Abstract
The cause of schizophrenia is unknown, but it has a significant genetic component. Pharmacologic studies, studies of gene expression in man, and studies of mouse mutants suggest involvement of glutamate and dopamine neurotransmitter systems. However, so far, strong association has not been found between schizophrenia and variants of the genes encoding components of these systems. Here, we report the results of a genomewide scan of schizophrenia families in Iceland; these results support previous work, done in five populations, showing that schizophrenia maps to chromosome 8p. Extensive fine-mapping of the 8p locus and haplotype-association analysis, supplemented by a transmission/disequilibrium test, identifies neuregulin 1 (NRG1) as a candidate gene for schizophrenia. NRG1 is expressed at central nervous system synapses and has a clear role in the expression and activation of neurotransmitter receptors, including glutamate receptors. Mutant mice heterozygous for either NRG1 or its receptor, ErbB4, show a behavioral phenotype that overlaps with mouse models for schizophrenia. Furthermore, NRG1 hypomorphs have fewer functional NMDA receptors than wild-type mice. We also demonstrate that the behavioral phenotypes of the NRG1 hypomorphs are partially reversible with clozapine, an atypical antipsychotic drug used to treat schizophrenia.
Introduction
Schizophrenia (MIM 181500) is a disabling brain disease affecting 0.5%–1% of the general population. It has a myriad of manifestations. These classically include one or more of the following: delusions, disordered thought, hallucinations, blunted emotions, paranoid ideation, and motor abnormalities such as stereotypic behaviors and catatonia (Liddle et al. 1994). More recently, other cognitive abnormalities—such as impaired memory, attention, and executive function—have been documented (Bilder 1996). Although there are medications that can alleviate some of these symptoms in patients, there is no cure available for schizophrenia, and most patients who respond to current treatments fail to cope fully in society.
Although conventional antipsychotics act by blocking dopamine receptors, there is considerable recent evidence linking defects in glutamatergic neurotransmission to the psychiatric manifestations of schizophrenia. Antagonists of glutamatergic signaling through the N-methyl-D-aspartate (NMDA) receptor (such as PCP, MK-801, and ketamine) induce psychosis in normal individuals and exacerbate manifestations of schizophrenia in patients. Recent studies have shown decreased binding to subunits of the glutamate receptor and decreased expression of subunits of the glutamate receptor in thalamic nuclei, hippocampus, and dorsal lateral prefrontal cortex of schizophrenic patients (Ibrahim et al. 2000; Gao et al. 2000). It is of interest that some of the atypical antipsychotics (such as clozapine) increase NMDA-receptor expression and directly facilitate glutamatergic transmission (Goff and Coyle 2001). Finally, mice with a reduced number of NMDA receptors exhibit behavior that overlaps that induced by PCP—hyperactivity, deficits in social interaction, and decreased prepulse inhibition (PPI); these are reversible with clozapine (Mohn et al. 1999). It is important to point out, however, that the glutamate dysfunction hypothesis is not incompatible with the dopamine hypothesis, because many reciprocal synaptic relationships exist between the two systems (Carlsson and Carlsson 1990).
Although the cause of schizophrenia is unknown, family and adoption studies suggest that schizophrenia has a significant genetic component (Cardno et al. 1999; Tsuang et al. 2001). As in most common diseases, the inheritance pattern is complex, and the penetrance is low. Unfortunately, there is no reliable biological marker for the disease. The many ways that schizophrenia manifests itself could be explained by locus heterogeneity, which also may have hampered progress in the search for schizophrenia genes. Given that neurotransmitter pathways are probably abnormal in schizophrenia, variants of genes encoding receptor subunits and transporters have been extensively tested for association to schizophrenia in many populations (O'Donovan and Owen 1999). Although there have been reports of weak association with variations of certain genes, no significant findings have been reproduced in independent cohorts. The associations reported so far, if real, represent only minor genetic contributions.
Numerous genomewide linkage scans have been reported for schizophrenia. There is modest evidence for linkage with several loci, including chromosome 1q, 2, 6, 8p, 13q, and 22q. The chromosome 8p locus (SCZD6 [MIM 603013]) shows suggestive linkage to schizophrenia in several different populations (Pulver et al. 1995; Kendler et al. 1996; Levinson et al. 1996; Blouin et al. 1998; Kaufmann et al. 1998; Shaw et al. 1998; Brzustowicz et al. 1999; Gurling et al. 2001). Information on the LOD scores and the P values in these studies is summarized by Brzustowicz et al. (1999).
Here, we present results of a genomewide scan for linkage to schizophrenia and a follow-up fine mapping at a locus on chromosome 8p. Haplotype analysis identifies neuregulin 1 (NRG1 [MIM 142445]) as a candidate gene for schizophrenia. Furthermore, we report functional studies supporting the notion that NRG1 plays a role in the pathogenesis of schizophrenia. First, we confirm previous work (Gerlai et al. 2000) showing that mice hypomorphic for NRG1 (with a different NRG1 mutation than previously phenotyped) exhibit behavioral abnormalities. Second, we report that mice hypomorphic for the NRG1 receptor, ErbB4, exhibit similar behavioral abnormalities. Third, we show that behavioral abnormalities of the NRG1 hypomorphs can be partially reversed with clozapine. Fourth, our binding studies show that mice hypomorphic for NRG1 have fewer functional NMDA receptors than do control mice, which is in keeping with observations made on brains from patients with schizophrenia (Ibrahim et al. 2000).
Subjects and Methods
Patients
This study was approved by appropriate ethics committees and the Data Protection Commission of Iceland (DPC). Informed consent was obtained from all patients and from their relatives whose blood and DNA samples were used in the linkage and association analysis. All personal identifiers associated with medical information and blood samples were encrypted by the DPC with a third-party encryption system, as described elsewhere (Gulcher et al. 2000). The genealogy database was encrypted by the DPC in the same manner. Taking part in the study were 476 patients—including 440 diagnosed with schizophrenia, 32 with schizoaffective disorder, and 4 with unspecified functional psychosis—as well as unaffected family members. Most probands were identified through referrals to the in- and outpatient services of all three psychiatric departments in Iceland (total population 285,000). Diagnoses were assigned according to Research Diagnostic Criteria (RDC) (Spitzer et al. 1978) through the use of the lifetime version of the Schizophrenia and Affective Disorders Schedule (SADS-L) (Spitzer and Endicott 1977). Psychiatrists who were blind to the genotyping data made consensus diagnoses.
Linkage Analysis
Large multiplex families were collected for the linkage scan. Pedigrees allowing for up to seven meiotic events between affected individuals were constructed, using a clustering algorithm, from an encrypted genealogy database that covers the entire Icelandic nation (Gulcher et al. 2000). A genomewide scan was performed using a framework map of 950 microsatellite markers and using protocols described elsewhere (Gretarsdottir et al. 2002). The marker order and positions for the framework mapping set were obtained from our high-resolution genetic map (Kong et al. 2002). We analyzed the data and determined statistical significance by applying a model independent affecteds-only allele-sharing method, implemented in the Allegro program, that calculates LOD scores on the basis of multipoint calculations (Gudbjartsson et al. 2000). Our baseline linkage analysis uses the Spairs scoring function (Kruglyak et al. 1996), the exponential allele-sharing model (Kong and Cox 1997), and a family-weighting scheme that is halfway, on the log scale, between weighting each affected pair equally and weighting each family equally. In the analysis, we treat all genotyped individuals who are not affected as “unknown.” Affecteds are those who are diagnosed, using the RDC criteria, with schizophrenia, schizoaffective disorder, or unspecified functional psychosis.
Physical Mapping
The BAC contig of the region of interest, 8p11-p21, was generated using the RCPI-11 Human BAC library. BAC identification and contig orders were determined by hybridization using available STS markers and microsatellite markers in the region, followed by successive rounds of hybridization using markers designed from BAC end sequences. BAC fingerprint data complemented these data. Fingerprints of positive clones (FPCs) were analyzed using the FPC database developed at the Wellcome Trust Sanger Institute. New microsatellite markers were discovered from cloning and by screening fragments from nebulized BACs.
