Human BCS1L cDNA was PCR-cloned to pBluescript (Stratagene) vector, using linker primers. IMAGE clone 44576 served as a template. Four hundred eighty base pairs of yeast BCS1 upstream sequence and 210 bp of yeast BCS1 downstream sequence were amplified by PCR from yeast genomic DNA and were cloned to the pBluescript plasmid containing the human BCS1L cDNA. The 232A→G mutagenesis in BCS1L cDNA, resulting in an S78G amino acid change, was performed using the QuickChange site-directed mutagenesis kit, according to the manufacturer’s protocol (Stratagene). The whole cassette, containing yeast BCS1 promoter, human BCS1L cDNA, and yeast BCS1 terminator, was then subcloned to the high-copy yeast expression vector pRS425 (Sikorski and Hieter 1989).
The yeast BCS1 gene was PCR-cloned, as one fragment containing a 480-bp upstream sequence, the coding region, and a 210-bp downstream sequence, to a pGEM-T Easy vector (Promega), using yeast genomic DNA as a template in PCR. The 343,344TC→GG mutagenesis, resulting in a S115G amino acid change (corresponding to S78G in human), was performed with a QuickChange site-directed mutagenesis kit. The gene was subcloned both to the high-copy yeast expression vector pRS425 and to the low-copy vector pRS315 (Sikorski and Hieter 1989). All constructs were confirmed by sequencing.
Yeast strain Y14211 (EUROSCARF, European S. cerevisiae archives for functional analysis), deleted for BCS1, was transformed with the yeast expression constructs described above. Transformations were performed by the lithium acetate procedure (Ito et al. 1983). Individual transformants were purified and checked for growth on YPEG plates (3% glycerol, 2% ethanol, 2% peptone, 1% yeast extract, and 2% agar). To control the growth on glucose, the same transformants were also streaked on YPD plates (2% glucose, 2% peptone, 1% yeast extract, and 2% agar) and on synthetic dextrose (SD) minimal medium plates without leucine (2% glucose; 0.67% yeast nitrogen base; 2% agar; 20 μg/ml histidine, uracil, and tryptophan; and 30μg/ml adenine and lysine).
Activity Measurements of Mitochondrial Enzyme Complexes
Mitochondria were isolated from muscle biopsy specimens of five patients with GRACILE syndrome and from several autopsy-derived tissues obtained from four patients 2–7 h postmortem. Age-matched control samples from patients without any mitochondrial diseases were available for muscle biopsy specimens and myocardial autopsy samples. The activities of respiratory chain enzymes (NADH:cytochrome c oxidoreductase [complex I + III], succinate:cytochrome c oxidoreductase [complex II + III], succinate:ubiquinone oxidoreductase [complex II], and cytochrome c oxidase [complex IV]) were measured spectrophotometrically (Majander et al. 1995). A modified spectrophotometrical assay (Rustin et al. 1994), including separate assessment of NADH:ubiquinone oxidoreductase (complex I) and ubiquinol:ferricytochrome-c oxidoreductase (complex III), was used to measure the enzyme activities from three deep-frozen liver samples and one muscle sample of the patients. The control tissues for these measurements were from children and adults who had died of unrelated disorders; the tissues had been stored at −80°C. In addition, respiratory chain enzyme activities were measured from the fibroblasts of two patients with GRACILE syndrome, as described elsewhere (Pitkänen and Robinson 1996).
Results
Localization and Structure of the BCS1L Gene
We first confirmed the localization of BCS1L to the critical 1.4-Mb GRACILE region by comparing the RH panel data of BCS1L with the RH data of the genetic markers constituting the shared founder haplotype in GRACILE chromosomes. The hybrid clones positive for BCS1L were exactly the same as those positive for the markers D2S2250 and D2S433, which demonstrated the strongest linkage disequilibrium in GRACILE chromosomes (Visapää et al. 1998), indicating that BCS1L is located in the immediate vicinity of these markers (fig. 1A).
Comparison of the different EST and mRNA sequences suggested that the 5′ end of the BCS1L transcript can be alternatively spliced. Fifteen EST or mRNA sequences found in public databases and Celera (GenBank accession numbers and Celera sequence numbers for the EST and mRNA sequences are provided in the Electronic-Database Information section of this article) represented six different splice variants. Two of the variants (GenBank accession numbers AF026849 and AF038195) were represented several times and had been found in clones originating from normal tissues, whereas the other four were rare and were reported only in EST libraries of cancer tissues or had a nonspecified tissue origin in the databases. In most transcripts, the first exon is small (20–120 bp), and the size of the first intron varies from 800 to 1,200 bp. The second exon is 369 bp, starts 49 bp before the start codon, and contains an in-frame stop codon 42 bp upstream from the start codon. This exon and all six exons thereafter are identical in all splice variants (fig. 1B). A few clones contained an additional short exon in the middle of the first intron. The exons span 3.4–3.8 kb in the genomic sequence, and the size of the spliced transcript varies from 1.4 kb to 1.5 kb. The coding region is 1,260 bp, encoding a polypeptide of 419 amino acids (Petruzzella et al. 1998).
