Abstract
Essential hypertension (EH) is a complex disorder that results from the interaction of a number of susceptibility genes and environmental factors. We studied an isolated Sardinian village (Talana) in which the prevalence of hypertension is comparable to that in most Western populations. Talana exhibits features, such as slow demographic growth, high inbreeding, a low number of founders, stable lifestyle and culture, and accurate genealogical records, that make it suitable for the study of complex disorders. Clinical assessment of the entire adult population (N=∼1,000) identified ∼100 hypertensive subjects. For our study, we selected the individuals with the most-severe EH (i.e., diastolic blood pressure >100 mm Hg), belonging to a single deep-rooted pedigree (12 generations), whose common ancestors lived in the 17th century. We performed a three-stage genomewide search using 36 affected individuals, by means of parametric linkage and allele-sharing approaches. LOD scores >1 were observed on chromosomes 1, 2, 13, 15, 17, and 19 (stage I). The most striking result was found in a 7.57-cM region on chromosome 2p24-p25. All five nonparametric linkage statistics estimated by the SimWalk2 program lie above the significance threshold of P<.008 for the whole region. Similar significance was obtained for 2p24-25 when parametric linkage (LOD score 1.99) and linkage disequilibrium mapping (P=.00006) were used, suggesting that a hypertension-susceptibility locus is located between D2S2278 and D2S168. This finding is strengthened by a recent report of linkage with marker D2S168 in a hypertensive sib-pair sample from China.
Introduction
Essential hypertension (EH) affects about one quarter of the adult population in industrialized countries and contributes to considerable morbidity and mortality from stroke, heart failure, coronary heart disease, and renal failure (Burt et al. 1995; Vasan et al. 2001; Family Blood Pressure Program Investigators 2002). EH is a complex disorder that results from the interaction of a number of susceptibility genes and environmental factors. Data from animal models and human population studies suggest that inherited genetic factors influence ∼50% of the variation in blood pressure (BP) level, but the number of contributing genes and their relative risks remain unknown (Kato et al. 1999, 2000; Wright et al. 1999b; Stoll et al. 2000; Sugiyama et al. 2001).
Three approaches have been employed to identify genes influencing hypertension: a search for genes showing Mendelian inheritance, candidate-gene evaluation, and genomewide scans (Timberlake et al. 2001). The investigation of Mendelian disorders affecting BP has led to the identification of several genes (Lifton et al. 1992; Shimkets et al. 1994; Hansson et al. 1995; Mune et al. 1995; Simon et al. 1997; Disse-Nicodeme et al. 2000), but the contribution of these rare conditions to BP variation in the general population is very small. In recent years, a growing list of >100 candidate genes has been proposed to influence BP, and evidence of linkage with hypertension has been reported for several of these (Jeunemaitre et al. 1992; Svetkey et al. 1997; Krushkal et al. 1998; O'Donnell et al. 1998; Kainulainen et al. 1999). However, none of these candidate genes has been shown to contribute substantially to BP variability in the general population. Genomewide scans can be used to identify chromosomal regions with unknown genes influencing BP (Hsueh et al. 2000; Levy et al. 2000; Pankow et al. 2000; Perola et al. 2000; Rice et al. 2000; Allayee et al. 2001; Hollenberg 2001; Zhu et al. 2001).
Recently, many investigators have recommended the study of founder populations for complex-trait mapping, with the expectation that fewer susceptibility genes will be segregating in a restricted gene pool (Terwilliger and Weiss 1998; Kruglyak 1999; Wright et al. 1999a; Peltonen et al. 2000; Shifman and Darvasi 2001). The majority of these populations are inbred, and, in some cases, accurate pedigree information is available, which allows the analysis of large extended pedigrees. These communities derive from a small number of founders, and a high rate of endogamous and consanguineous marriages may reduce the number of susceptibility genes and increase genetic homogeneity (Peltonen et al. 2000; Angius et al. 2001). Furthermore, since individuals are exposed to a common environment and a relatively uniform lifestyle, nongenetic variability is minimized, and the noise caused by other etiological determinants is therefore reduced.
We studied an isolated Sardinian village (Talana) in which the prevalence of hypertension is ∼10% in the adult population, which is comparable to many Western populations. We performed a multistep genomewide search (GWS), selecting 35 related individuals with EH from a deep-rooted pedigree. Sample selection was optimized on the basis of the genealogical distance between individuals and their position in the extended family. The strategy was based on a search for genomic segments shared identical-by-descent (IBD) by affected individuals originating from a common ancestor (Houwen et al. 1994; Nikali et al. 1995; Hovatta et al. 1999).
Subjects and Methods
Population and Genealogical Data
We studied an isolated village, Talana, in central Sardinia, with a population of ∼1,200 inhabitants, characterized by slow population growth, high endogamy, and high inbreeding. Characterization of maternal and paternal lines allowed us to establish that 80% of the present-day population is descended from 8 paternal and 11 maternal ancestral lineages (Angius et al. 2001). Geographical and cultural isolation have generated a great deal of homogeneity in lifestyle and eating habits, providing a uniform environmental and genetic context for studies of complex diseases. Precise records of births, deaths, marriages, and people's origins have been registered since 1640 in the parochial Quinque Libri and in the municipal archives. All data were transferred into appropriate databases, allowing the creation of genealogical trees for any individual in the village.
Study Subjects
After an accurate check of BP in ∼1,000 adults (three BP measures were averaged to derive diastolic BP [DBP] and systolic BP [SBP]), we identified 100 hypertensive subjects. BP was measured in the seated position after 10 min of rest, through use of a mercury sphygmomanometer by experienced and certified examiners. A detailed questionnaire was filled out, and a physical examination was performed. Blood samples were drawn from all subjects, for serological and biochemical assays and for DNA extraction. Affection status was established on the basis of the following severe criteria: (i) an SBP ⩾150 mm Hg and/or DBP ⩾95 mm Hg if the patient was out of therapy or taking one antihypertensive medication, or an SBP ⩾145 mm Hg and/or DBP ⩾90 mm Hg if the patient was currently taking two or more antihypertensive medications for at least one year; (ii) onset of hypertension before age 60 years; and (iii) an accurate anamnesis, as well as the evaluation of renal and hepatic functionality, plasmatic electrolytes, lipid profile, and hemocrome, to exclude secondary forms of hypertension. Exclusion criteria included the use of other drugs, such as cortisone-based medications or symphaticotonics; the use (by women) of estroprogestinic hormones; and the presence of nephrolithiasys, vascular diseases, and diabetes. All individuals participating in the study signed informed consent forms, and all samples were taken in accordance with the Declaration of Helsinki (World Medical Association Web site).