Identification of At-Risk Haplotypes
To handle missing genotypes and uncertainty with phase, our own implementation of a likelihood approach, using the expectation-maximization (EM) algorithm (Dempster et al. 1977) as a computational tool, was applied to estimate the haplotype frequencies. Under the null hypothesis, the affected individuals and controls are assumed to have identical frequencies of all haplotypes. Under the alternative hypothesis, the candidate at-risk haplotype is allowed to have a higher frequency in affected individuals than controls, while the ratios of the frequencies of all other haplotypes are assumed to be the same in both groups. Likelihoods are maximized separately under both hypotheses and a corresponding 1-df likelihood ratio statistic is used to evaluate statistical significance. While our own computer program was developed to fit our chosen models, and to handle missing genotypes and haplotypes with many markers efficiently, our use of the EM-algorithm is very similar to methods used by others (Excoffier and Slatkin 1995; Hawley and Kidd 1995; Long et al. 1995). Although applied in a slightly different setting, the 1-df model we use is essentially that used by Clayton and Jones (1999).
Transmission/Disequilibrium Test
The transmission/disequilibrium test (TDT) was performed on probands for whom both parents are genotyped, and one or both parents are heterozygous for the at-risk haplotype (Spielman et al. 1993).
Sequencing of the NRG1 Locus and Identification of Exons and Transcripts
BACs covering the minimum tiling path of the region of interest were analyzed by shotgun cloning and sequencing (GenBank accession numbers AF491780 and TPA BK000383). Dye terminator (ABI PRISM BigDye) chemistry was used for fluorescent automated DNA sequencing. ABI Prism 377 sequencers were used to collect data, and the Phred/Phrap/Consed and Polyphred software packages were used to assemble sequences. We identified syntenic mouse BACs (library RPCI-23), and, by BAC walking, a contig across the NRG1 locus was made. The methods described above were used to subclone and sequence eight syntenic BAC clones from the mouse. The mouse sequence was used to identify more exons and potential regulatory elements. Both 3′ and 5′ RACE (rapid amplification of cDNA ends) were performed using the Marathon-Ready cDNA from Clontech Laboratories and cDNA libraries made at deCODE genetics. cDNA libraries from whole brain, fetal brain, and testis were screened.
Exon trapping was performed using Exon trapping system (Live Technologies). Primers were designed for amplification of candidate exons from cDNA libraries, touchdown PCRs were performed, and products were verified by sequencing.
Search for SNPs
Exons were screened by direct sequencing from a PCR template. Exon sequences and sequences 2 kb upstream of each transcription start site from 184 patients were analyzed for SNP detection. Conserved regions within the 1.5-Mb NRG1 locus sequence (potential regulatory elements) showing ⩾80% mouse:human identity over ⩾100 bp were screened in 94 patients. SNPs were scored using a fluorescent-based method (Chen et al. 1999).
Mouse Phenotyping
NRG1 transmembrane-domain–knockout mice were generated using a targeting vector in which most of exon 11, which encodes the transmembrane domain, and some of the immediate downstream intron were replaced with a neomycin resistance gene cassette, preceded by an oligonucleotide carrying stop codons in each reading frame and a polyadenylation sequence. The targeting vector was electroporated into J1 embryonic stem (ES) cells (129/terSv), and founding chimeras were outcrossed to C57Bl/6 mice to establish the line. Heterozygous mice were healthy and fertile, although homozygous embryos died of cardiac defects around E10.5–E11.5. These mice will be described more fully elsewhere (R. P. Harvey, D. Lai, and M. Zhou, unpublished data). ErbB4 hypomorphic mice heterozygous for a null allele of the gene were generated by replacement of the coding region of exon 2 with a reporter gene (Gassmann et al. 1995). Heterozygous null NRG1 and ErbB4 mice were bred at Charles River Laboratories by crossing to a C57Bl/6 background. Six weeks prior to behavioral testing, male mice and litter-mate control mice for each line were shipped to the testing laboratory at PsychoGenics, where they were housed in groups of three to five related mice per cage.
The open-field study was conducted when the male mice were 5–6 mo of age. Group-housed mice were brought into the experimental room and were allowed to acclimate for 1 h prior to testing. Each mouse was placed for 30 min in a square open-field box (17 × 17 × 12 inches). Up to eight animals were tested at one time, with one animal in each of eight arenas, under low lighting conditions (provided by a 15-W red lamp). The automated infrared beam array system measured locomotion in the center and periphery of the test arena. Activity data were collected in 5-min intervals over the 30-min open-field session and were analyzed with a series of repeated-measures analysis of variance (ANOVA), with session interval as a within-subject factor and genotype as a between-subject factor. Clozapine (1 mg/kg in 1% Tween 20, pH 6.0) was injected intraperitoneally (i.p.) 25 min before behavioral testing. Total activity data from the study with clozapine were analyzed with a two-tailed Student's t test. Experimentally naive mice were used for these experiments. It seemed that handling the mice or habituation to the testing conditions changed the level of hyperactivity or the sensitivity to clozapine on repeated testing.
The cross maze consisted of an eight-arm radial arm maze with four of the arms blocked. The maze was placed on the floor of a dimly lit room (one 25-W lamp) that had contrasting cues on the wall and on the outside of the blocked arms of the maze. Each arm measured 14 inches long × 3 inches wide, and the center hub measured ∼8.5 inches in diameter. Wild-type and heterozygous mice were individually placed and tested in the cross maze for 8 min. Global activity was reflected in the total number of visits to the different arms. A visit to an arm was recorded when the body of the mouse crossed the dividing line between the hub and the arm. The number of visits to each arm and the order of arm visits were recorded. Percentages of triple and quadruple alternations were calculated, where a triple alternation was the visit of three different arms without entry into any arm already visited, and quadruple alternation was the visit to four arms without re-entry (e.g., A-B-C-D, A-D-C-B, etc., where the letters refer to the arms of the maze). The percentage was obtained by comparison of the number of triple or quadruple alternations against the number of possible triple or quadruple visits.
For assessment of PPI, group-housed mice were brought into the experimental room and were allowed to acclimate for a minimum of 1 h prior to testing. Mice were individually placed in a startle enclosure in the startle chamber with a background white noise of 70 dB and were left undisturbed for 10 min. Then a 16-min session was started that consisted of 56 trials. Each trial started with a 50-ms null period, followed by a 20-ms prepulse white noise of 72, 74, or 78 dB. After a 100-ms delay, the startle stimulus was presented (a 40-ms 120 dB white noise), followed by a 290-ms recording time. The total duration of the trial was 500 ms. Eight types of trials were given: prepulse (72, 74, or 78 dB) plus startle (10 trials per prepulse intensity), prepulse (72, 74, or 78 dB) alone (4 trials per prepulse intensity), startle alone (10 trials), and no stimulation (4 trials). The variable intertrial interval averaged 15 s (range 10–20 s). In the no-stimulation trials, baseline measurements were taken. In the startle-alone trials, the basic auditory startle was measured, and, in the prepulse-plus-startle trials, the amount of inhibition of normal startle was measured and was expressed as a percentage of the basic startle. In the prepulse-alone trials, the normal response to a weak noise was measured as a control. Data from trials in which the latency to the peak amplitude of the startle was >80 ms were excluded from the analysis. Animals with a mean startle <100 units were also excluded from the analysis.
[3H] MK-801 Binding Sites
[3H]-dizocilpine (MK-801) binding in NRG1 hypomorphic mice (Gerlai et al. 2000) and control mice was studied as follows: Wild-type (n=18), and NRG1 mutant mice (n=16) forebrains were homogenized individually at 4°C in 25 volumes of Tris-HCl 50 mM, EDTA 10 mM, pH 7.1 buffer with a polytron (10,000 rpm, 30 s). The homogenate was centrifuged at 48,000 g for 10 min, and the pellet was rehomogenized as above and was incubated for 10 min at 37°C. After centrifugation, the pellet was homogenized as above, and the homogenate was frozen at −80°C. [3H]-dizocilpine saturation isotherms were obtained by incubation of various amounts of the radioligand (0.1–100 nM, final concentration) in the presence of 10 mg brain membranes for 2 h at room temperature in a Tris-HCl 5 mM, glycine 100 μM, glutamate 100 μM, pH 7.4 binding buffer. The nonspecific binding was measured in the presence of 100 μM 1-[1-(2-thienyl)cyclohexyl]piperidine (TCP). After incubation, the membranes were filtered on GF/B glass-fiber filters preincubated for 1 h in a polyethylenimine 0.1% solution. The filters were washed three times with 3 ml of cold binding buffer, and the radioactivity bound to the membranes was measured by liquid scintillation counting. The binding parameters KD and Bmax were obtained from the fit to the data of the equation of a rectangular hyperbola (one-site model) by nonlinear regression and were analyzed by ANOVA.