Sequence Analysis of the BCS1L Gene
Table 2 and figure 1B summarize the results of the sequence analysis. All Finnish patients with GRACILE syndrome were homozygous for a 232A→G mutation in the second exon of BCS1L, which causes an S78G amino acid change. All parents were heterozygous for this mutation, and the siblings and half-siblings of the patients were either homozygous for the normal allele or heterozygous. The two sequenced Finnish patients with GRACILE syndrome had no other nucleotide changes in the BCS1L genomic sequence (GenBank accession number AF516670). The coding region of BCS1L was also sequenced from the RT-PCR product from the liver RNA of one Finnish patient and two control individuals. Neither defective splicing nor any nucleotide changes other than the 232A→G mutation were detected in the patient. Of the 494 Finnish control individuals and 50 other white individuals screened for the S78G mutation, 1 Finnish control individual was a heterozygous carrier of this mutation, and all other control individuals were homozygous for the normal allele.
Table 2.
BCS1L Mutations Detected in the Finnish Patients with GRACILE Syndrome and in Three British Patients[Note]
|
NucleotideChange |
|||||
| Patient and Mutation Type | Homozygous/Heterozygous | Location | GenomicDNAa | cDNA | Predicted Consequence |
| Finnish patients with GRACILE syndrome: | |||||
| Missense | Homozygous | Exon 2 | 232A→G | 232A→G | S78G |
| British patient 1: | |||||
| Nonsense | Heterozygous | Exon 2 | 166C→T | 166C→T | R56STOP |
| Missense | Heterozygous | Exon 7 | 1986T→C | 980T→C | V327A |
| British patient 2: | |||||
| Pathogenicity not confirmed | Heterozygous | Intron 1 | −588T→A | … | ? |
| Splice site | Heterozygous | Intron 2, donor | 321G→T | … | Polypeptide truncation |
| British patient 3: | |||||
| Missense | Heterozygous | Exon 2 | 232A→G | 232A→G | S78G |
| Missense | Heterozygous | Exon 3 | 529G→A | 431G→A | R144Q |
Note.— Other nucleotide changes observed both in patients and control individuals include −376C→T in intron 1, which is a previously unknown SNP, and 1295T→C in intron 5, which is a known SNP in the NCBI database (rs#2303561).
Sequencing of BCS1L in the three British patients (Morris et al. 1995) revealed three missense mutations, one premature stop codon, one splice-site mutation, and one intronic nucleotide change. Patient 1 was a compound heterozygote for a premature stop codon at amino acid position 56 in the second exon, as well as for a V327A missense mutation in the seventh exon. Patient 2 had a heterozygous splice-donor mutation changing the first G of the second intron to a T, as well as a T→A heterozygous single-nucleotide change in the middle of the first intron, 588 bp upstream from the start codon of the BCS1L. Patient 3 was a compound heterozygote for two missense mutations: the Finnish S78G mutation in exon 2 and an R144Q mutation in exon 3. None of these mutations, including the intronic T→A change of patient 2, was found in 140 control individuals, 90 of whom were Finnish and 50 of whom were of other white ancestry. Two SNPs were observed both in the British infants and in two sequenced control individuals (table 2).
Northern Blot Analysis of Patients with GRACILE Syndrome
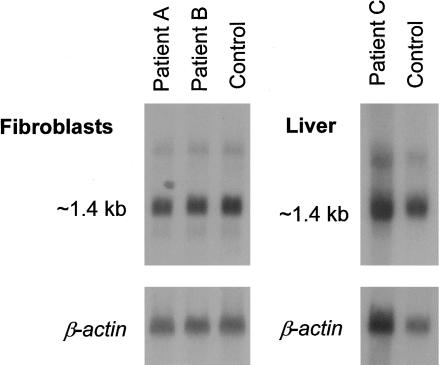
Northern blot analysis of BCS1L was performed in the Finnish patients with GRACILE syndrome, to monitor the steady-state expression level of the gene and possible differences in the size of the transcript. An ∼1.4-kb transcript was detected in both patient and control fibroblasts, as well as in patient and control liver (fig. 2). There was no obvious difference in the steady-state expression level between patients and control individuals. Another faint signal of ∼2.7 kb was observed in both patients and control individuals.
Figure 2.
Northern blot analysis of BCS1L in Finnish patients with GRACILE syndrome. For the fibroblast filter, each line was loaded with 9 μg poly-A-RNA; for the liver filter, each line was loaded with 6 μg poly-A-RNA. The same filters were rehybridized with β-actin cDNA, to control for the total mRNA quantity.
Effect of the S78G Mutation on the Expression, Targeting, and Stability of BCS1L Protein in Transiently Transfected COS-1 Cells
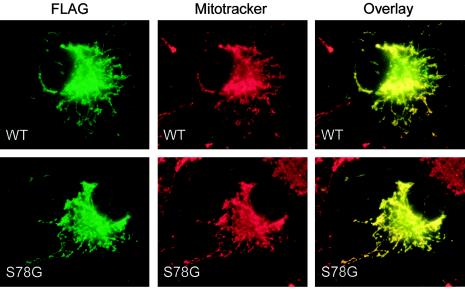
It has previously been shown that human BCS1L protein is targeted unmodified to the inner mitochondrial membrane in vitro (Petruzzella et al. 1998). To study the effect of the S78G mutation on the synthesis, targeting, and stability of the BCS1L protein, we subcloned the coding region of both wild-type and mutant cDNA into pCMV5 expression vector with a C-terminal FLAG tag. Intracellular targeting of the wild-type and mutant BCS1L polypeptides was studied in the transiently transfected COS-1 cells, by immunofluorescence microscopy. The staining with the anti-FLAG antibody completely overlapped with the mitochondrial staining, both in the cells transfected with the wild-type construct and in the cells transfected with the mutant construct (fig. 3), indicating that the S78G mutation does not influence the mitochondrial targeting of the BCS1L protein in vitro.