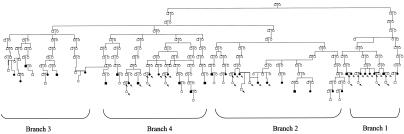
Within the EH-affected group, we selected 35 of the individuals with the most-severe EH, of whom 69% were undergoing treatment and the remainder showed a DPB >100 mm Hg, with a mean age at onset of 54.5±7.8 years and no lifetime history of smoking (table 1). All 35 selected individuals belong to a single deep-rooted 12-generation pedigree whose founding ancestors lived in the 17th century (fig. 1).
Table 1.
Clinical Characteristics of the Study Population
|
Mean ± SD |
||||||||
| Population | DBP(mm Hg) | SBP(mm Hg) | Age atExamination(years) | Height(cm) | Weight(kg) | BMI(kg/m2) | % with Treatmenta | Lifetime History of Smoking |
| Affected: | ||||||||
| Men (n=12) | 96.67 ± 8.8 | 161.67 ± 12.67 | 61.58 ± 20.06 | 163.25 ± 6.45 | 73.58 ± 11.20 | 27.58 ± 3.78 | 50 | No |
| Women (n=23) | 93.96 ± 10.34 | 162.39 ± 16.51 | 64.96 ± 8.91 | 151.00 ± 6.46 | 63.13 ± 10.04 | 27.70 ± 4.10 | 79 | No |
| Combined (n=35) | 94.89 ± 9.82 | 162.14 ± 15.11 | 63.80 ± 13.57 | 155.20 ± 8.68 | 66.71 ± 11.45 | 27.66 ± 3.94 | 69 | No |
| Unaffected: | ||||||||
| Men (n=19) | 80.83 ± 6.51 | 145.83 ± 18.87 | 64.00 ± 19.77 | 162.71 ± 5.09 | 71.64 ± 8.42 | 26.96 ± 2.01 | … | … |
| Women (n=23) | 83.61 ± 8.61 | 139.35 ± 19.15 | 57.61 ± 21.64 | 151.75 ± 3.24 | 69.18 ± 17.65 | 30.11 ± 7.88 | … | … |
| Combined (n=42) | 82.45 ± 7.97 | 142.00 ± 19.51 | 60.81 ± 20.68 | 157.58 ± 7.01 | 64.41 ± 10.58 | 25.80 ± 2.84 | … | … |
| Talana (n=772) | 81.87 ± 33.11 | 137.62 ± 19.03 | 47.50 ± 20.92 | 155.06 ± 8.55 | 63.64 ± 14.09 | 26.35 ± 4.93 | … | … |
Proportion taking antihypertensive medications for ⩾1 year.
Figure 1.
Twelve-generation pedigree used in the linkage and Simwalk2 analyses. Because of computational constraints, the pedigree was broken into four smaller branches, as indicated in the figure. Family members analyzed in stage I are indicated by arrows. Blackened symbols denote affected individuals.
Genotyping
Genomic DNA was extracted from 7 ml of EDTA-treated blood, as described by Ciulla et al. (1988). Multiplex fluorescent–based genotyping was performed using ABI PRISM linkage mapping set, version 2 (Perkin Elmer). PCR was performed according to standard protocols. Microsatellite products were loaded on an ABI PRISM 3100 DNA Analyzer (PE Biosystems), and data were processed by GENESCAN version 3.1 and GENOTYPER version 2.5 software. Additional markers in stage II and stage III were selected from The Genome Database.
Multistep GWS
Stage I
In the first stage, we selected only 16 affected individuals and their close relatives (33 total genotyped samples), belonging to the pedigree referred to above. The individuals analyzed in stage I are indicated by arrows in figure 1. Because of computational constraints, the full family was analyzed by breaking it into four smaller branches. Four hundred polymorphic microsatellite markers, at an average intermarker distance of ∼10 cM, were used for genotyping. Genetic maps were derived from the Marshfield map (Center for Medical Genetics Web site), and allele frequencies were calculated using 25 unrelated individuals from the same village. All pedigree branches were analyzed using both parametric linkage and nonparametric IBD-sharing approaches. Regions yielding suggestive or nominal evidence for both parametric linkage (LOD score >1) and nonparametric allele sharing (P<.05) were followed up in the II stage. As suggested by Lander and Kruglyak (1995), the P<.0001 level was considered statistically significant, a P value of .0001–.01 was considered suggestive, and a P value of .01–.05 was considered nominal evidence for linkage.
Stage II
In this stage, we investigated potentially interesting regions from the stage I screening. We genotyped 35 distantly related affected subjects and their close relatives (77 total genotyped samples) for all markers showing a positive signal in stage I and for additional microsatellite markers at an average intermarker distance of 2 cM. The sample included all 33 individuals already examined in stage I (fig. 1). In the selection of additional markers, we chose those with the highest heterozygosity. Allele frequencies were estimated by genotyping 25 healthy unrelated individuals from the same village and from the pedigree, which included all affected and unaffected individuals, to check the accuracy of results.
Stage III: Linkage disequilibrium (LD) mapping
The genomic region spanning >7.57 cM located on 2p24-p25 was analyzed in detail, adding 10 supplementary markers: D2S2164, D2S2207, D2S2326, D2S423, D2S2278, D2S328, D2S2200, D2S262, D2S2199, and D2S131. The average distance between all analyzed markers on 2p24-25 is ∼0.97 cM.
Statistical Analyses
GWS data were analyzed using two statistical approaches: a model-dependent method of linkage analysis and a nonparametric method to compute allele-sharing statistics. Two-point LOD scores were calculated by FASTLINK version 4.1P (Cottingham et al. 1993; Schaffer et al. 1994) in an affected-only strategy, under the assumption that all unaffected individuals are phenotypically unknown, since unaffected subjects do not provide reliable information on the underlying disease-locus genotype for a complex disease. Dominant and recessive models were examined under the assumption of disease allele frequencies (D) of 0.01 for the dominant model and 0.1 for the recessive model. A reduced penetrance of 50% was assumed. Although the choice of genetic model is arbitrary and, presumably, incorrect, the LOD-score method can still be robust when a small number of different models are assumed (Hodge et al. 1997; Greenberg et al. 1998; Durner et al. 1999). For linkage analysis, it was necessary to break the entire pedigree down into four smaller branches. Pedigree size is a severe limitation for multipoint statistics, where IBD-sharing analysis is usually restricted to pedigrees of moderate size and structure. To take advantage of the power provided by our complex and deep-rooted pedigree structure, the SimWalk2 program was utilized. This program is based on Markov chain–Monte Carlo sampling and simulated annealing algorithms, and it performs multipoint analyses in a reasonable length of time (Sobel and Lange 1996). SimWalk2 reports five nonparametric statistics and their empirical P values, which measure the degree of clustering (and its significance), among the affected individuals, of marker alleles originating from the pedigree founders. All the marker information was used in this multipoint computation of allele-sharing statistics. In addition, D statistics were selected over other nonparametric statistics calculated by SimWalk2, because they are generally more powerful when the model of inheritance is unknown. The D statistic is a general statistic indicating whether a few founder alleles are overrepresented among the affected individuals, and it represents the extent of allele sharing among all affected pairs, as measured by their IBD kinship coefficient.