Results
Schizophrenia Mapped to Chromosome 8p12-p21
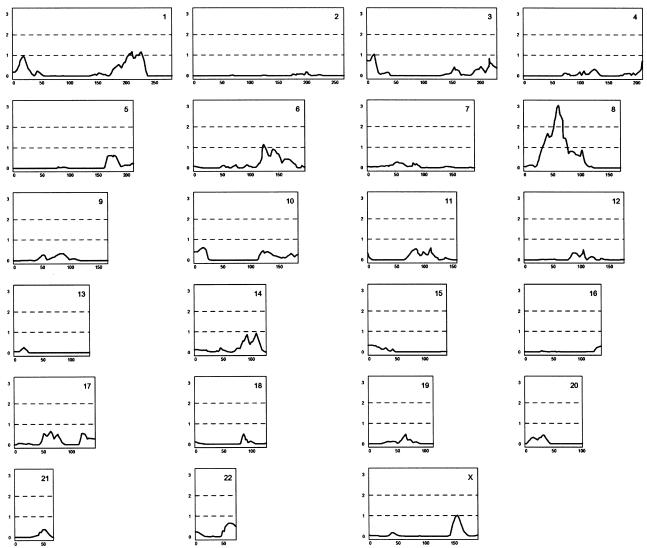
The 33 families used for linkage consist of 110 patients, 105 of whom were available for genotyping. The highest initial multipoint LOD score was 3.06, on chromosome 8p at marker D8S532 (fig. 1). Linkage to 8p has been reported by a number of groups, working with different populations (Pulver et al. 1995; Kendler et al. 1996; Levinson et al. 1996; Blouin et al. 1998; Shaw et al. 1998; Kaufmann et al. 1998; Brzustowicz et al. 1999; Gurling et al. 2001). The region we find to have the highest LOD score on 8p is 10–15 cM centromeric to most previously reported linkage to 8p. Low marker density, low resolution of linkage studies, and uncertainty with maps in many of the older studies may account for this difference. The work reported here was done utilizing a high-resolution genetic map that was not available to the other studies (Kong et al. 2002). It is, however, possible that this is a distinct locus.
Figure 1.
Genomewide scan of 110 patients with schizophrenia from 33 families. The 110 patients included 106 who fulfilled the Research Diagnostic Criteria (RDC) schizophrenia diagnosis, three patients diagnosed with unspecified functional psychosis, and one diagnosed with schizoaffective disorder. With the assistance of our genealogical database, the patients were clustered into 33 families, and relations between patients are as distant as second cousins. The patients and their relatives were genotyped using a framework set of 950 microsatellite markers. The X-axis gives the genetic distance (in cM) along the chromosome, and the Y-axis gives the LOD score. The data were analyzed using multipoint allele-sharing, without specification of an inheritance model, using Allegro (Gudbjartsson et al. 2000).
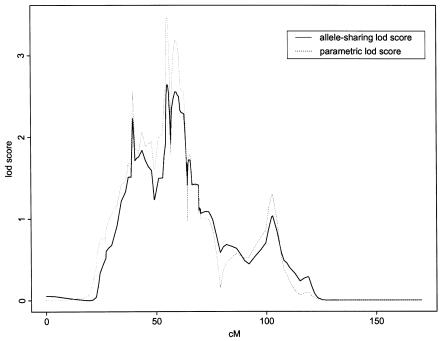
The 8p12-p21 locus was physically mapped using BACs. The primary goal with the BAC map was to achieve a high-resolution ordering (100–150 kb) of all polymorphic markers in a 30-cM region and to facilitate the search for new polymorphic markers. After adding extra markers within the linkage peak, thus increasing the information content, the peak allele-sharing LOD score for the 33 families declined from 3.06 to 2.53 at D8S278 (fig. 2). The corresponding multipoint parametric LOD score was 3.48 when an affecteds-only multiplicative model was used (i.e., an allele frequency of 0.03 and a six-fold risk, relative to the wild-type, for every at-risk allele carried). These results constitute only suggestive linkage to chromosome 8p, but, along with results of previous studies, they convinced us to search for haplotypes in this region to assess for association with schizophrenia.
Figure 2.
Fine mapping of the schizophrenia locus on chromosome 8p, with 50 additional markers mapped to the 30-cM locus. Since correct marker order and genetic distance is essential for multipoint genetic analysis, we determined the order of markers by generating a physical map of 30 cM in 8p12-p21, using BAC clones, and we determined the genetic distances on our high-resolution genetic map, on the basis of 1,200 meiotic events (Kong et al. 2002). The solid line represents the allele-sharing LOD score, and the dashed line represents the parametric LOD score. The additional markers increased the information content from 0.7 to >0.9. The X-axis gives the genetic distance (in cM) along the chromosome, and the Y-axis gives the LOD score.
Haplotype Analysis Identifies NRG1 as a Candidate Gene for Schizophrenia
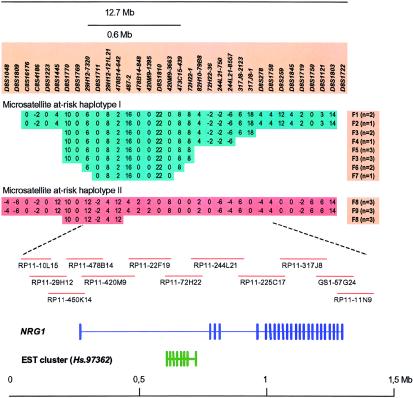
We increased the density of microsatellite markers further at the 8p locus, to achieve an average density of 1 marker every 75 kb within a 5 cM region centered on D8S278. For the linkage families, we examined all patient haplotypes shared identical by descent by multiple patients within a family, and then we compared these haplotypes across families. Two such haplotypes, each appearing in multiple families, are shown in figure 3: microsatellite haplotypes I and II. Shared haplotypes between families carrying microsatellite haplotype I pointed us to an ∼600-kb region (fig. 3). To validate the relevance of the two haplotypes identified and to further narrow down the region, 373 additional patients were collected and genotyped, in addition to the patients used in the linkage analysis. Many of these additional patients had sporadic disease, or their affected relatives were not alive, but a fraction of the patients were related. In parallel, we identified, from our physical map, BACs covering the at-risk haplotypes (fig. 3) and sequenced them by a shotgun approach, covering a total of 1.2 Mb, to search for genes within the region (GenBank AF491780). Within the boundaries of the at-risk microsatellite haplotypes, we found a 5′ exon from the NRG1 gene and an EST cluster with unknown function, Hs.97362 (fig. 3). We have submitted the complete sequence of the large NRG1 gene to GenBank (accession number TPA BK000383).
Figure 3.
Microsatellite at-risk haplotypes on chromosome 8p and known genes within a region showing haplotype sharing between 9 of the 33 linkage families. Extensive sharing of two microsatellite haplotypes between patients from the linkage families is shown at the top. Haplotypes were reconstructed by application of the Allegro program (Gudbjartsson et al. 2000), using available family members to derive the phase. Key markers in the haplotypes are shown, and the size of the region is indicated. Families carrying the haplotypes are labeled F1–F9, and the number of affected individuals in each family carrying that haplotype is given in parentheses. Maximum haplotype sharing between families is 9.5 Mb for haplotype I and 11.4 Mb for haplotype II. Shared haplotypes between families narrow the region of interest to 600 kb between markers 29H12-7320 and 473C15-439, indicated by a bar (microsatellite haplotype I). The location of a BAC contig covering 1.5 Mb of the locus region is indicated. The sequence of GS1-57G24 was obtained from the public domain (GenBank accession number AF128834), but we sequenced the other BACs shown. The positions of NRG1 and an EST cluster of unknown function (Hs.97362) are schematically shown in relation to the BACs. Exons are indicated by vertical bars.