Figure 3.
Subcellular localization of the wild-type and S78G mutant BCS1L polypeptides in transiently transfected COS-1 cells. The BCS1L polypeptides were detected with anti-FLAG antibody (green), and the distribution was compared with mitochondrial staining (red). Colocalization is indicated in yellow. Magnification = 630×.
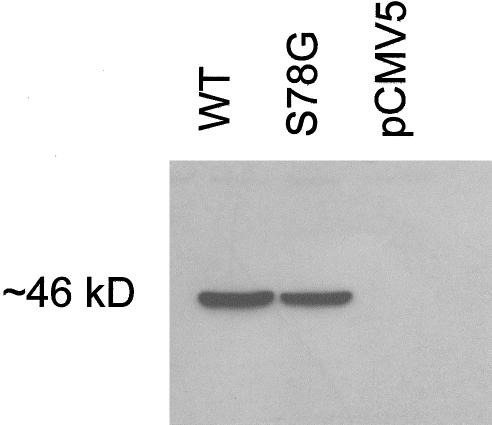
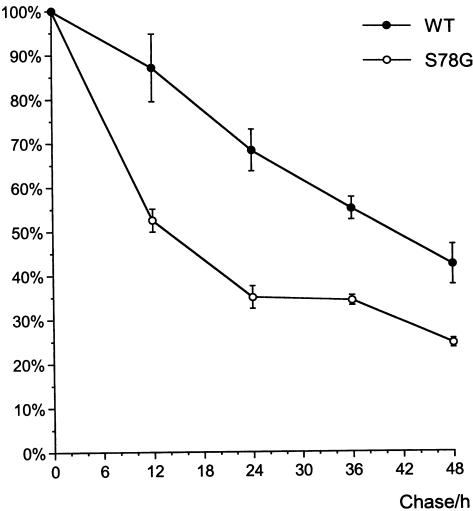
Western blotting of the transiently transfected cell lysates revealed a single ∼46-kD polypeptide. There was no obvious difference in size or intensity between the signal from the wild-type and mutant polypeptides (fig. 4), suggesting that the S78G mutation does not affect the synthesis or posttranslational modifications of the BCS1L polypeptide. Pulse-chase experiments were performed to monitor the stability of the mutant BCS1L protein. The half-life of the FLAG-tagged wild-type BCS1L polypeptide was 41 h, whereas that of the S78G mutant was only 14 h, suggesting that the S78G mutation decreases the stability of the BCS1L polypeptide in vitro (fig. 5).
Figure 4.
Expression of the wild-type and S78G mutant BCS1L polypeptides. COS-1 cells transfected with the wild-type and mutant BCS1L constructs were subjected to western blot analysis using anti-FLAG antibody. Each lane was loaded with 25 μg protein. pCMV5 = cells transfected with the vector only.
Figure 5.
Stability of the wild-type and S78G mutant BCS1L polypeptides. Transiently transfected COS-1 cells were pulse-labeled for 1 h and were chased for 0, 12, 24, 36, and 48 h. The BCS1L polypeptides were immunoprecipitated, were separated on 10% SDS-PAGE, and were visualized by fluorography. The radioactive bands were quantitated by densitometry. The results are presented as a percentage of values at 0 h chase. The values on the curve are average values of duplicate experiments, with error bars showing the SD.
Yeast Complementation Studies
Yeast BCS1p protein has been shown to be essential for the assembly of the mitochondrial respiratory chain complex III (Nobrega et al. 1992; Cruciat et al. 1999). Yeast strains deleted for the BCS1 gene cannot grow on glycerol, since, on nonfermentable carbon sources, yeast cells are dependent on oxidative respiration. On glucose, BCS1-deletion strains are able to grow by utilizing anaerobic metabolism. Human BCS1L protein is highly homologous to yeast BCS1p protein (50% identity), and human BCS1L can partially rescue a yeast strain deleted for BCS1 (de Lonlay et al. 2001). This facilitated the use of yeast complementation studies in monitoring the functional effect of the S78G mutation.
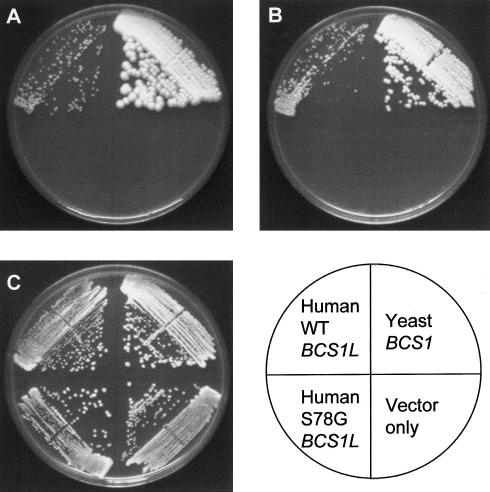
We tested the effect of the S78G mutation on the ability of human BCS1L to restore oxidative respiration in a BCS1 deletion strain. The BCS1-deletion yeast strain Y14211 was transformed with the wild-type and 232A→G (S78G) mutant BCS1L cDNAs in a high-copy yeast expression plasmid, pRS425. As a positive control, we also transformed the same strain with the wild-type yeast BCS1 cDNA in the same vector, and we used the plain vector as a negative control. The purified transformants were streaked on YPEG (ethanol-glycerol), YPD, and SD plates without leucine (vector selection) and were incubated at 30°C and 37°C. On YPD and SD-leu plates, all the transformants grew equally well at both temperatures, indicating that all the transformants were viable on glucose. On YPEG plates, the yeast BCS1 transformants were clearly growing after 48 h incubation. The wild-type human BCS1L transformants grew, but considerably more slowly, showing the first signs of growth after 4 d at 37°C and after 6 d at 30°C. The S78G mutant and the vector-only transformants did not show any growth, even after incubation for 2 wk, suggesting that the S78G mutation causes a functional defect in the BCS1L protein (fig. 6).