The SimWalk2 program was used for haplotype analysis to estimate the most likely set of fully typed maternal and paternal haplotypes of marker loci for each individual in the pedigree. The program runs over the data several times, to find the optimal haplotype configuration.
The degree of relationship and the level of clustering between affected individuals were evaluated by counting meiotic steps and estimating kinship coefficients (ψ). The mean value of ψ=.04 indicates that, on average, the relatedness of affected individuals within the full pedigree is less than that of first cousins (ψ=1/16=.0625). The lack of dense clustering (ψ=.04) indicates that splitting the full pedigree into smaller branches would maximize the power of the linkage analysis, since extensive amounts of information on segregation and allele sharing are gained by using small pedigrees that include related affected individuals, so as to obtain a higher clustering of affected individuals (i.e., a higher ψ value).
An LD mapping statistic test, ancestral haplotype reconstruction (AHR), which is designed to find LD via haplotype analysis in samples drawn from population isolates, was also applied (Service et al. 1999). AHR compares the distribution of haplotypes in affected individuals with the distribution expected for individuals bearing a disease mutation descended from a common ancestor. This test was applied using windows spanning three markers. Three main parameters are estimated under the null hypothesis: the time since a common founder (g); the percentage of chromosomes, in affected individuals, that have descended from this founder (α); and the position (x) of the disease locus. The assumption of linkage equilibrium under the null-hypothesis likelihood can be problematic for dense marker spacings (e.g., <1 cM) and/or for very young populations, because of the potential for “background” LD (BLD) between markers—that is, LD unrelated to a disease phenotype. When strong BLD exists in the absence of a disease locus, it may produce false-positive results (Service et al. 1999). Because of the strong and extended LD that we observed in the Talana population (Angius et al. 2001), we tried to overcome this problem through use of a modified version of the AHR program that includes LD between markers under the null hypothesis (McPeek and Strahs 1999; Escamilla et al. 2001).
Exact power estimation and assessment of genomewide significance through simulation is impractical for the whole pedigree, given the complexity of the genealogical structure. Therefore, we estimated the probability of obtaining specific LOD score values through simulation of the four smaller branches. Using the average marker heterozygosity (∼0.70) estimated from the 400–microsatellite marker set used in the genomewide search, we generated 1,000 replicates of the family data, assuming no linkage under the dominant model, using the SLINK program (Weeks et al. 1990). For all replicates, we then calculated the number of times that the LOD scores exceeded a specific threshold under the null hypothesis.
Allele Frequency Estimates
In a large pedigree such as the one presented here, it is important to estimate accurate allele frequencies for linkage analysis (Boehnke 1991; Ott 1992; Freimer et al. 1993; Nechiporuk et al. 1993; Lindholm et al. 2001). To check the accuracy of the LOD score results, two sets of Talana control allele frequencies were used. The PEDMANAGER program, version 0.9 was utilized to calculate allele frequencies and to check Mendelian inconsistencies in the pedigree data. The first set of allele frequencies was obtained from 25 unrelated individuals from the village. This was done because a control sample from an outbred population would not allow a reliable estimate of common alleles frequencies for Talana.
The second set of allele frequencies was derived from the genotyped pedigree, including all affected and unaffected individuals (77 genotyped individuals); this group is biased toward false-negative results, because it includes a large number (i.e., 35) of affected individuals.
In some cases, allele frequency estimates were significantly different when evaluated in the two samples (i.e., unrelated individuals versus within those within the pedigree). However, the second set of allele frequencies was used to check only its effect on LOD scores.
For some of the additional microsatellite markers used in stage II, allele frequencies were derived from the entire Talana population.
Results
Stage I
The stage I genome scan results are given in table 2, which indicates markers showing evidence of suggestive linkage to hypertension (LOD score >1, nominal P<.05). Among the 13 identified regions, six loci are particularly interesting, because similar results were obtained using both two-point parametric linkage and multipoint nonparametric statistics. These six loci were located on chromosomes 1, 2, 13, 15, 17, and 19. A significant nonparametric result (nominal P=.005) was obtained for markers on 2p24-25 and was also confirmed by two-point parametric linkage, under both the dominant (LOD score 1.46) and the recessive (LOD score 1.53) models. The regions identified in stage I were further evaluated with additional microsatellite markers spaced at an average distance of ∼2 cM, in all stage I individuals and in additional affected and control members of the same extended family.
Table 2.
Results from Stage I
|
Two-PointLOD Score |
||||||
| Chromosome | Marker Region | Interval (cM) | Peak Marker | Allele-SharingP Valuea | DominantModel | RecessiveModel |
| 1 | D1S214–D1S450 | 9.4–15.9 | D1S450 | .0402 | ||
| 1 | D1S484 | NS | 1.18 | 1.68 | ||
| 2 | D2S2211–D2S168 | 9.2–21.7 | D2S162 | .0050 | 1.46 | 1.53 |
| 3 | D3S1277 | NS | 1.10 | |||
| 8 | D8S270–D8S1784 | 99.9–110.9 | D8S1784 | .0407 | ||
| 9 | D9S171 | NS | 1.90 | 1.65 | ||
| 10 | D10S192 | .0407 | ||||
| 11 | D11S901 | .0379 | ||||
| 12 | D12S364 | .0389 | ||||
| 13 | D13S156–D13S173 | 48.9–88.2 | D13S265 | .0398 | ||
| 13 | D13S173 | NS | 1.78 | 2.23 | ||
| 15 | D15S153–D15S205 | 69.2–78.6 | D15S205 | .0398 | 1.67 | 2.21 |
| 17 | D17S849–D17S831 | 0–6.0 | D17S849 | .0631 | ||
| 17 | D17S938 | NS | 1.46 | 2.10 | ||
| 18 | D18S464–D18S53 | 30.9–39.9 | D18S53 | .0417 | ||
| 19 | D19S414–D19S220 | 41.8–49.7 | D19S220 | .0398 | ||
| 19 | D19S414 | NS | 3.10 | 3.1 | ||
Allele-sharing statistics obtained using SIMWALK2 (Sobel and Lange 1996). NS = not significant (P>.05).
