Abstract
Smith-McCort dysplasia is a rare autosomal recessive osteochondrodysplasia characterized by short limbs and a short trunk with a barrel-shaped chest. The radiographic phenotype includes platyspondyly, generalized abnormalities of the epiphyses and metaphyses, and a distinctive lacy appearance of the iliac crest. We performed a genomewide scan in a consanguineous family from Guam and found evidence of linkage to loci on chromosome 18q12. Analysis of a second, smaller family was also consistent with linkage to this region, producing a maximum combined two-point LOD score of 3.04 at a recombination fraction of 0 for the marker at locus D18S450. A 10.7-cM region containing the disease gene was defined by recombination events in two affected individuals in the larger family. Furthermore, all affected children in the larger family were homozygous for a subset of marker loci within this region, defining a 1.5-cM interval likely to contain the defective gene. Analysis of three small, unrelated families with Dyggve-Melchior-Clausen syndrome, a radiographically identical disorder with the additional clinical finding of mental retardation, provided evidence of linkage to the same region, a result consistent with the hypothesis that the two disorders are allelic.
Smith-McCort dysplasia (SMC [MIM 223800]) is a rare autosomal recessive osteochondrodysplasia characterized by short limbs and a short trunk with a barrel-shaped chest (Spranger et al. 1976). The phenotype is not usually recognized at birth, but, by ∼18 mo of age, difficulty with feeding and deformity of the chest become apparent. Subsequently, poor musculature is noted, along with the evolution of short stature. With increasing age, patients may develop dorsal kyphosis, lumbar lordosis and scoliosis, genu valgus or varus deformity of the knees, and limited joint movement (Smith and McCort 1958; Dyggve et al. 1962; Kaufman et al. 1971; Spranger et al. 1976).
Radiographs show generalized abnormalities of the epiphyses and metaphyses. The extremities are short, and the hands show delayed and irregular ossification of the carpal bones, with accessory ossification centers in the metacarpals and phalanges (Spranger et al. 1976). There is platyspondyly, and the vertebral bodies exhibit a double-humped appearance with a central constriction. Odontoid hypoplasia, leading to atlanto-axial instability, is a common feature. The pelvis is small, with a distinctive and diagnostic lacy appearance of the iliac crest.
SMC (Spranger et al. 1976) is similar to Dyggve-Melchior-Clausen syndrome (DMC) (Dyggve et al. 1962), a related autosomal recessive osteochondrodysplasia. The radiographic findings in the two conditions are identical; however, patients with DMC also exhibit mental retardation, whereas there is no developmental delay in SMC (Spranger et al. 1976). SMC and DMC have been classified together in the International Nomenclature of Constitutional Disorders of Bone (International Working Group on Constitutional Diseases of Bone 1998) and share the same MIM number, reflecting the hypothesis that the two disorders are allelic. However, this hypothesis has not been presented in the published literature, in which the opposite statement has been made (Spranger et al. 1976; Beighton 1990).
Histological findings in SMC and DMC are also identical. Deposition of bone tissue in a wavy pattern at the osteochondral junction appears to explain the lacy appearance of the margins of the iliac crest seen on x-ray (Horton and Scott 1982; Nakamura et al. 1997). Growth plates show abnormal endochondral ossification with very poor chondrocyte columnization and with clusters of chondrocytes that include degenerating cells (Horton and Scott 1982; Nakamura et al. 1997). Electron microscopy of chondrocytes demonstrates dilated cisternae of rough endoplasmic reticulum that contains granular material in both SMC and DMC (Engfeldt et al. 1983; Nakamura et al. 1997).
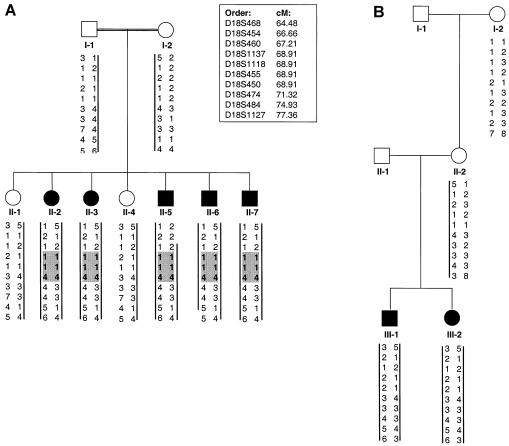
Linkage studies were performed in two families from Guam with SMC (fig. 1). The phenotype in both families shows typical features of SMC and has been described in a separate publication (Burns et al., in press). The parents in family SMC1 (International Skeletal Dysplasia Registry [ISDR] number R94-126) are both of Chamorro descent and are consanguineous; however, the exact degree of consanguinity could not be determined. The mother (II-2) in family SMC2 (ISDR number R98-116) is also of Chamorro ancestry, but no information was available on the ethnic background of the father (SMC2 II-1). No common ancestry could be found between the two families over the past three generations.
Figure 1.
Pedigrees of families from Guam with SMC dysplasia. Blackened symbols indicate affected individuals, and vertical bars indicate the haplotypes from the linked region on chromosome 18q that segregate with the disease. A, Family SMC1. The recombination interval was defined by recombination events observed for II-5 and II-6. Shaded areas indicate region of autozygosity. B, Family SMC2. The maternal disease haplotype is identical in the two families between markers D18S486 and D18S474.
After informed consent was provided, blood was obtained for preparation of genomic DNA. A genomewide scan was performed using DNA from all nine individuals from family SMC1, with 361 polymorphic microsatellite markers selected from ABI Prism Linkage Mapping Sets LD-20 and HD-5. Marker order and the genetic distances between markers were determined using the Marshfield and the University of California at Santa Cruz human genome maps.