Figure 6.
Yeast complementation experiment with BCS1L. The BCS1 deletion yeast strain was transformed with the wild-type and mutant BCS1L constructs in a high-copy yeast expression plasmid pRS425. The wild-type yeast BCS1 cDNA in the same vector was used as a positive control, and the plain vector was used as a negative control. A, YPEG (ethanol-glycerol) plate incubated at 30°C for 11 d. B, YPEG plate incubated at 37°C for 8 d. C, YPD (glucose) plate incubated at 30°C for 3 d.
We also introduced a S115G mutation (corresponding to the S78G mutation in human) into the yeast BCS1 gene and transformed the Y14211 yeast strain with the wild-type and S115G mutant yeast BCS1 gene in the low-copy yeast expression vector pRS315. The mutant transformants grew on YPEG plates as well as the wild-type transformants did, indicating that the S115G amino acid change does not cause any significant defect in the yeast BCS1p protein.
Complex III Enzyme Activity Measurements
The decreased complex III activities (37% of control values in patient 1 and 15% of control values in patients 2 and 3) in the skeletal muscle of the British patients have been described elsewhere in detail (Birch-Machin et al. 1989; Morris et al. 1995). The Finnish patients with GRACILE syndrome have had normal complex III activities in isolated mitochondria of all tissues studied (Fellman et al. 1998), but, because of an evident discrepancy between the English and Finnish findings, we confirmed the findings in detail.
All the respiratory chain enzyme activities measured in isolated mitochondria from skeletal muscle biopsies of the Finnish patients were within the normal range (table 3). From their autopsy-derived tissues, the myocardial samples showed activities higher than in the autopsy controls (table 4). However, the complex III–dependent activities were relatively less increased than the complex IV activity. Activities similar to those in the myocardium were found in liver, brain, and kidney specimens, for which sufficient controls were not available. Since these activities represent the values of indirect measurements of complex III, deduced from the combined activities with complex I and complex II, we also assessed complex III separately in homogenized tissue samples. In two deep-frozen (−70°C) liver tissue samples, all the mitochondrial enzyme activities were low, indicating a tissue preservation problem. However, in liver and muscle homogenates of one patient (specimens stored in liquid nitrogen), the activities were generally within the normal range (table 5). The liver showed normal complex III activity, whereas in the muscle complex III activity was moderately decreased. In proportion to citrate synthase (CS) activity, the complex III activity of the muscle specimen was reduced by 1.5 SD, compared with the controls. In addition, no deficiencies in respiratory chain enzyme activities (complexes I + III, II + III, and IV) were detected in fibroblast cell lines of patients with GRACILE syndrome.
Table 3.
Respiratory Chain Enzyme Activity in Muscle Biopsy Samples from Finnish Patients with GRACILE Syndrome
|
Activity (Ratio to CS)b(nmol/min/mg mitochondrial protein) |
|||||
| Samplea | Complex I + III | Complex II + III | Complex II | Complex IV | CS |
| Patient B | 65 (.13) | 89 (.18) | 188 (.37) | 1,169 (2.31) | 506 |
| Patient C | 76 (.11) | 154 (.23) | 195 (.29) | 3,093 (4.54) | 682 |
| Patient D | 84 | 81 | 249 | 1,815 | NDc |
| Patient E | 132 (.21) | 163 (.26) | 101 (.16) | 1,929 (3.08) | 626 |
| Patient F | 218 (.27) | 61 (.07) | 154 (.19) | 2,219 (2.72) | 816 |
| Control (n=7)d | 140 ± 70 (.14) | 197 ± 88 (.20) | 162 ± 51 (.17) | 2,521 ± 759 (2.60) | 969 ± 243 |
Samples are isolated mitochondria from muscle biopsy specimens.
Activity measurement performed as described by Majander et al. (1995).
ND = not determined.
Data are mean ± SD (ratio to CS calculated from controls' mean).
Table 4.
Respiratory Chain Enzyme Activity in Autopsy Samples of Myocardium from Finnish Patients with GRACILE Syndrome
|
Activityb(nmol/min/mg mitochondrial protein) |
|||
| Samplea | Complex I+III/Complex II | Complex II+III/Complex II | Complex IV/Complex II |
| Patient C | 2.9 | 2.1 | 28 |
| Patient D | .4 | .6 | 4.7 |
| Patient E | 1.4 | 1.4 | 32 |
| Patient F | .6 | .2 | 19 |
| Control (n=5)c | 1.1 ± .4 | 1.6 ± .7 | 8.8 ± 2.6 |
Samples are isolated mitochondria from autopsy samples obtained 2–7 h after death.
Activity measurements performed as described by Majander et al. (1995).
Data are mean ± SD.
Table 5.