Stage II
The results of the stage II linkage analysis are shown in table 3. Most of the stage I results were not confirmed in the second stage. Following Lindholm et al. (2001), we used two different sets of allele frequencies for the calculations, to evaluate how allele frequencies affect linkage evidence.
Table 3.
Results from Stage II[Note]
|
Two-Point LOD Scoresfor Allele Frequencies from |
||||||||
| Control Population |
Pedigree |
|||||||
| Chromosome | Marker Region | Interval(cM) | Peak Marker | Allele-SharingP Valuea | DominantModel | RecessiveModel | DominantModel | RecessiveModel |
| 1 | D1S214–D1S 450 | 0–6.6 | D1S450 | .0459 | 1.59 | 1.11 | NS | NS |
| 1 | D1S1655–D1S1551 | 97.6–109.8 | D1S2841 | .0137 | 1.60 | .97 | NS | NS |
| 2 | D2S162–D2S149 | 9.1–23.0 | D2S287 | .0047 | 1.86 | 1.52 | 2.08 | 1.63 |
| 13 | … | … | D13S1306 | .0631 | 1.90 | 1.95 | 1.99 | 2.07 |
| 13 | … | … | D13S173 | NS | 1.31 | 1.23 | NS | NS |
| 15 | … | … | D15S205 | NS | 3.13 | 3.26 | NS | NS |
| 17 | D17S831–D17S1298 | 5.9–10.1 | D17S938 | NS | 2.32 | 2.59 | NS | NS |
Note.— NS = not significant (P>.05).
Allele sharing statistics obtained with SIMWALK2 (Sobel and Lange 1996).
Positive stage I linkage results on chromosomes 15 and 17 were not supported by nonparametric statistical results (table 3). The multipoint allele-sharing results for chromosomes 15 and 17 were not significant (nominal P>.05). Results showing suggestive linkage to chromosome 1 were supported by nonparametric statistical results (nominal P=.0459 for D1S450, and nominal P=.0137 for D1S2841), but two-point LOD scores under the recessive model are <1. On chromosome 13, the initial linkage findings for marker D13S173 were not confirmed (table 2), but interesting results were obtained by parametric analysis with marker D13S1306 (LOD scores of 1.90 and 1.95, for the dominant and recessive models, respectively, under the affected-only method). The D13S1306 marker was added in stage II and shows a suggestive LOD score when allele frequencies both from control subjects and from the pedigree are used, as well as a P value of .0631, which is close to significance (table 3), from nonparametric analysis.
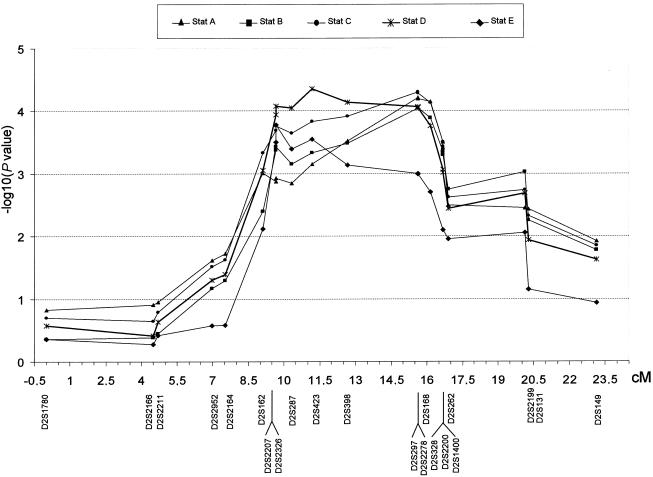
On chromosome 2, two-point LOD scores of 1.86 (dominant model) and 1.52 (recessive model) were observed in stage I and, using allele frequencies estimated from the pedigree, we confirmed these results in stage II (LOD scores of 2.08 and 1.63, under the dominant and the recessive model, respectively; table 3). In this genomic region, the interval D2S162–D2S149, which spans ∼13.9 cM, showed strong evidence of linkage (nominal P=.0047) when nonparametric statistics were used, indicating clustering of founder alleles among the affected individuals (Sobel and Lange 1996). Additional markers, with an average marker density of ∼0.97 cM (21 markers analyzed), were used to verify the results on chromosome 2. Figure 2 shows the results of all SimWalk2 nonparametric statistical tests for stage II. All five statistics lie above the significance threshold of P<.008 in the 7.57-cM region from D2S162 and D2S1400 and, in particular, the D statistic is above the significance threshold of P<.0009 from D2S162 and D2S1400, with the maximum value on D2S423 (P=.0004) (fig. 2). Within this region, the most interesting linkage result was observed for marker D2S2278 (LOD score 1.99 under the dominant model, and LOD score 1.5 under the recessive model). These linkage scores were not influenced by allele frequency estimates within the pedigree.
Figure 2.
Nonparametric statistics estimated by SimWalk2 (Sobel and Lange 1996). Statistic A is the number of different founder alleles contributing alleles to the affected individuals. Statistic B is the maximum number of alleles among the affected individuals descended from any one founder allele. Statistic C is the “entropy” of the marker alleles among the affected individuals. Statistic D is the extent of allele sharing among all affected pairs, as measured by their IBD kinship coefficient. Statistic E is the “NPL_all” statistic, as implemented in GeneHunter.
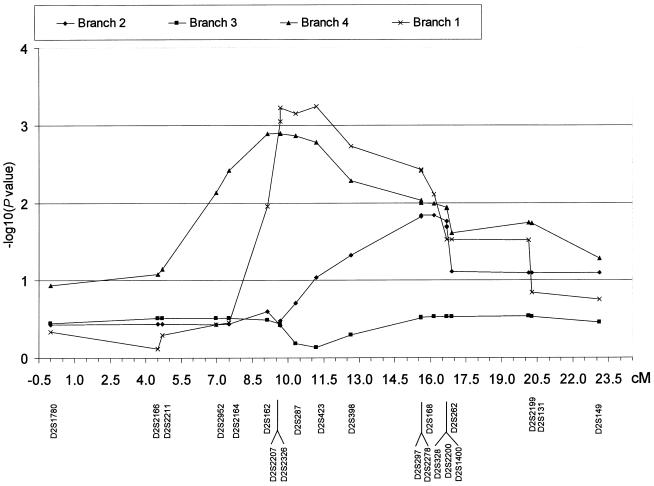
To evaluate possible genetic heterogeneity within the different branches of the pedigree, D statistics were examined for each branch through use of markers from the 2p24-p25 region. As shown in figure 3, not all branches are equally informative for multipoint analysis. Branch 3 is completely uninformative, whereas branches 1 and 4 show similar significance levels, with a maximum between D2S162 and D2S168 (fig. 3). The graph of branch 2 shows its maximum between D2S297 and D2S1400, with a partial overlap with branches 1 and 4.