Furthermore, we sequenced the syntenic mouse locus, 1.2 Mb in size, to help identify exons and regulatory elements. In figure 3, the exon-intron structure of the two genes is shown schematically, together with the overlapping at-risk microsatellite haplotype I. In the search for novel exons and genes, we used RT-PCR, 3′ and 5′ RACE, and exon trapping from BACs covering the NRG1 gene and 200 kb of the upstream sequence. Novel exons and splice variants were identified for both genes (H. Stefansson, V. Steinthorsdottir, J. R. Gulcher, and K. Stefansson, unpublished data).
In an attempt to identify causal alleles, we have screened all known and novel exons of NRG1 (n=25) and EST cluster Hs.97362 (n=8) and have identified 15 nonsynonymous SNPs for NRG1 and 3 in the EST cluster Hs.97362. We have identified two synonymous SNPs and seven SNPs in the untranslated part of NRG1 and one synonymous SNP and four SNPs in untranslated regions of EST cluster Hs.97362. A total of >1,200 SNPs have been identified in the entire NRG1 sequence (see deCODE Genetics Web site). All coding SNPs and a number of SNPs in promoter regions were genotyped for 394 unrelated control individuals and 478 patients. Furthermore, a number of SNPs identified in conserved regions were also scored. A total of 58 SNPs were genotyped for all patients, and, in a subset of patients (n=94) and control individuals (n=124), an additional 123 SNPs were scored for association. A few SNP alleles showed mild but significant single-marker associations, but they do not change amino acids or splice sites.
Haplotype analysis was performed as described in the “Subjects and Methods” section. Unless otherwise stated, P values reported for association tests are one-sided, without adjustment for multiple comparisons, and are based on the EM algorithm. Also, the likelihoods are computed treating all affected individuals as independent. If the affected list includes related individuals, the estimates of haplotype frequencies are still valid, whereas the P values calculated would be slightly anticonservative. We indicate all such P values with an asterisk (*). From the list of all affected individuals, a smaller list of independent affected individuals is created, blind to the genotypes, by eliminating first- and second-degree relatives. Although the P values computed from such a reduced list is valid, the frequency of a haplotype that is shared by many affected relatives tends to be underestimated. We have also used other methods, including the Allegro program (Gudbjartsson et al. 2000), to construct the most likely haplotypes on the basis of patients and relatives, with comparable results.
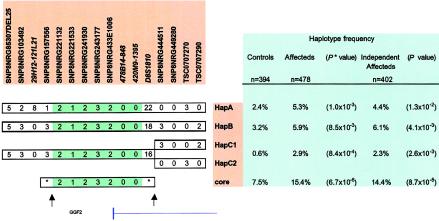
No single SNP marker or microsatellite marker was very statistically significant or had very high estimated relative risk. The SNP giving the best uncorrected single-marker association was SNP8NRG221533 (P=.003). However, a core haplotype consisting of five SNPs (including SNP8NRG221533) and two microsatellite markers was identified (P value between 6.7 × 10−6* and 8.7 × 10−5) (fig. 4). Four of the SNP markers are upstream of the NRG1 gene, but the fifth one (SNP8NRG433E1006) is coding, changing an arginine to a glycine in the 5′ exon of glial growth factor 2 (GGF2), which is one of many splice forms of NRG1. However, the common variant, which is glycine (found in 88% allelic frequency in case individuals and 85% in control individuals), is associating with the core at-risk haplotype and therefore not likely by itself to be the causative allele. Information on sequence of SNPs and microsatellites within the NRG1 sequence can be found at the deCODE Genetics Web site. The core haplotype covers 290 kb and represents a block of linkage disequilibrium containing the first 5′ exon of NRG1, encoding for the amino terminus of GGF2, and upstream sequences. Close to 90% of this core haplotype can be accounted for by four extended haplotypes involving 16 markers (fig. 4). Interestingly, whereas HapA in figure 4 corresponds to the extended microsatellite haplotype I in figure 3 identified in the linkage families, HapB and HapC (1 and 2), apart from having common alleles for microsatellite markers 478B14-848 and 420M9-1395, tend to have different alleles for many of the microsatellite markers (not all shown in fig. 4). Indeed, at various intermediate stages before we realized that they share a core, HapA, HapB, and HapC (1 and 2) were each considered as independent at-risk haplotypes showing significant excess in the patients relative to the control individuals (fig. 4). The estimates of their risks, relative to the wild type, are also comparable for HapA and HapB (relative risk ∼2) and are higher for HapC, but the estimate for HapC is based on small numbers (fig. 4). Hence, we believe that this core haplotype is capturing an ancestral at-risk haplotype that is represented by a number of extended microsatellite/SNP haplotypes in the current population. For the seven markers, that define the core at-risk haplotype in figure 4, there are only 15 distinct haplotypes with frequencies > 1% (frequencies estimated under the null hypothesis with the data from the patients and controls combined), together accounting for 56% of the haplotypes in the population. There are 65 distinct haplotypes with estimated frequencies >0.1%, together accounting for 94% of haplotypes.
Figure 4.
Microsatellite and SNP at-rick haplotypes at the 5′ end of the NRG1 gene. Four haplotypes, defined by 12 SNPs and four microsatellite markers, were individually found in excess in the schizophrenia patients with similar relative risk. The common core for these haplotypes, defined by five SNPs and two microsatellite markers, is shown at the bottom. The frequencies for each haplotype in all affected individuals, independent affected individuals, and control individuals are indicated in the panel on the right. The only known exon within the boundaries of the core at-risk haplotype is the 5′ exon of GGF2, as indicated in the figure. The distance between the markers flanking possible recombination breakpoints (arrows) is 290 kb, corresponding to 0.16–0.45 Mb on the scale at the bottom of figure 3.
Figure 4 shows the likely locations of historic recombination breakpoints and also reveals that the microsatellite marker D8S1810 has probably mutated since the at-risk SNP haplotype was formed. Haplotypes derived using information from relatives of patients and controls agreed with these haplotypes derived using the likelihood approach.
The core at-risk haplotype has an estimated frequency of 7.5% in the general population and of 15.4% among all patients with schizophrenia. When a multiplicative model is assumed, affected individuals are estimated to have 2.2 times the risk of the wild type for each at-risk haplotype evaluated. To supplement the results from the case-control study, we performed the transmission/disequilibrium test (Spielman et al. 1993) and found that, for parents who were heterozygous with respect to the core haplotype, there were 33 transmissions to the affected offspring and 17 nontransmissions (P=.016). Although these results are only marginally significant, because of the small sample size (parents were not available for genotyping in many cases), it is heartening that the ratio of transmissions to nontransmissions is close to 2:1, which is consistent with the relative risk of 2.2 estimated on the basis of the case-control data.
It is worth noting that the region of interest exhibits extensive linkage disequilibrium. The core haplotype of seven markers can be identified by only three markers, one SNP and two microsatellites. Specifically, if a haplotype includes alleles 1, 0, and 0 for SNP8NRG221533, 478B14-848, and 420M9-1395, respectively, then there is little uncertainty that it has the corresponding alleles for the other four markers. Moreover, HapA, HapB, and HapC in figure 4 each can be captured by only five markers, the three identifying the core plus microsatellite markers 29H12-121L21 and D8S1810. Also, HapA is a good surrogate for the extended microsatellite haplotype I in figure 3. The core at-risk haplotype does not overlap with EST cluster Hs.97362, suggesting that NRG1 is the more likely candidate gene in this region. The EST cluster is 103 kb centromeric to marker TSC0707290 shown in figure 4. None of the five SNPs defining the core haplotype is likely to be the causative SNP, since they do not individually capture the same degree of association as the core haplotype, they are, therefore, more likely in linkage disequilibrium with a causative allele, within the boundaries of the core at-risk haplotype. Although we have screened 20 kb of the 290-kb core at-risk haplotype for SNPs, including the known exon and regulatory elements within the core, there may remain regulatory elements we have not identified, containing a functional variant that may affect transcription, RNA splicing, RNA stability, RNA transport, or translation. Alternatively, the functional variant may include more than one polymorphic site, and a haplotype may be necessary to account for the susceptibility, as has been suggested for other complex diseases (Drysdale et al. 2000; Horikawa et al. 2000). Finally, this may represent large or small inversions, deletions, or duplications that have not been uncovered by our mutational analysis so far.