Respiratory Chain Enzyme Activity in Autopsy Samples of Liver and Muscle from a Finnish Patient with GRACILE Syndrome
|
Activity (Ratio to CS)b(nmol/min/mg mitochondrial protein) |
||||||
| Samplea | Complex I | Complex II | Complex II + III | Complex III | Complex IV | CS |
| Liver: | ||||||
| Patient C | 11 (.12) | 67 (.74) | 15 (.17) | 25 (.28) | 5.5 (.06) | 90 |
| Control (n=13)c | 19 [8–37] (.23) | 110 [58–160] (1.33) | 28 [13–42] (.34) | 36 [14–68] (.43) | 46 [26–70] (.55) | 83 [41–117] |
| Muscle: | ||||||
| Patient C | 32 (.11) | 54 (.19) | 47 (.16) | 28 (.10) | 147 (.52) | 285 |
| Control (n=30)c | 26 [13–44] (.12) | 47 [23–86] (.22) | 43 [20–75] (.20) | 57 [21–118] (.27) | 126 [75–205] (.59) | 214 [140–334] |
Samples are from tissue stored in liquid nitrogen.
Activity measurements of tissue homogenates performed as described by Rustin et al. (1994).
Data are mean [5%–95% range] (ratio to CS calculated from controls' mean).
Discussion
Our results strongly suggest that GRACILE syndrome in the Finnish patients is caused by an S78G amino acid change in the BCS1L protein. The protein has previously been characterized as an essential factor for the assembly of respiratory chain complex III. We show here that, surprisingly, the S78G mutation does not seem to affect complex III activity considerably, but the patients have iron overload. This finding suggests a completely new and unexplored function for BCS1L in cellular iron metabolism.
We found the same BCS1L mutation in all Finnish patients with GRACILE syndrome, as well as five mutations (including the Finnish mutation) in three British patients with similar symptoms. The clinical characteristics of both the Finnish and British patients included severe fetal growth retardation, lactic acidosis, aminoaciduria, and death before the age of 5 mo. The Finnish patients with GRACILE syndrome did not have any neurological problems or clearly decreased complex III activity, in contrast to the British patients, but the Finnish patients had marked hepatic iron overload, associated with abnormal levels of proteins involved in iron transfer and storage, and free plasma iron. Four different mutations in BCS1L have also been identified elsewhere, in five patients from four unrelated Turkish families (de Lonlay et al. 2001). The symptoms in these patients varied, the essential findings being metabolic acidosis, tubulopathy, hepatic features, and severe neurological problems. Growth retardation was less severe than in the Finnish and British patients. Three of the patients died at the age of 3 mo to 2 years, one was 9 years old, and one was lost to follow-up at the age of 5 mo. Like the British infants, all the Turkish patients had complex III deficiency. Unfortunately, iron metabolism was not studied in either the British or the Turkish patients.
The S78G mutation segregated without exception with the disease phenotype in the Finnish families with GRACILE syndrome, and only one carrier was detected in 544 control individuals, which supports the pathogenic role of the mutation. The causative role was further supported by the finding of the same mutation, in heterozygous form, in a British patient, who also had a R144Q mutation in her other allele. In other British patients, we identified an early stop mutation, a splice-site mutation, and a V327A missense mutation. S78 is highly conserved across the species, and the predicted effect of the serine-to-glycine change on the polypeptide is considerable. The smallest amino acid, glycine enables the polypeptide backbone to make turns that are not possible with any other residue, and it also lacks the hydroxyl group and the polarity of serine. Our in vitro experiments in COS-1 cells suggest that the S78G mutation does not affect targeting or posttranslational modifications of the BCS1L protein but results in decreased stability of the protein. The functional significance of most other mutations is also obvious: the premature stop codon is a truncating mutation and can be considered pathogenic even without functional evidence. The R144Q mutation changes a basic and charged arginine into acidic and uncharged glutamine. R144 is conserved in mouse, and, in both Drosophila melanogaster and S. cerevisiae, it is replaced by lysine, another basic and charged amino acid, being relatively well conserved. The V327A change appears to be the least dramatic of the current mutations, since both valine and alanine are small, nonpolar amino acids without functional groups. V327 is not strictly conserved either, since in S. cerevisiae it is replaced by threonine, a polar amino acid. However, since this mutation was not found in 140 control individuals and since the early stop mutation found in the other allele of the same patient is an obligatory mutation, we consider this V327A substitution to be a likely mutation as well. It is located in the conserved ATP-binding AAA-sequence motif of the BCS1L polypeptide, which could explain the pathogenicity of this relatively moderate amino acid change (fig. 1B and 1C).
In British patient 2, we identified a heterozygous splice-donor mutation and a heterozygous T→A nucleotide change in the middle of the first intron. The pathogenicity of these nucleotide changes has not yet been proven at the RNA level. However, since the phenotype of patient 2 was identical to that of patient 3, we consider BCS1L mutations to be a likely cause underlying the symptoms of patient 2 as well. Since the splice-site mutation abolishes a classical splice-donor consensus sequence, it is likely to result in truncation of the protein. The importance of the nucleotide change in the first intron is more questionable. It was not present in 140 control individuals, implying that it does not represent a common SNP. However, the 20 bp surrounding the nucleotide variant are not conserved in mouse (Celera mouse sequence GA_x5J8B7W5GLS), so the functional importance of the region is not supported by phylogenetic conservation. We did not identify any other possible mutations in the DNA of patient 2, but we cannot exclude the possibility of a regulatory mutation located far upstream.