Figure 3.
Statistic D results for each analyzed branch of the pedigree in the whole region shown in figure 2
We also estimated the probability of obtaining specific LOD score values through simulation of the four smaller branches. A LOD score ⩾2 was observed in 13.7% of the replicates obtained under no linkage, with a 95% CI of 13.23%–14.30%. A LOD score ⩾3 was observed in 4.24% of the replicates obtained under no linkage, with a 95% CI of 3.92%–4.56%.
Although the empirical P value estimated from simulation of obtaining a LOD score of 2 is not significant (P=.14), the concordance with results obtained using nonparametric linkage analyses (P<.0009) supports the evidence for the region being on 2p24-p25. Furthermore, in the simulation, a specific genetic model for the trait has to be assumed, and the empirical P value estimates are valid only under that specific model. Therefore, the results of the simulation should be judged cautiously.
Stage III: LD Mapping
Haplotypes were estimated using the SimWalk2 program. These haplotypes were analyzed using the AHR program in three overlapping windows of three markers. The AHR results for the 21 markers are shown in table 4. These data suggest a possible disease locus at D2S2207 (peak LOD score 2.76, equivalent to P=.00174) and between D2S2278 and D2S168 (peak LOD score 4.21, equivalent to P=.00006). Marker D2S168 gave a peak LOD score of 3.02, equivalent to a P value of .00095. This localization between D2S2278 and D2S168 is strongly supported by allele-sharing and parametric linkage results (fig. 2; table 3).
Table 4.
AHR results from Three-Marker Intervals on Chromosome 2p24-p25[Note]
| Three-Marker Interval and Genetic Distances | Peak LODScore | Location of Peak Score(Recombination Fraction)a | Estimateof g | Estimateof α |
| D2S1780–4.05 cM–D2S2166–.2 cM–D2S2211 | .18 | .017680 from D2S1780 | 1,000 | .44 |
| D2S2166–.2 cM–D2S2211–2.27 cM–D2S2952 | 1.09 | .018066 from D2S2211 | 800 | .86 |
| D2S2211–2.27 cM–D2S2952–.54 cM–D2S2164 | .48 | .004519 from D2S2211 | 400 | .46 |
| D2S2952–.54 cM–D2S2164–1.61 cM–D2S162 | 1.06 | At D2S2164 | 10 | .12 |
| D2S2164–1.61 cM–D2S162–.54 cM–D2S2207 | .19 | .004301 from D2S162 | 20 | .04 |
| D2S162–.54 cM–D2S2207–.00 cM–D2S2326 | 2.76 | At D2S2207 | 10 | .08 |
| D2S2207–.00 cM–D2S2326–.65 cM–D2S287 | 1.39 | .005173 from D2S2326 | 10 | .08 |
| D2S2326–.65 cM–D2S287–.88 cM–D2S423 | 2.05 | .005173 from D2S2326 | 10 | .08 |
| D2S287–.88 cM–D2S423–1.47 cM–D2S398 | 1.12 | At D2S287 | 30 | .12 |
| D2S423–1.47 cM–D2S398–2.95 cM–D2S297 | 1.37 | .023052 from D2S398 | 10 | .12 |
| D2S398–2.95 cM–D2S297–.00 cM–D2S2278 | 1.09 | .005865 from D2S398 | 500 | .76 |
| D2S297–.00 cM–D2S2278–.54 cM–D2S168 | 4.21 | .004301 from D2S2278 | 10 | .10 |
| D2S2278–.54 cM–D2S168–.54 cM–D2S328 | 2.67 | At D2S2278 | 10 | .08 |
| D2S168–.54 cM–D2S328–.00 cM–D2S2200 | 3.02 | At D2S168 | 10 | .08 |
| D2S328–.00 cM–D2S2200–.00 cM–D2S1400 | 1.93 | .000001 from D2S328 | 10 | .10 |
| D2S2200–.00 cM–D2S1400–.00 cM–D2S262 | 2.12 | .000001 from D2S1400 | 1,000 | .14 |
| D2S1400–.00 cM–D2S262–3.45 cM–D2S2199 | .50 | At D2S1400 | 30 | .14 |
| D2S262–3.45 cM–D2S2199–.15 cM–D2S131 | .58 | .006853 from D2S262 | 300 | .56 |
| D2S2199–.15 cM–D2S131–2.84 cM–D2S149 | 1.61 | .022212 from D2S131 | 10 | .14 |
Note.— Study sample consisted of 35 individuals with EH and 42 unaffected individuals.
Position (x) of the disease locus with the distance (recombination fraction) from the marker.
Discussion
We report the results of a three-stage linkage study that aimed to localize susceptibility genes for EH in a genetic isolate, the Talana population. We studied a large 12-generation pedigree comprising 35 EH-affected individuals, through use of different approaches. Parametric linkage analysis can be very powerful for study of the Talana village, because affected individuals all belong to a genetically isolated population and common ancestors can be unambiguously identified (Angius et al. 2001; Ombra et al. 2001). Large pedigrees that include distantly related affected individuals provide power for allele-sharing detection, since the prior probability of sharing any genome segment is small for distant relatives, and evidence of IBD sharing indicates the presence of a genetic factor contributing to the trait (Houwen et al. 1994; Nikali et al. 1995; Hovatta et al. 1999). On the basis of their genealogical distance and position in the Talana hypertension kindred, a subset of affected individuals sharing common ancestors were carefully selected for a multistep GWS. The number of samples required for the initial GWS was optimized by initially selecting only 16 affected individuals and their close relatives (33 in total), and a 400–microsatellite marker scan was performed (stage I). The approach is based on an economic selection of samples, which takes advantage of the power provided by pedigree structure and the reduced genetic heterogeneity resulting from high inbreeding.
In stage II, selected genomic regions were investigated further by genotyping an expanded sample of 77 subjects, adding additional microsatellite markers to the selected regions at ∼2-cM intervals. To evaluate how allele frequencies affect the linkage results, two different sets of allele frequencies were used in stage II for the calculations, either from 25 unrelated individuals from Talana village or from the entire pedigree, including all affected and unaffected individuals. The second set of allele frequencies was used to check only its effect on LOD scores. For some additional microsatellite markers used in stage II, the allele frequencies were derived from the entire Talana population.
In stage I, significant results were obtained for loci on chromosomes 1, 2, 13, 15, 17, and 19, with the most striking results on chromosome 2, with a nonparametric nominal P value of .0050. In stage II, linkage to genomic regions on chromosomes 1, 2, 13, 15, and 17 was further supported by parametric analysis, although, when allele frequencies were estimated within the pedigree, the LOD scores decreased on chromosomes 1, 15, and 17. This highlights the importance of using accurate estimates of allele frequencies in the analysis of extended pedigrees (Lindholm et al. 2001).