Microsatellite haplotype II (fig. 3) was found in substantially higher frequency in patients from the linkage families than in control individuals (data not shown). The markers identifying this haplotype overlap with those of the core haplotype shown in figure 4, but the alleles are different. This haplotype is rare in control individuals and in the patients who were not used in the linkage analysis. Hence, we can currently neither confirm nor reject this haplotype as conferring increased risk.
On the basis of the estimated frequencies and relative risks reported above, the core haplotype has a population attributed risk of 16% when the multiplicative model is used. It accounts for a 9% increase in risk for siblings of an affected individual. Hence, its contribution to the familial risk of schizophrenia, which has been reported to have λs close to 8.6 (Risch 1990), is small and cannot fully explain the linkage results we and others obtain for this region. Although part of the reason could be that there are other schizophrenia susceptibility genes in the 8p region contributing to the LOD scores and results from linkage analyses have a tendency to overestimate the contribution of the gene (Goring et al. 2001), we believe there must be other at-risk alleles/haplotypes of NRG1, probably rarer but possibly with higher penetrance, yet to be found (e.g., microsatellite haplotype II). Hence, the overall contribution of NRG1 to schizophrenia may be substantially higher than the estimate reported here.
NRG1 and ErbB4 Mutant Mice Display Behavioral Abnormalities
Schizophrenia is one of the diseases affecting higher cortical functions for which it has been difficult to develop an animal model that comes close to the human disease. Indeed, many of the clinical manifestations of schizophrenia affect mental processes that draw a line of distinction between humans and other animals. Conventional antipsychotic drugs can be evaluated in rodents, using behavioral assays sensitive to dopaminergic tone (such as locomotion), yet these have no clear correspondence in the schizophrenic patient. The rank order of potency of such drugs shows a relatively good correlation between inhibition of dopamine-receptor binding and clinical benefit (Creese et al. 1976). In addition to altering dopaminergic function, the newer class of atypical antipsychotic drugs (e.g., clozapine) have potent effects on other neurotransmitter systems, including serotonin.
NRG1 and its receptors, ErbB2, ErbB3, and ErbB4, are essential genes for development (Gassmann et al. 1995). Homozygous null embryos die at 10.5–11 d of gestation or soon after birth. Heterozygous null mice, in contrast, are viable and live to adulthood (Gerlai et al. 2000). NRG1 hypomorphic mice show hyperactivity in a number of tests, including the novel open field and alternating-Y maze, whereas ErbB2 and ErbB3 heterozygous null mice are behaviorally normal. The NRG1 mutant mice used in these studies were generated by a disruption of the gene targeted so that the epidermal growth factor (EGF)–like domain is missing (Erickson et al. 1997). To replicate the abnormal behavioral results of the reported NRG1 mutant mouse, we carried out behavioral studies on a second line of NRG1 heterozygous mice (in which the targeted deletion was of exon 11, encoding the NRG1 transmembrane domain [R. P. Harvey, D. Lai, and M. Zhou, unpublished data]). We also studied a line of ErbB4 heterozygous null mice (Gassmann et al. 1995). Quantitation of NRG1 and ErbB4 mRNA in hippocampus, performed using RT-PCR, confirmed that 50% reduction of message from the targeted genes was achieved in both lines of mice.
The NRG1 and ErbB4 heterozygous null mice bred well, developed normally, and showed generally normal behavior. The lines were maintained by back crossing onto C57Bl/6 but were not congenic. When evaluated in the novel open-field test performed under dim red light, both types of mice were significantly more active than their wild-type litter-mate controls, with whom they had been housed since weaning (figs. 5a and 5b). Because mice are neophobic and find open spaces aversive, normal mice will prefer to stay close to the walls (thigmotaxis), and, indeed, there was no difference between NRG1 or ErbB4 hypomorphs and their normal litter-mate controls in measures of anxiety, such as time spent in the center of the test arena. Hyperactivity in the novel open-field test was more robust in the NRG1 hypomorphs than in the ErbB4 hypomorphs. Hyperactivity of the NRG1 hypomorphs also was seen in a four-arm–cross maze test. This is an ethologically based test that assesses exploratory activity in a novel environment but does not involve reward delivery. The total number of entries into one of the four arms of the maze was increased (37 ± 1.7 vs. 30 ± 1.6, F-test; P=.01). The mice were otherwise behaviorally normal, with no difference between genotypes in patterns of quadruple or triple alternations or clockwise or counterclockwise patterns of entry. That the hyperactivity was more robust in the NRG1 hypomorphs than in the ErbB4 mutant mice may indicate that the ligand is limiting in the NRG1/ErbB4 signaling pathway.
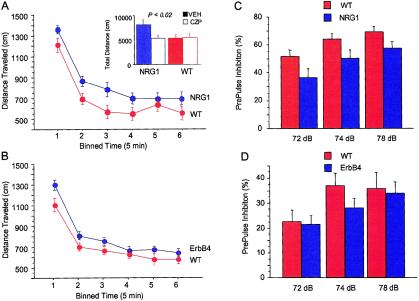
Figure 5.
Locomotion and PPI in NRG1 and ErbB4 hypomorphic mice. NRG1 and ErbB4 hypomorphic mice were significantly more active, according to different measures that reflect locomotion and exploration. Here, we show distance traveled in a novel open-field test for the two lines of mice: NRG1 hypomorphic mice (A) and ErbB4 hypomorphic mice (B). Data have been binned into 5-min intervals over the 30-min observation period. Distance traveled was significantly increased in both NRG1 and ErbB4 male mice in comparison to litter-mate control mice (n=21 NRG1 mice and 22 litter-mate control mice, P=.04; n=39 ErbB4 mice and 22 litter-mate control mice, P=.03). Open-field activity was monitored as described. Error bars indicate the SEM. A, insert, hyperactivity of the NRG1 mutant mice was reversed by clozapine. The NRG1 mice and litter-mate control mice were injected i.p. with either clozapine (1 mg/kg) or vehicle, 25 min prior to testing (n=10 NRG1 mice and 10 control mice, P=.02). Open-field activity was monitored as described in the “Subjects and Methods” section. PPI was recorded for NRG1 (C) and ErbB4 (D) hypomorphic mice, using a conditioning, prepulse noise burst of 72, 74, or 78 dB. NRG1 hypomorphic mice have impaired PPI in comparison to normal, litter-mate control mice (F-test, P=.03), whereas ErbB4 mutant mice were not significantly impaired (F-test, P=.056).
Hyperactivity is seen in a number of mutant mice, particularly those showing an increase in dopaminergic tone, such as the dopamine transporter (DAT) null mice (Glickstein and Schmauss 2001), the muscarinic acetylcholine–receptor M1 subtype null mice (Gerber et al. 2001), and N-methyl-D-aspartatereceptor subunit NR1 hypomorphic mice (NR1 mice) that express only 10% of the normal level of NR1 (Mohn et al. 1999). In both the DAT and NR1 mutants, the hyperactivity is reversible with antipsychotic drugs. Drugs such as clozapine are sedating at high doses, but, at doses of ⩽1 mg/kg, they will reverse the hyperactivity in mutant or pharmacologic models of schizophrenia but will have no effect on spontaneous activity (O'Neill and Shaw 1999). To explore reversibility of the hyperactivity of the NRG1 hypomorphs in the two behavioral assays, we tested the effect of clozapine at a dose of 1 mg/kg on the NRG1 hypomorphs and their normal litter-mate controls. Clozapine at the dose chosen reversed the increased activity of the NRG1 hypomorphic mice in the novel open-field test (t-test, P<.02) (see insert in fig. 5a). Clozapine had no effect on the spontaneous activity of normal mice in the novel open field and thus was not sedating at that dose. In the cross maze test, clozapine at 1 mg/kg also reversed the hyperactivity of the NRG1 hypomorphs. The total number of entries by the NRG1 hypomorphs was reduced to baseline in the clozapine-treated animals (from 36.9 ± 4 in vehicle-treated animals to 27 ± 2 in clozapine-treated animals; P=.03), whereas clozapine had no significant effect on the spontaneous activity of normal litter-mate control mice in the cross-maze test.