The carrier frequency of the S78G mutation observed among the Finnish control individuals was lower than expected. The incidence of GRACILE syndrome is ⩾1/47,000 in Finland, predicting a carrier frequency of ∼1/110. The ancestors of the families with GRACILE syndrome originated from rural areas in eastern and central Finland (Fellman et al. 1998), and most of the families still live in these regions. Therefore, the carrier frequency in eastern and central Finland is estimated to be higher, ∼1/80, whereas in western Finland it is predicted to be very low. Thus, we should have identified about five carriers among our 494 control individuals, 70% of whom originated from the eastern and central parts of Finland. The GRACILE mutation most probably represents a relatively new mutation, enriched in the restricted regional subisolates of eastern and central Finland. We have earlier demonstrated quite significant regional differences in our DNA array-based identification of the carriers of Finnish mutations, reflecting the effect of multiple population bottlenecks during the inhabitation of the country (Pastinen et al. 2001).
The sequence information of BCS1L in databases suggests alternative splicing of BCS1L mRNA. These splice variants would not affect the structure of the translated BCS1L polypeptide, since the second exon containing the start codon and all six exons thereafter are identical in all splice variants. It is also unlikely that the alternative 5′ UTRs would affect the translation initiation, because an in-frame stop codon lies 42 bp before the start codon, making an earlier translation initiation impossible. The use of a methionine further downstream as an alternative start codon is not likely either, since the next methionine is residue 48, which is located downstream from the only transmembrane domain of the protein. Alternative splicing of the 5′ UTR could, however, enable tissue-specific up- or downregulation of BCS1L expression. The occurrence of various splice variants in different tissue and cell types and their importance for the regulation of gene expression remains to be verified by experimental tools.
The only known function of BCS1L protein is the chaperone function in the assembly of complex III in the respiratory chain. This has been studied in detail in yeast (Nobrega et al. 1992; Cruciat et al. 1999), in which the BCS1p protein has been shown to act as an ATP-dependent chaperone maintaining the precomplex in a competent state for the assembly of Rieske FeS and Qcr10p proteins. In human, BCS1L has a related function, since both our British patients and the Turkish patients with BCS1L mutations (de Lonlay et al. 2001), unequivocally had a complex III defect. However, in the Finnish patients with GRACILE syndrome, complex III activity was not severely affected. This would suggest that the Finnish S78G mutation disturbs an unknown BCS1L function that does not involve the assembly of complex III but probably has a role in iron metabolism. Interestingly, the Finnish patients with GRACILE syndrome did not have any neurological problems, in contrast to the British and Turkish patients, who had variable neurological symptoms and findings, including muscular hypotonia, encephalopathy, seizures, and Leigh syndrome, which are typical for mitochondrial respiratory chain defects.
Since it was surprising that the complex III activity seemed normal in the Finnish patients, we used all available material and methods to confirm it. For respiratory chain enzyme activity measurements, fresh biopsy samples generally give the most reliable results, and we had appropriate age-matched controls available for muscle biopsy specimens. Enzyme activity measurements of the muscle biopsy samples of the Finnish patients were within the normal range (table 3), and oxygen consumption studies have also previously implied normal respiratory chain function in fresh muscle samples (Fellman et al. 1998). In one patient, analysis of the biopsy sample showed normal enzyme activities, but direct complex III measurement of the autopsy muscle sample was in the lower part of the control range (table 5). If this reduction is not a postmortem artifact due to a tissue preservation problem and reduced enzyme activities, then the mild decrease may have remained undetected in the indirect complex III analyses of the biopsy specimen. Therefore, our data suggest that complex III is not markedly affected in Finnish patients but cannot exclude a mild defect. Partial reduction in the complex III activity alone would not be likely to cause the severe clinical picture of the Finnish patients, since the Turkish patients (de Lonlay et al. 2001) with clearly decreased complex III activities survived longer and were less growth retarded than were the Finnish patients. However, even a mild complex III defect might contribute to the lactic acidosis and other symptoms.
Nuclear gene defects causing mitochondrial disorders often have a strictly tissue-specific expression (Papadopoulou et al. 1999), and, in GRACILE syndrome, the most relevant tissue for expressing complex III deficiency is the liver, since it is the major organ of dysfunction. The enzyme activity measurements of the liver specimens of the Finnish patients with GRACILE syndrome do not support complex III deficiency. The complex III activity in the liver homogenate of one patient (table 5) was comparable to that of the control individuals. Enzyme activity measurements of the isolated mitochondria of the liver specimens of four patients, obtained at autopsy shortly after death, also supported this finding by showing the presence of complex III activity, although, because there were so few control individuals, fully reliable conclusions regarding these samples are not possible. Thus, we consider it unlikely that the Finnish patients with GRACILE syndrome have a liver-specific complex III defect severe enough to result in the fatal course of the disease.
How could the S78G mutation disturb the other function of BCS1L while leaving complex III activity practically unaffected? The mutation is located between the transmembrane domain and the AAA domain of the BCS1L polypeptide. The function of this region remains to be characterized and could potentially be associated with this as-yet-uncharacterized function of BCS1L (fig. 1B and 1C). Our pulse-chase experiments also showed that the S78G mutation accelerates degradation of the BCS1L polypeptide in vitro. If this were also to result in decreased BCS1L protein levels in tissues in vivo, without the mutation interfering in protein conformation, then the reduced protein amount in the mitochondria could possibly be sufficient to perform complex III assembly, whereas the other function could be compromised.