The most striking result was obtained in a 7.57-cM region on chromosome 2p24-p25. All five nonparametric statistics estimated by SimWalk2 lie above the significance threshold of P<.008 in the whole region between D2S162 and D2S1400 and, in particular, the D statistic settles above the significance threshold of P<.0009, with its maximum value on D2S423 (P=.0004). The result on chromosome 2p24-p25 is not inflated by the set of allele frequencies used for initial computations. When evidence from haplotypes is included, affected individuals show only small shared haplotypes that segregate with the disease in pedigree branches (data not shown). It was not possible to identify one haplotype shared between all affected individuals. One possible explanation lies in haplotype heterogeneity within the pedigree, which is probably due to the extended kindred. To evaluate heterogeneity within the pedigree, the D-statistic results for each analyzed branch were examined for markers in the region 2p24-p25. We chose the D statistic over other nonparametric statistics calculated by SimWalk2 because it is generally more powerful when the mode of inheritance is unknown (Weeks and Lange 1988; Garner et al. 2001). Not all branches were equally informative for allele-sharing analysis. Branch 3 is completely uninformative, but statistics for branches 1, 2, and 4 lie above the significance level of P<.015 between D2S297 and D2S168. The overlap of the D statistics between branches 1, 2, and 4 indicates a clustering of the founder alleles among distantly related affected individuals. The analysis of nonparametric results from all subpedigrees permitted the identification of a 7.57-cM genomic region. A more detailed analysis of the overlap between statistics results in branches 1, 2, and 4 narrowed the localization to between markers D2S297and D2S168.
In stage III, proposed susceptibility regions were further evaluated by LD mapping, which narrowed the 7.57-cM region further through use of the AHR test statistic. (Service et al. 1999; Escamilla et al. 2001). The AHR results suggested a possible hypertension locus localized between markers D2S2278 and D2S168, which are 0.54 cM apart. A maximum peak LOD score of 4.21 (nominal P=.00006) was obtained at a recombination fraction of 0.004 at marker D2S2278, and a LOD score of 3.02 (nominal P=.00095) was obtained at marker D2S168. AHR analysis also gives an estimate of the time elapsed (g) since a common founder for the analyzed data. Although it is possible that a susceptibility mutation shared by hypertensive probands predates the genealogical reconstruction (i.e., the 17th century), a g value of 10 generations was obtained for all markers showing significant linkage results.
The concordance of results on chromosomal region 2p24-p25 obtained using parametric and nonparametric linkage analyses and LD mapping suggests a hypertension susceptibility locus located between D2S2278 and D2S168. This hypothesis is further strengthened by a recent report by Zhu et al. (2001), who found evidence for linkage (two-point nonparametric linkage Z score 1.89; P=.031) with marker D2S168 in a GWS of affected sib pairs from the Shanghai area. However, they did not replicate this result in the second stage, through use of an independent sib-pair sample, despite adding additional markers close to D2S168. In our study, we used a denser marker map (∼0.97-cM spacing), thus allowing a better estimate of recombination close to D2S168. Given the complexity of the inheritance of hypertension and possible phenotype/environment interactions complicating the detection of underlying genetic factors, the evidence for nonparametric linkage in the same locus in Italian and Chinese families makes this a region of interest. No hypertension candidate genes have previously been reported within or in the vicinity of the identified locus. In this genomic region, there are seven predicted genes that apparently are not involved in the regulation of BP or the development of hypertension. Further studies are needed to characterize and complete the sequence, in order to identify other possible candidate genes.
In the small Talana population, LD has been shown to span a relatively wide interval on the X chromosome, suggesting the possibility of using an LD approach to identify IBD regions associated with complex traits, using a genomewide search at a low marker density (Wright et al. 1999a; Angius et al. 2001, 2002; Ombra et al. 2001). When the large amount of information available from the extended genealogy was used, only 16 individuals with EH were selected from the pedigree for the initial GWS for linkage. A small number of genomic regions showed suggestive evidence of linkage to essential hypertension. Both parametric and nonparametric linkage approaches identified a region on chromosome 2p24-p25 that may harbor a locus for susceptibility to essential hypertension. An allele-sharing statistic and LD-based mapping, as implemented in AHR, proved to be sensitive methods for detecting a disease locus, whereas parametric linkage results were affected by different sets of allele frequencies used for calculations. The results provide significant evidence for a susceptibility locus on chromosome 2p24-p25, and, although this locus has been identified in a genetic isolate, it may also play a role in EH in other populations.
Acknowledgments
We thank Dr. Alan F. Wright for helpful discussion, as well as Dr. James L. Weber and the National Heart, Lung, and Blood Institute Mammalian Genotyping Service. We thank Susan K. Service for permission to use the AHR software for the necessary LD mapping analysis, as well as for helpful suggestions. We thank Dr. Alessandra Pala and Ms. Antonella Sanna for technical assistance. We are very grateful to the Talana population for their collaboration. This research was supported by Telethon (Italy) grant number E1185.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- Genome Database, The, http://www.gdb.org/
- World Medical Association, http://www.wma.net/e/policy/17-c_e.html (for the Helsinki Declaration)
References
- Allayee H, de Bruin TW, Michelle Dominguez K, Cheng LS, Ipp E, Cantor RM, Krass KL, Keulen ET, Aouizerat BE, Lusis AJ, Rotter JI (2001) Genome scan for blood pressure in Dutch dyslipidemic families reveals linkage to a locus on chromosome 4p. Hypertension 38:773–778 [DOI] [PubMed] [Google Scholar]