PPI
Although locomotory assays in rodents may have validity as pharmacologic models for the evaluation of antipsychotic drugs affecting dopaminergic tone, their direct clinical correlate in the schizophrenic patient is unclear. On the other hand, a number of studies indicate that some schizophrenic patients have impaired PPI. This is a psychometric measure of sensory gating that can be evaluated in similar fashion in rodents and humans (Braff and Geyer 1990). We find that PPI is impaired in the NRG1 hypomorphic mice and less so in the ErbB4 mutant mice. The auditory/perceptual systems of NRG1 and ErbB4 hypomorphic mice are generally intact, since both show normal acoustic startle responses to a single noise burst of 120 dB. With respect to PPI, however, NRG1 hypomorphic mice were impaired in comparison to normal litter-mate controls (F-test, P=.03) (fig. 5c). However, the ErbB4 hypomorphs did not show significant difference in PPI (F-test, P=.056) (fig. 5d). The defect in PPI in NRG1 hypomorphic mice was not reversed by clozapine at a dose of 1 mg/kg, although these studies need to be expanded.
16% Fewer Functional NMDA Receptors in the NRG1 Mutant Mice
We performed MK-801 binding studies of forebrain homogenates from NRG1 hypomorphs generated by Gerlai et al. (2000) and control mice. The pKD values were analyzed by ANOVA and did not reveal any differences between the wild-type and the mutant mice. Bmax values (pmoles/mg protein) in the mutants were found to differ significantly (one-sided P=.0068) from those in the wild-type mice, suggesting that there are 16% fewer functional NMDA receptors in the mutant animals overall (table 1). This is in keeping with reports suggesting a role for NRG1 in regulation of NMDA subunit expression (Ozaki et al. 1997).
Table 1.
[3H] MK801 Binding[Note]
| Bmax values(moles/mg protein) |
||||
| Mouse | Count | Mean | SD | P |
| NRG1 knockout | 16 | 1.23 × 10−12 | 2.22 × 10−13 | 6.8 × 10−3 |
| Wild type | 18 | 1.43 × 10−12 | 2.14 × 10−13 | |
Note.— Concentrations of [3H] MK801 in homogenates from forebrains of NRG1 knockout and wild-type mice. One-sided P value is given.
It is also of interest here that there appears to be reduction in the numbers of functional NMDA receptors in certain regions of brains from schizophrenic individuals (Ibrahim et al. 2000; Goff and Coyle 2001). We feel, however, that we have to caution that this does not necessarily mean that the principal pathogenic alteration in schizophrenia lies in the glutamate system.
Discussion
NRG1 Provides a Way of Unifying a Large Body of Conflicting Evidence
Through the work described above, we have identified the NRG1 gene as a strong candidate for a gene playing a role in the pathogenesis of schizophrenia. We present three lines of evidence in support of this role for NRG1.
The first is genetic evidence, consisting of suggestive linkage of schizophrenia to chromosome 8p in Icelandic families, supported by suggestive linkage in other populations (Pulver et al. 1995; Kendler et al. 1996; Levinson et al. 1996; Blouin et al. 1998; Shaw et al. 1998; Kaufmann et al. 1998; Brzustowicz et al. 1999; Gurling et al. 2001). This evidence is further supported by highly significant association of overlapping haplotypes that contain only one known gene within the overlap, namely NRG1. The population attributed risk for the identified core haplotype is 16%, which is a substantial contribution to the public health burden. The weakness in the genetic evidence is that we have not yet found a clear pathogenic mutation, which may, however, be par for the course in the genetics of common diseases.
The second line of evidence is that mice hypomorphic for each of two mutations in NRG1 and one mutation in a receptor for NRG1 display behavior that overlaps, in part, with mouse models for schizophrenia, and this is reversed, in part, with clozapine in a NRG1 mutant line.
The third line of evidence is that the number of NMDA receptors in the NRG1 hypomorphs is reduced which is in keeping with observations made on brains from schizophrenia patients. Thus, these results argue that variants of the NRG1 gene contribute to the pathogenesis of schizophrenia in some patients, probably through a decrease in NRG1 signaling. The overlap in behavioral phenotype between the NRG1 and ErbB4 hypomorphic mice, and the lack of a similar behavioral phenotype in ErbB2 or ErbB3 mice (Gerlai et al. 2000), argues that the defect is primarily neuronal. Although each line of evidence is not conclusive, they constitute, when put together, a strong case for NRG1 as a culprit in the pathogenesis of schizophrenia. Furthermore, the NRG1 knockout mice provide a tool to explore this hypothesis further, on the basis of our understanding of the genetics of the disease.
The NRG1 gene was identified as a schizophrenia gene by using a combination of a linkage and association approaches, based on microsatellite markers and then SNPs once microsatellite at-risk haplotypes were identified. Although SNP haplotypes are in general more stable and may capture the ancestral haplotype better than the overriding microsatellites, at-risk haplotypes in our study would not have been identified in a systematic manner without using the more cost-effective and informative microsatellite markers. In fact, our core at-risk haplotype is best represented by a combination of two stable microsatellites and one SNP.
Despite finding >1,200 SNPs in the NRG1 gene and scoring 181 for association, we failed to find a clear functional polymorphism that captures the same degree of association as the schizophrenia at-risk core haplotype. SNPs screened were found within exons, promoters, splice sites, and regions of the gene most homologous between mouse and human. Our core at-risk haplotype represents a block of linkage disequilibrium that can be represented by just a few SNP and microsatellite markers. We continue to screen the large introns and flanking regions of the gene for associated variations, especially within the regions most conserved with the mouse.
NRG1 has not previously been considered in the context of schizophrenia. However, it is a fascinating candidate gene for the disease. A unique strength of the hypothesis that a defect in neuronal NRG1 signaling underlies some cases of schizophrenia is that it brings together the disparate clinical and pathological features of this disease into a common pathway. NRG1 isoforms (also called “ARIA,” “GGF2,” “NDF,” “SMDF,” and “heregulin,” depending on the isoform and tissue in which they were first identified) are a group of proteins that arise from alternative splicing of a single primary transcript (Fischbach and Rosen 1997; Wang et al. 2001). NRG1 isoforms are expressed in many tissues, including the CNS, and these isoforms clearly have a developmental role, as indicated by knockout mice displaying severe developmental anomalies in the heart and the nervous system (Liu et al. 1998; Gerlai et al. 2000). An NRG1 isoform originally discovered on the presynaptic membrane of the motor neuron (as ARIA) was found to be the factor inducing expression and localization of acetylcholine receptors (AchR) at the neuromuscular synapse. NRG1 isoforms influence gliogenesis and neuronal migration during development of the brain, and, in the adult nervous system, NRG1 appears to have a marked impact on the expression and activation of several neurotransmitter receptors, including NMDA glutamate receptors (Ozaki et al. 1997; Ibrahim et al. 2000), possibly Ca2+-activated K+ channels (Chu et al. 1995; Cameron et al. 2001), and it also facilitates neurotransmitter release from GABAergic interneurons.
The transmembrane forms of NRG1 are present within synaptic vesicles, including those containing glutamate. After exocytosis, NRG1 is in the presynaptic membrane, where the ectodomain of NRG1 may be cleaved off. The ectodomain then migrates across the synaptic cleft and binds to and activates a member of the EGF-receptor family on the postsynaptic membrane. This has been shown to increase the expression of certain glutamate-receptor subunits. In addition, ErbB4, the predominant receptor for NRG1 on CNS neurons, is colocalized with NMDA receptors in the postsynaptic density 95 complex (Garcia et al. 2000; Huang et al. 2000). In the CNS, the NMDA subunits, NR2A and NR2B, are heavily phosphorylated at tyrosine residues (Lau and Huganir 1995), and this posttranslational modification enhances receptor activity by altering the kinetic properties of the channel (Lau and Huganir 1995). Garcia et al. (2000) pointed out the possibility that the activity-dependent activation of ErbB4 receptors by NRG1 may regulate synaptic plasticity by recruiting tyrosine kinases that regulate NMDA receptor function. Thus, NRG1 appears to signal for glutamate-receptor subunit expression, localization, and /or phosphorylation facilitating subsequent glutamate transmission. Therefore, the activity-dependent effects of NRG1 and ErbB4 on glutamatergic transmission efficiency may be one mechanism of synaptic plasticity. Abnormalities in synaptic plasticity may lead to the abnormalities in thought processes and cognition seen in schizophrenia that is out of proportion to the subtle decrease in glutamate receptors.