Rieske protein, which is joined to precomplex III by BCS1p in yeast, contains an FeS cluster. FeS proteins are essential in electron transport systems and in numerous other cellular functions in the mitochondria, cytosol, and nucleus. It is an intriguing question whether BCS1L has a more general role in the biosynthesis or transport of these proteins. Biosynthesis of the FeS clusters is accomplished in mitochondria, and a dozen mitochondrial proteins involved in the process have so far been identified in S. cerevisiae (Muhlenhoff and Lill 2000).
Our yeast complementation studies showed that aerobic energy production in S78G transformants was defective. This could be due either to complex III deficiency, a defect in the respiratory chain in general, or iron toxicity. However, the wild-type human BCS1L was only partially able to complement the BCS1-deletion yeast, and, therefore, the consequences of the S78G mutation can appear more serious in yeast complementation experiments than they are in vivo. No significant defects in respiratory chain complexes other than complex III have been reported in patients with BCS1L mutations, although a slight, nonspecific decrease in complex I has been observed in Finnish patients (Fellman et al. 1998), and two British infants had slightly decreased complex IV activity in skeletal muscle (Morris et al. 1995).
Until now, DNA diagnosis of GRACILE syndrome has been possible in Finland, based on linkage analysis in families with at least one affected child (Fellman et al. 2002). Now the Finnish S78G mutation can be identified directly, which enables DNA diagnosis and genetic counseling even for families without a previously affected baby. For non-Finnish patients, sequence analysis of the BCS1L gene is a diagnostic option.
Acknowledgments
We thank Sari Pitkänen for the complex III activity measurements of the fibroblast cell lines. We thank Anna Majander, Kari Raivio, Anu Jalanko, Giancarlo Costaguta, and Todd Lorenz for their valuable methodological advice. We thank Eija Hämäläinen, Anne Jokiaho, Tuula Manninen, Maija Parkkonen, Carina von Schantz, and Arja Terola for their skillful technical assistance. This study was financially supported by the UCLA School of Medicine, the Academy of Finland, the Helsinki Biomedical Graduate School, the Ulla Hjelt Fund, the Maud Kuistila Foundation, the Finnish medical society Duodecim, and the Paulo Foundation.
Footnotes
Nucleotide sequence data reported herein are available in the DDBJ/EMBL/GenBank databases; for details, see the Electronic-Database Information section of this article.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Celera, http://www.celera.com/ (for BCS1L mRNA sequences hCT1955111 and hCT7339 and for the mouse genomic sequence GA_x5J8B7W5GLS)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for BCS1L genomic sequence [accession number AF516670], genomic clone RP11-1077K22 [accession number AC079810, gi 15147206], and BCS1L mRNA and EST sequences [accession numbers BI091793, BG615931, XM_002588, AF026849, NM_004328, AL526509, BG740684, AF038195, BG536545, BC007500, BC000416, AL530106, and BE729532])
- Human Genome at NCBI, http://www.ncbi.nlm.nih.gov/genome/guide/human/ (for MapViewer and SNP database [rs#2303561])
- NCBI BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for GRACILE syndrome [MIM 603358])
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/
References
- Andersson S, Davis DL, Dahlback H, Jornvall H, Russell DW (1989) Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem 264:8222–8229 [PubMed] [Google Scholar]
- Andreu AL, Hanna MG, Reichmann H, Bruno C, Penn AS, Tanji K, Pallotti F, Iwata S, Bonilla E, Lach B, Morgan-Hughes J, DiMauro S (1999) Exercise intolerance due to mutations in the cytochrome b gene of mitochondrial DNA. N Engl J Med 341:1037–1044 [DOI] [PubMed] [Google Scholar]
- Birch-Machin MA, Shepherd IM, Watmough NJ, Sherrat HS, Bartlett K, Darley-Usmar VM, Milligan DW, Welch RJ, Aynsley-Green A, Turnbull DM (1989) Fatal lactic acidosis in infancy with a defect of complex III of the respiratory chain. Pediatr Res 25:553–559 [DOI] [PubMed] [Google Scholar]
- Cruciat CM, Hell K, Fölsch H, Neupert W, Stuart RA (1999) Bcs1p, an AAA-family member, is a chaperone for the assembly of the cytochrome bc1 complex. EMBO J 18:5226–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lonlay P, Valnot I, Barrientos A, Gorbatyuk M, Tzagoloff A, Taanman JW, Benayoun E, Chretien D, Kadhom N, Lombes A, de Baulny HO, Niaudet P, Munnich A, Rustin P, Rötig A (2001) A mutant mitochondrial respiratory chain assembly protein causes complex III deficiency in patients with tubulopathy, encephalopathy and liver failure. Nat Genet 29:57–60 [DOI] [PubMed] [Google Scholar]
- Fellman V, Rapola J, Pihko H, Varilo T, Raivio KO (1998) Iron-overload disease in infants involving fetal growth retardation, lactic acidosis, liver haemosiderosis, and aminoaciduria. Lancet 351:490–493 [DOI] [PubMed] [Google Scholar]
- Fellman V, Visapää I, Vujic M, Wennerholm UB, Peltonen L (2002) Antenatal diagnosis of hereditary fetal growth retardation with aminoaciduria, cholestasis, iron overload, and lactic acidosis in the newborn infant. Acta Obstet Gynecol Scand 81:398–402 [DOI] [PubMed] [Google Scholar]