- Angius A, Bebbere D, Petretto E, Falchi M, Forabosco P, Maestrale GB, Casu G, Persico I, Melis PM, Pirastu M (2002) Not all isolates are equal: linkage disequilibrium analysis on Xq13.3 reveals different patterns in Sardinian sub-populations. Hum Genet 111:9–15 [DOI] [PubMed] [Google Scholar]
- Angius A, Melis PM, Morelli L, Petretto E, Casu G, Maestrale GB, Fraumene C, Bebbere D, Forabosco P, Pirastu M (2001) Archival, demographic and genetic studies define a Sardinian sub-isolate as a suitable model for mapping complex traits. Hum Genet 109:198–209 [DOI] [PubMed] [Google Scholar]
- Boehnke M (1991) Allele frequency estimation from data on relatives. Am J Hum Genet 48:22–25 [PMC free article] [PubMed] [Google Scholar]
- Burt VL, Whelton P, Roccella EJ, Brown C, Cutler JA, Higgins M, Horan MJ, Labarthe D (1995) Prevalence of hypertension in the US adult population: results from the Third National Health and Nutrition Examination Survey, 1988–1991. Hypertension 25:305–313 [DOI] [PubMed] [Google Scholar]
- Ciulla TA, Sklar RM, Hauser SL (1988) A simple method for DNA purification from peripheral blood. Anal Biochem 174:485–488 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Disse-Nicodeme S, Achard JM, Desitter I, Houot AM, Fournier A, Corvol P, Jeunemaitre X (2000) A new locus on chromosome 12p13.3 for pseudohypoaldosteronism type II, an autosomal dominant form of hypertension. Am J Hum Genet 67:302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner M, Vieland VJ, Greenberg DA (1999) Further evidence for the increased power of LOD scores compared with non-parametric methods. Am J Hum Genet 64:281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla MA, McInnes LA, Service SK, Spesny M, Reus VI, Molina J, Gallegos A, Fournier E, Batki S, Neylan T, Matthews C, Vinogradov S, Roche E, Tyler DJ, Shimayoshi N, Mendez R, Ramirez R, Ramirez M, Araya C, Araya X, Leon PE, Sandkuijl LA, Freimer NB (2001) Genome screening for linkage disequilibrium in a Costa Rican sample of patients with bipolar-I disorder: a follow-up study on chromosome 18. Am J Med Genet 105:207–213 [DOI] [PubMed] [Google Scholar]
- Family Blood Pressure Program Investigators (2002) Multi-center genetic study of hypertension: The Family Blood Pressure Program (FBPP). Hypertension 39:3–9 [DOI] [PubMed] [Google Scholar]
- Freimer NB, Sandkuijl LA, Blower SM (1993) Incorrect specification of marker allele frequencies: effects on linkage analysis. Am J Hum Genet 52:1102–1110 [PMC free article] [PubMed] [Google Scholar]
- Garner C, McInnes LA, Service SK, Spesny M, Fournier E, Leon P, Freimer NB (2001) Linkage analysis of a complex pedigree with severe bipolar disorder, using a Markov chain Monte Carlo method. Am J Hum Genet 68:1061–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet 63:870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson JH, Nelson-Williams C, Suzuki H, Schild L, Shimkets R, Lu Y, Lu Y, Canessa C, Iwasaki T, Rossier B, Lifton RP (1995) Hypertension caused by a truncated epithelial sodium channel g-subunit: genetic heterogeneity of Liddle syndrome. Nat Genet 11:76–82 [DOI] [PubMed] [Google Scholar]
- Hodge SE, Abreu PC, Greenberg DA (1997) Magnitude of type I error when single-locus linkage analysis is maximized over models: a simulation study. Am J Hum Genet 60:217–227 [PMC free article] [PubMed] [Google Scholar]
- Hollenberg NK (2001) A genome-wide search for susceptibility loci to human essential hypertension. Curr Hypertens Rep 3:7–8 [PubMed] [Google Scholar]
- Houwen RHJ, Baharloo S, Blankenship K, Raeymaekers P, Juyn J, Sandkuijl LA, Freimer NB (1994) Genome screening by searching for shared segments: mapping a gene for benign recurrent intrahepatic cholestasis. Nat Genet 8:380–386 [DOI] [PubMed] [Google Scholar]
- Hovatta I, Varilo T, Suvisaari J, Terwilliger JD, Ollikainen V, Araja¨ rvi R, Juvonen H, Kokko-Sahin ML, Va¨ isa¨ nen L, Mannila H, Lönnqvist J, Peltonen L (1999) A genomewide screen for schizophrenia genes in an isolated Finnish subpopulation, suggesting multiple susceptibility loci. Am J Hum Genet 65:1114–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh WC, Mitchell BD, Schneider JL, Wagner MJ, Bell CJ, Nanthakumar E, Shuldiner AR (2000) QTL influencing blood pressure maps to the region of PPH1 on chromosome 2q31-34 in Old Order Amish. Circulation 101:2810–2816 [DOI] [PubMed] [Google Scholar]
- Jeunemaitre X, Soubrier F, Kotelevtsev YV, Lifton RP, Williams CS, Charru A, Hunt SC, Hopkins PN, Williams RR, Lalouel JM (1992) Molecular basis of human hypertension: role of angiotensinogen. Cell 71:169–180 [DOI] [PubMed] [Google Scholar]
- Kainulainen K, Perola M, Terwilliger J, Kaprio J, Koskenvuo M, Syvanen AC, Vartiainen E, Peltonen L, Kontula K (1999) Evidence for involvement of the type 1 angiotensin II receptor locus in essential hypertension. Hypertension 33:844–849 [DOI] [PubMed] [Google Scholar]
- Kato N, Hyne G, Bihoreau MT, Gauguier D, Lathrop GM, Rapp JP (1999) Complete genome searches for quantitative trait loci controlling blood pressure and related traits in four segregating populations derived from Dahl hypertensive rats. Mamm Genome 10:259–265 [DOI] [PubMed] [Google Scholar]
- Kato N, Tamada T, Nabika T, Ueno K, Gotoda T, Matsumoto C, Mashimo T, Sawamura M, Ikeda K, Nara Y, Yamori Y (2000) Identification of quantitative trait loci for serum cholesterol levels in stroke-prone spontaneously hypertensive rats. Arterioscler Thromb Vasc Biol 20:223–229 [DOI] [PubMed] [Google Scholar]
- Kruglyak L (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22:139–144 [DOI] [PubMed] [Google Scholar]
- Krushkal J, Xiong M, Ferrell R, Sing CF, Turner ST, Boerwinkle E (1998) Linkage and association of adrenergic and dopamine receptor genes in the distal portion of the long arm of chromosome 5 with systolic blood pressure variation. Hum Mol Genet 7:1379–1383 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Levy D, DeStefano AL, Larson MG, O'Donnell CJ, Lifton RP, Gavras H, Cupples LA, Myers RH (2000) Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the Framingham heart study. Hypertension 36:477–483 [DOI] [PubMed] [Google Scholar]
- Lifton RP, Dluhy RG, Powers M, Rich GM, Gutkin M, Fallo F, Gill JR Jr, Feld L, Ganguly A, Laidlaw JC (1992) Hereditary hypertension caused by chimaeric gene duplications and ectopic expression of aldosterone synthase. Nat Genet 2:66–74 [DOI] [PubMed] [Google Scholar]