There are many pharmacologic and mouse mutant data that support the glutamate dysfunction hypothesis, and glutamate-receptor expression and binding may be decreased in schizophrenia patients (Huang et al. 2000). However, studies looking for variations in receptor subunits have not shown clear association to schizophrenia. Hypofunction of NRG1 in schizophrenia, could explain, in part, the apparent deficiency in glutamate-receptor expression and binding described within in some parts of brains of schizophrenia patients (Huang et al. 2000). NRG1 has additional roles in AchR expression. Abnormalities in PPI, the sensory gating of which may be mediated by nicotinic AchR in the hippocampus (Freedman et al. 1995; Holt et al. 1999) as well as the high rate of nicotine addiction in schizophrenic patients could be secondary to NRG1 abnormalities. Thus, it is also possible that the NRG1 abnormality causes schizophrenia through a mechanism in which the glutamate system plays a minor role.
Our behavioral data on the NRG1 and ErbB4 mouse mutants provide additional evidence for a role of NRG1 in schizophrenia. We replicate the work done by Gerlai et al. (2000) on a different NRG1 mutant and we show that both the NRG1 and the ErbB4 hypomorph mice are hyperactive, a phenotype that overlaps with behaviors induced by PCP in normal mice. Clozapine reduced the hyperactivity in the NRG1 mice as it does in PCP-treated mice and mice with reduced numbers of NMDAR1 (NR1)–receptor subunits (Mohn et al. 1999). The clozapine reversal, therefore, further supports the possibility that the hyperactivity observed in the NRG1 mice is related to schizophrenic phenotypes. In addition, the NRG1 mice showed a defect in PPI, a psychomotor defect seen in some schizophrenic patients that can be evaluated in animals in a similar fashion.
Heterozygous ErbB2 and ErbB3 knockouts do not exhibit behavior abnormalities (Gerlai et al. 2000), but ErbB4 is the predominant neuronal receptor for NRG1 in the CNS (Steiner et al. 1999). It is not clear, at this point, how NRG1 contributes to schizophrenia susceptibility, and it is likely that additional factors, such as environmental stress or variants of interacting genes, may be required to unmask the disease. Nevertheless, the availability of NRG1 and ErbB4 hypomorphic mice offers new opportunities for exploration of genetic and environmental interactions that may contribute to the pathogenesis of schizophrenia and for the development of novel therapies.
In summary, we have presented linkage, case-control haplotype-association, and TDT analyses that support the notion that NRG1 contributes to schizophrenia. In addition, we present behavioral and pharmacological data on mouse mutants of NRG1 and ErbB4, as well as glutamate-receptor–binding data supporting its proposed role in schizophrenia.
If NRG1 is a schizophrenia susceptibility gene, its impact is unlikely to be limited to the Icelandic population, since schizophrenia has been linked to 8p in other populations. NRG1 provides a way of unifying a large body of evidence coming from many directions, suggesting that multiple neurotransmitter systems and their receptors are involved in schizophrenia by representing a common denominator upstream of neurotransmitter expression and activation. Furthermore, it may provide support for the view that schizophrenia is caused by disregulation of synaptic plasticity in the adult.
Acknowledgments
We thank the participating patients and their families, and we thank Hjördís Pálsdóttir, Hallbera Leifsdóttir, Þuríður Þórðardóttir, Guðrun Jóhannesdóttir, and Tómas Þór Ágústsson for assisting with the sample collection.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Children's Hospital Oakland, BACPAC Resources, http://www.chori.org/bacpac/ (for RCPI-11 human BAC library)
- deCODE Genetics, http://www.decode.com/nrg1/markers/ (for SNPs and microsatellite markers in the NRG1 locus sequence)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for NRG1 [AF491780 and TPA BK000383])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NRG1 [MIM 142445], schizophrenia [MIM 181500], and SCZD6 [MIM 603013])
- Phred/Phrap/Consed home page, http://bozeman.mbt.washington.edu/index.html
- Polyphred, http://www.codoncode.com/polyphred/
References
- Bilder RM, Corcoran R, Frith CD (1996) Neuropsychology and neurophysiology in schizophrenia. Curr Opin Psychiatry 9:57–62 [Google Scholar]
- Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, et al (1998) Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet 20:70–73 [DOI] [PubMed] [Google Scholar]
- Braff DL, Geyer MA (1990) Sensorimotor gating and schizophrenia: human and animal model studies. Arch Gen Psychiatry 47:181–188 [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Honer WG, Chow EW, Little D, Hogan J, Hodgkinson K, Bassett AS (1999) Linkage of familial schizophrenia to chromosome 13q32. Am J Hum Genet 65:1096–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JS, Dryer L, Dryer SE (2001) β-Neuregulin-1 is required for the in vivo development of functional Ca2+-activated K+ channels in parasympathetic neurons. Proc Natl Acad Sci USA 98:2832–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardno AG, Marshall EJ, Coid B, Macdonald AM, Ribchester TR, Davies NJ, Venturi P, Jones LA, Lewis SW, Sham PC, Gottesman II, Farmer AE, McGuffin P, Reveley AM, Murray RM (1999) Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Arch Gen Psychiatry 56:162–168 [DOI] [PubMed] [Google Scholar]
- Carlsson M, Carlsson A (1990) Interactions between glutamatergic and monoaminergic systems within the basal ganglia—implications for schizophrenia and Parkinson's disease. Trends Neurosci 13:272–276 [DOI] [PubMed] [Google Scholar]
- Chen X, Levine L, Kwok PY (1999) Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res 9:492–498 [PMC free article] [PubMed] [Google Scholar]
- Chu GC, Moscoso LM, Sliwkowski MX, Merlie JP (1995) Regulation of the acetylcholine receptor subunit gene by recombinant ARIA: an in vitro model for transynaptic gene regulation. Neuron 14:329–339 [DOI] [PubMed] [Google Scholar]
- Clayton D, Jones H (1999) Transmission/disequilibrium tests for extended marker haplotypes. Am J Hum Genet 65:1161–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creese I, Burt DR, Snyder SH (1976) Dopamine receptor binding predicts clinical and pharmacological potencies of antischizophrenic drugs. Science 192:481–483 [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc B 39:1–38 [Google Scholar]
- Drysdale CM, McGraw DW, Stack CB, Stephens JC, Judson RS, Nandabalan K, Arnold K, Ruano G, Liggett SB (2000) Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci USA 97:10483–10488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson SL, O'Shea KS, Ghaboosi N, Loverro L, Frantz G, Bauer M, Lu LH, Moore MW (1997) ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2-and heregulin-deficient mice. Development 124:4999–5011 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12:921–927 [DOI] [PubMed] [Google Scholar]
- Fischbach GD, Rosen KM (1997) ARIA: a neuromuscular junction neuregulin. Annu Rev Neurosci 20:429–458 [DOI] [PubMed] [Google Scholar]
- Freedman R, Hall M, Adler LE, Leonard S (1995) Evidence in postmortem brain tissue for decreased numbers of hippocampal nicotinic receptors in schizophrenia. Biol Psychiatry 38:22–33 [DOI] [PubMed] [Google Scholar]