- Fölsch H, Guiard B, Neupert W, Stuart RA (1996) Internal targeting signal of the BCS1 protein: a novel mechanism of import into mitochondria. EMBO J 15:479–487 [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley JA, Anitori R, Burton MD, Quan F, Buist NRM, Kennaway NG (2000) Mitochondrial encephalomyopathy and complex III deficiency associated with a stop-codon mutation in the cytochrome b gene. Am J Hum Genet 67:1400–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F, Chatzoglou E, Frachon P, de Baulny HO, Laforet P, Jardel C, Godinot C, Lombes A (2001) Functional characterization of novel mutations in the human cytochrome b gene. Eur J Hum Genet 9:510–518 [DOI] [PubMed] [Google Scholar]
- Majander A, Rapola J, Sariola H, Suomalainen A, Pohjavuori M, Pihko H (1995) Diagnosis of fatal infantile defects of the mitochondrial respiratory chain: age dependence and postmortem analysis of enzyme activities. J Neurol Sci 134:95–102 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH (2002) CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res 30:281–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi N, Miki T, Senbongi H, Yokoi N, Yano H, Miyazaki M, Nakajima N, Iwanaga T, Yokoyama Y, Shibata T, Seino S (2000) MTABC3, a novel mitochondrial ATP-binding cassette protein involved in iron homeostasis. J Biol Chem 275:17536–17540 [DOI] [PubMed] [Google Scholar]
- Morris AAM, Taylor RW, Birch-Machin MA, Jackson MJ, Coulthard MG, Bindoff LA, Welch RJ, Howell N, Turnbull DM (1995) Neonatal Fanconi syndrome due to deficiency of complex III of the respiratory chain. Pediatr Nephrol 9:407–411 [DOI] [PubMed] [Google Scholar]
- Mount SM (1982) A catalogue of splice junction sequences. Nucleic Acids Res 10:459–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff U, Lill R (2000) Biogenesis of iron-sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Biochim Biophys Acta 1459:370–382 [DOI] [PubMed] [Google Scholar]
- Nobrega FG, Nobrega MP, Tzagoloff A (1992) BCS1, a novel gene required for the expression of functional Rieske iron-sulfur protein in Saccharomyces cerevisiae. EMBO J 11:3821–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio R, Nevanlinna HR, Perheentupa J (1973) Hereditary diseases in Finland: rare flora in rare soil. Ann Clin Res 5:109–141 [PubMed] [Google Scholar]
- Papadopoulou LC, Sue CM, Davidson MM, Tanji K, Nishino I, Sadlock JE, Krishna S, Walker W, Selby J, Glerum DM, Coster RV, Lyon G, Scalais E, Lebel R, Kaplan P, Shanske S, De Vivo DC, Bonilla E, Hirano M, DiMauro S, Schon E (1999) Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat Genet 23:333–337 [DOI] [PubMed] [Google Scholar]
- Pastinen T, Perola M, Ignatius J, Sabatti C, Tainola P, Levander M, Syvänen AC, Peltonen L (2001) Dissecting a population genome for targeted screening of disease mutations. Hum Mol Genet 10:2961–2972 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Jalanko A, Varilo T (1999) Molecular genetics of the Finnish disease heritage. Hum Mol Genet 8:1913–1923 [DOI] [PubMed] [Google Scholar]
- Petruzzella V, Tiranti V, Fernandez P, Ianna P, Carrozzo R, Zeviani M (1998) Identification and characterization of human cDNAs specific to BCS1, PET112, SCO1, COX15, and COX11, five genes involved in the formation and function of the mitochondrial respiratory chain. Genomics 54:494–504 [DOI] [PubMed] [Google Scholar]
- Pitkänen S, Robinson BH (1996) Mitochondrial complex I deficiency leads to increased production of superoxide radicals and induction of superoxide dismutase. J Clin Invest 98:345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapola J, Heikkilä P, Fellman V (2002) Pathology of lethal fetal growth retardation syndrome with aminoaciduria, iron overload, and lactic acidosis (GRACILE). Pediatr Pathol Mol Med 21:183–193 [DOI] [PubMed] [Google Scholar]
- Rustin P, Chretien D, Bourgeron T, Gerard B, Rötig A, Saudubray JM, Munnich A (1994) Biochemical and molecular investigations in respiratory chain deficiencies. Clin Chim Acta 228:35–51 [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syvänen AC, Sajantila A, Lukka M (1993) Identification of individuals by analysis of biallelic DNA markers, using PCR and solid-phase minisequencing. Am J Hum Genet 52:46–59 [PMC free article] [PubMed] [Google Scholar]
- Wibrand F, Ravn K, Schwartz M, Rosenberg T, Horn N, Vissing J (2001) Multisystem disorder associated with a missense mutation in the mitochondrial cytochrome b gene. Ann Neurol 50:540–543 [DOI] [PubMed] [Google Scholar]
- Visapää I, Fellman V, Lanyi L, Peltonen L (2002) ABCB6 (MTABC3) excluded as the causative gene for the growth retardation syndrome with aminoaciduria, cholestasis, iron overload, and lactacidosis. Am J Med Genet 109:202–205 [DOI] [PubMed] [Google Scholar]
- Visapää I, Fellman V, Varilo T, Palotie A, Raivio KO, Peltonen L (1998) Assignment of the locus for a new lethal neonatal metabolic syndrome to 2q33-37. Am J Hum Genet 63:1396–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]