- Lindholm E, Ekholm B, Shaw S, Jalonen P, Johansson G, Pettersson U, Sherrington R, Adolfsson R, Jazin E (2001) A schizophrenia-susceptibility locus at 6q25, in one of the world's largest reported pedigrees. Am J Hum Genet 69:96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek MS, Strahs A (1999) Assessment of linkage disequilibrium by the decay of haplotype sharing, with application to fine-scale genetic mapping. Am J Hum Genet 65:858–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mune T, Rogerson FM, Nikkila H, Agarwal AK, White PC (1995) Human hypertension caused by mutations in the kidney isozyme of 11 b-hydroxysteroid dehydrogenase. Nat Genet 10:394–399 [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Fain P, Kort E, Nee LE, Frommelt E, Polinsky RJ, Korenberg JR, Pulst SM (1993) Linkage of familial Alzheimer disease to chromosome 14 in two large early-onset pedigrees: effects of marker allele frequencies on lod scores. Am J Med Genet 48:63–66 [DOI] [PubMed] [Google Scholar]
- Nikali K, Soumalainen A, Terwilliger JD, Koskinen T, Weissenbach J, Peltonen L (1995) Random search for shared chromosomal regions in four affected individuals: the assignment of a new hereditary ataxia locus. Am J Hum Genet 56:1088–1095 [PMC free article] [PubMed] [Google Scholar]
- O'Donnell CJ, Lindpaintner K, Larson MG, Rao VS, Ordovas JM, Schaefer EJ, Myers RH, Levy D (1998) Evidence for association and genetic linkage of the angiotensin-converting enzyme locus with hypertension and blood pressure in men but not women in the Framingham Heart Study. Circulation 97:1766–1772 [DOI] [PubMed] [Google Scholar]
- Ombra MN, Forabosco P, Casula S, Angius A, Maestrale GB, Petretto E, Casu G, Colussi G, Usai E, Melis P, Pirastu M (2001) Identification of a new candidate locus for uric acid nephrolithiasis in a genetic isolate. Am J Hum Genet 68:1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1992) Strategies for characterizing highly polymorphic markers in human gene mapping. Am J Hum Genet 51:283–290 [PMC free article] [PubMed] [Google Scholar]
- Pankow JS, Rose KM, Oberman A, Hunt SC, Atwood LD, Djousse L, Province MA, Rao DC (2000) Possible locus on chromosome 18q influencing postural systolic blood pressure changes. Hypertension 36:471–476 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K (2000) Use of population isolates for mapping complex traits. Nat Rev Genet 1:182–190 [DOI] [PubMed] [Google Scholar]
- Perola M, Kainulainen K, Pajukanta P, Terwilliger JD, Hiekkalinna T, Ellonen P, Kaprio J, Koskenvuo M, Kontula K, Peltonen L (2000) Genome-wide scan of predisposing loci for increased diastolic blood pressure in Finnish siblings. J Hypertens 18:1579–1585 [DOI] [PubMed] [Google Scholar]
- Rice T, Rankinen T, Province MA, Chagnon YC, Perusse L, Borecki IB, Bouchard C, Rao DC (2000) Genome-wide linkage analysis of systolic and diastolic blood pressure: the Quebec Family Study. Circulation 102:1956–1963 [DOI] [PubMed] [Google Scholar]
- Schäffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Service SK, Lang DW, Freimer NB, Sandkuijl LA (1999) Linkage-disequilibrium mapping of disease genes by reconstruction of ancestral haplotypes in founder populations. Am J Hum Genet 64:1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S and Darvasi A (2001) The value of isolated populations. Nat Genet 28:309–310 [DOI] [PubMed] [Google Scholar]
- Shimkets RA, Warnock DG, Bositis CM, Nelson-Williams C, Hansson JH, Schambelan M, Gill JR Jr, Ulick S, Milora RV, Findling JW (1994) Liddle's syndrome: heritable human hypertension caused by mutations in the b-subunit of the epithelial sodium channel. Cell 79:407–414 [DOI] [PubMed] [Google Scholar]
- Simon DB, Bindra RS, Mansfield TA, Nelson-Williams C, Mendonca E, Stone R, Schurman S, Nayir A, Alpay H, Bakkaloglu A, Rodriguez-Soriano J, Morales JM, Sanjad SA, Taylor CM, Pilz D, Brem A, Trachtman H, Griswold W, Richard GA, John E, Lifton RP (1997) Mutations in the chloride channel gene, CLCNKB, cause Bartter's syndrome type III. Nat Genet 17:171–178 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Stoll M, Kwitek-Black AE, Cowley AW Jr, Harris EL, Harrap SB, Krieger JE, Printz MP, Provoost AP, Sassard J, Jacob HJ (2000) New target regions for human hypertension via comparative genomics. Genome Res 10:473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama F, Churchill GA, Higgins DC, Johns C, Makaritsis KP, Gavras H, Paigen B (2001) Concordance of murine quantitative trait loci for salt-induced hypertension with rat and human loci. Genomics 71:70–77 [DOI] [PubMed] [Google Scholar]
- Svetkey LP, Chen YT, McKeown SP, Preis L, Wilson AF (1997) Preliminary evidence of linkage of salt sensitivity in black Americans at the b2-adrenergic receptor locus. Hypertension 29:918–922 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Weiss KM (1998) Linkage disequilibrium mapping of complex disease: fantasy or reality? Curr Opin Biotechnol 9:578–594 [DOI] [PubMed] [Google Scholar]
- Timberlake DS, O'Connor DT, Parmer RJ (2001) Molecular genetics of essential hypertension: recent results and emerging strategies. Curr Opin Nephrol Hypertens 10:71–79 [DOI] [PubMed] [Google Scholar]
- Vasan RS, Larson MG, Leip EP, Evans JC, O'Donnell CJ, Kannel WB, Levy D (2001) Impact of high-normal blood pressure on the risk of cardiovascular disease. N Engl J Med 345:1291–1297 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Lange K (1988) The affected-pedigree-member method of linkage analysis. Am J Hum Genet 42:315–326 [PMC free article] [PubMed] [Google Scholar]
- Weeks DE, Ott J, Lathrop GM (1990) SLINK: a general simulation program for linkage analysis. Am J Hum Genet Suppl 47:A204 [Google Scholar]
- Wright AF, Carothers AD, Pirastu M (1999a) Population choice in mapping genes for complex diseases. Nat Genet 23:397–404 [DOI] [PubMed] [Google Scholar]
- Wright FA, O'Connor DT, Roberts E, Kutey G, Berry CC, Yoneda LU, Timberlake D, Schlager G (1999b) Genome scan for blood pressure loci in mice. Hypertension 34:625–630 [DOI] [PubMed] [Google Scholar]
- Zhu DL, Wang HY, Xiong MM, He X, Chu SL, Jin L, Wang GL, Yuan WT, Zhao GS, Boerwinkle E, Huang W (2001) Linkage of hypertension to chromosome 2q14-q23 in Chinese families. J Hypertens 19:55–61 [DOI] [PubMed] [Google Scholar]