- Gao XM, Sakai K, Roberts RC, Conley RR, Dean B, Tamminga CA (2000) Ionotropic glutamate receptors and expression of N-methyl-D-aspartate receptor subunits in subregions of human hippocampus: effects of schizophrenia. Am J Psychiatry 157:1141–1149 [DOI] [PubMed] [Google Scholar]
- Garcia RA, Vasudevan K, Buonanno A (2000) The neuregulin receptor ErbB-4 interacts with PDZ-containing proteins at neuronal synapses. Proc Natl Acad Sci USA 97:3596–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, Lemke G (1995) Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 378:390–394 [DOI] [PubMed] [Google Scholar]
- Gerber DJ, Sotnikova TD, Gainetdinov RR, Huang SY, Caron MG, Tonegawa S (2001) Hyperactivity, elevated dopaminergic transmission, and response to amphetamine in M1 muscarinic acetylcholine receptor-deficient mice. Proc Natl Acad Sci USA 98:15312–15317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlai R, Pisacane P, Erickson S (2000) Heregulin, but not ErbB2 or ErbB3, heterozygous mutant mice exhibit hyperactivity in multiple behavioral tasks. Behav Brain Res 109:219–227 [DOI] [PubMed] [Google Scholar]
- Glickstein SB, Schmauss C (2001) Dopamine receptor functions: lessions from knockout mice. Pharmacol Ther 91:63–83 [DOI] [PubMed] [Google Scholar]
- Goff DC, Coyle JT (2001) The emerging role of glutamate in the pathophysiology and treatment of schizophrenia. Am J Psychiatry 158:1367–1377 [DOI] [PubMed] [Google Scholar]
- Goring HH, Terwilliger JD, Blangero J (2001) Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet 69:1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretarsdottir S, Sveinbjornsdottir S, Jonsson HH, Jakobsson F, Einarsdottir E, Agnarsson U, Shkolny D, et al (2002) Localization of a susceptibility gene for common forms of stroke to 5q12. Am J Hum Genet 70:593–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 [DOI] [PubMed] [Google Scholar]
- Gulcher JR, Kristjansson K, Gudbjartsson H, Stefansson K (2000) Protection of privacy by third-party encryption in genetic research in Iceland. Eur J Hum Genet 8:739–742 [DOI] [PubMed] [Google Scholar]
- Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, Petursson H, Curtis D (2001) Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet 68:661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley M, Kidd K (1995) HAPLO: a program using the EM algorithm to estimate the frequencies of multi-site haplotypes. J Hered 86:409–411 [DOI] [PubMed] [Google Scholar]
- Holt DJ, Herman MM, Hyde TM, Kleinman JE, Sinton CM, German DC, Hersh LB, Graybiel AM, Saper CB (1999) Evidence for a deficit in cholinergic interneurons in the striatum in schizophrenia. Neuroscience 94:21–31 [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, et al (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nature Genet 26:163–175 [DOI] [PubMed] [Google Scholar]
- Huang YZ, Won S, Ali DW, Wang Q, Tanowitz M, Du QS, Pelkey KA, Yang DJ, Xiong WC, Salter MW, Mei L (2000) Regulation of neuregulin signaling by PSD-95 interacting with ErbB4 at CNS synapses. Neuron 26:443–455 [DOI] [PubMed] [Google Scholar]
- Ibrahim HM, Hogg AJ Jr, Healy DJ, Haroutunian V, Davis KL, Meador-Woodruff JH (2000) Ionotropic glutamate receptor binding and subunit mRNA expression in thalamic nuclei in schizophrenia. Am J Psychiatry 157:1811–1823 [DOI] [PubMed] [Google Scholar]
- Kaufmann CA, Suarez B, Malaspina D, Pepple J, Svrakic D, Markel PD, Meyer J, Zambuto CT, Schmitt K, Matise TC, Harkavy Friedman JM, Hampe C, Lee H, Shore D, Wynne D, Faraone SV, Tsuang MT, Cloninger CR (1998) NIMH Genetics Initiative Millenium Schizophrenia Consortium: linkage analysis of African-American pedigrees. Am J Med Genet 81:282–289 [PubMed] [Google Scholar]
- Kendler KS, MacLean CJ, O'Neill FA, Burke J, Murphy B, Duke F, Shinkwin R, Easter SM, Webb BT, Zhang J, Walsh D, Straub RE (1996) Evidence for a schizophrenia vulnerability locus on chromosome 8p in the Irish Study of High-Density Schizophrenia Families. Am J Psychiatry 153:1534–1540 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander, ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lau LF, Huganir RL (1995) Differential tyrosine phosphorylation of N-methyl-D-aspartate receptor subunits. J Biol Chem 270:20036–20041 [DOI] [PubMed] [Google Scholar]
- Levinson DF, Wildenauer DB, Schwab SG, Albus M, Hallmayer J, Lerer B, Maier W, et al (1996) Additional support for schizophrenia linkage on chromosomes 6 and 8: a multicenter study. Am J Med Genet 67:580–594 [DOI] [PubMed] [Google Scholar]
- Liddle PF, Carpenter WT, Crow T (1994) Syndromes of schizophrenia: classic literature. Br J Psychiatry 165:721–727 [DOI] [PubMed] [Google Scholar]
- Liu X, Hwang H, Cao L, Buckland M, Cunningham A, Chen J, Chien KR, Graham RM, Zhou M (1998) Domain-specific gene disruption reveals critical regulation of neuregulin signaling by its cytoplasmic tail. Proc Natl Acad Sci USA 95:13024–13029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Williams RC, Urbanek M (1995) An E-M algorithm and testing strategy for multiple-locus haplotypes. Am J Hum Genet 56:799–810 [PMC free article] [PubMed] [Google Scholar]
- Mohn AR, Gainetdinov RR, Caron MG, Koller BH (1999) Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell 98:427–436 [DOI] [PubMed] [Google Scholar]
- O'Donovan MC, Owen MJ (1999) Candidate-gene association studies of schizophrenia. Am J Hum Genet 65:587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MF, Shaw G (1999) Comparison of dopamine receptor antagonists on hyperlocomotion induced by cocaine, amphetamine, MK-801 and the dopamine D1 agonist C-APB in mice. Psychopharmacology 145:237–250 [DOI] [PubMed] [Google Scholar]
- Ozaki M, Sasner M, Yano R, Lu HS, Buonanno A (1997) Neuregulin-beta induces expression of an NMDA-receptor subunit. Nature 390:691–694 [DOI] [PubMed] [Google Scholar]
- Pulver AE, Lasseter VK, Kasch L, Wolyniec P, Nestadt G, Blouin JL, Kimberland M, Babb R, Vourlis S, Chen HM, Lalioti M, Morris MA, Karayiorgou M, Ott J, Meyers D, Antonarakis SE, Housman D, Kazazian HH (1995) Schizophrenia: a genome scan targets chromosomes 3p and 8p as potential sites of susceptibility genes. Am J Med Genet 60:252–260 [DOI] [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet 46:222–228 [PMC free article] [PubMed] [Google Scholar]
- Shaw SH, Kelly M, Smith AB, Shields G, Hopkins PJ, Loftus J, Laval SH, Vita A, De Hert M, Cardon LR, Crow TJ, Sherrington R, DeLisi LE (1998) A genome-wide search for schizophrenia susceptibility genes. Am J Med Genet 81:364–376 [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–516 [PMC free article] [PubMed] [Google Scholar]
- Spitzer R, Endicott J (eds) (1977) The schedule for affective disorders and schizophrenia, lifetime version. 3rd ed. New York State Psychiatric Institute, New York [Google Scholar]
- Spitzer RL, Endicott J, Robins E (1978) Research diagnostic criteria: rationale and reliability. Arch Gen Psychiatry 35:773–782 [DOI] [PubMed] [Google Scholar]
- Steiner H, Blum M, Kitai ST, Fedi P (1999) Differential expression of ErbB3 and ErbB4 neuregulin receptors in dopamine neurons and forebrain areas of the adult rat. Exp Neurol 159:494–503 [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Stone WS, Faraone SV (2001) Genes, environment and schizophrenia. Br J Psychiatry Suppl 40:18–24 [DOI] [PubMed] [Google Scholar]
- Wang JY, Miller SJ, Falls DL (2001) The N-terminal region of neuregulin isoforms determines the accumulation of cell surface and released neuregulin ectodomain. J Biol Chem 276:2841–2851 [DOI] [PubMed] [Google Scholar]