Multiplex PCR analyses, using groups of three to five markers, were performed, and the products were separated by electrophoresis, using an ABI 373 DNA sequencer. Alleles were scored manually, using GENESCAN and GENOTYPER software (Applied Biosystems). Two-point LOD scores were calculated using either the MLINK program of the LINKAGE software package or the location-score option of the MENDEL software, version 4 (Lange et al. 2001), using an autosomal recessive disease model, under the assumption of an allele frequency of 0.0001 and complete penetrance. For missing parental-marker data, alleles were inferred from the genotypes of their offspring.
Evidence suggestive of linkage was obtained for the markers at three adjacent loci (18S468, D18S450, and D18S474) on chromosome 18q12. Because of discrepancies between the genetic and physical maps in this region, we used sequence and STS data from the National Center for Biotechnology Information and the Stanford Human Genome Center to assemble a contig and to independently determine locus order. Genotypes were then determined, in both SMC families, for markers at additional loci in this region. Two-point LOD scores are shown in table 1. The highest total LOD score, 3.04 at θ=0.00, was obtained for the marker at locus D18S450. Across the genome, no other markers were found with LOD scores >1.5.
Table 1.
Two-Point LOD Scores at Chromosome 18q Loci for Families with SMC
| LOD Score at θ = |
|||||||
| Markerand Family | .00 | .01 | .05 | .1 | .2 | .3 | .4 |
| D18S468: | |||||||
| SMC1 | −.35 | .61 | 1.11 | 1.17 | .94 | .58 | .20 |
| SMC2 | .50 |
.49 |
.43 |
.36 |
.22 |
.10 |
.27 |
| Total | .15 | 1.10 | 1.54 | 1.53 | 1.16 | .68 | .47 |
| D18S454: | |||||||
| SMC1 | −1.25 | −.29 | .22 | .29 | .19 | .09 | .02 |
| SMC2 | .30 |
.29 |
.24 |
.18 |
.10 |
.04 |
.01 |
| Total | −.95 | .00 | .46 | .47 | .29 | .13 | .03 |
| D18S460: | |||||||
| SMC1 | .00 | .00 | .00 | .00 | .00 | .00 | .00 |
| SMC2 | .46 |
.45 |
.39 |
.33 |
.20 |
.09 |
.02 |
| Total | .46 | .45 | .39 | .33 | .20 | .09 | .02 |
| D18S1137: | |||||||
| SMC1 | 2.65 | 2.61 | 2.39 | 2.12 | 1.54 | .91 | .30 |
| SMC2 | .30 |
.29 |
.24 |
.18 |
.10 |
.04 |
.01 |
| Total | 2.95 | 2.90 | 2.63 | 2.30 | 1.64 | .95 | .31 |
| D18S1118: | |||||||
| SMC1 | .85 | .84 | .78 | .71 | .53 | .32 | .10 |
| SMC2 | .16 |
.16 |
.14 |
.11 |
.07 |
.03 |
.01 |
| Total | 1.01 | 1.00 | .92 | .82 | .60 | .35 | .11 |
| D18S455: | |||||||
| SMC1 | 2.65 | 2.61 | 2.39 | 2.12 | 1.54 | .91 | .30 |
| SMC2 | .33 |
.32 |
.26 |
.21 |
.11 |
.05 |
.01 |
| Total | 2.98 | 2.93 | 2.65 | 2.33 | 1.65 | .96 | .31 |
| D18S450: | |||||||
| SMC1 | 2.65 | 2.61 | 2.39 | 2.12 | 1.54 | .91 | .30 |
| SMC2 | .39 |
.38 |
.33 |
.27 |
.17 |
.08 |
.02 |
| Total | 3.04 | 2.99 | 2.72 | 2.39 | 1.71 | .99 | .32 |
| D18S474: | |||||||
| SMC1 | 1.45 | 1.43 | 1.32 | 1.18 | .88 | .54 | .19 |
| SMC2 | .20 |
.20 |
.17 |
.14 |
.08 |
.04 |
.01 |
| Total | 1.65 | 1.63 | 1.49 | 1.32 | .96 | .58 | .20 |
| D18S484: | |||||||
| SMC1 | 1.45 | 1.43 | 1.32 | 1.18 | .88 | .54 | .19 |
| SMC2 | .50 |
.49 |
.43 |
.36 |
.22 |
.10 |
.02 |
| Total | 1.95 | 1.92 | 1.75 | 1.54 | 1.10 | .64 | .21 |
| D18S1127: | |||||||
| SMC1 | −1.55 | −.57 | .42 | .23 | .28 | .19 | .64 |
| SMC2 | .50 |
.49 |
.43 |
.36 |
.22 |
.10 |
.02 |
| Total | −1.05 | −.08 | .85 | .59 | .50 | .29 | .66 |
Recombination mapping defined a 10.7-cM region likely to contain the disease gene: a centromeric boundary at D18S454 was established by a recombination in individual SMC1 II-5, and a telomeric boundary at D18S1127 was established by a recombination in individual SMC1 II-6 (fig. 1). Since the parents in family 1 are both of Chamorro descent and acknowledged consanguinity, we expected evidence of autozygosity in their affected children. Genotypes for additional markers within the linked region demonstrated that the affected children were homozygous at three adjacent markers (D18S1137, D18S1118, and D18S455) within the interval defined by recombination events, thereby allowing the region expected to contain the disease gene to be narrowed to ∼1.5 cM (fig. 1). This inference was further supported by the observation that the mother in family SMC2, who is also of Chamorro descent, has the same disease haplotype between the markers at loci D18S468 and D18S474 (fig. 1). These data also suggest the possibility that the Chamorro population has multiple SMC carriers with this mutant founder chromosome.
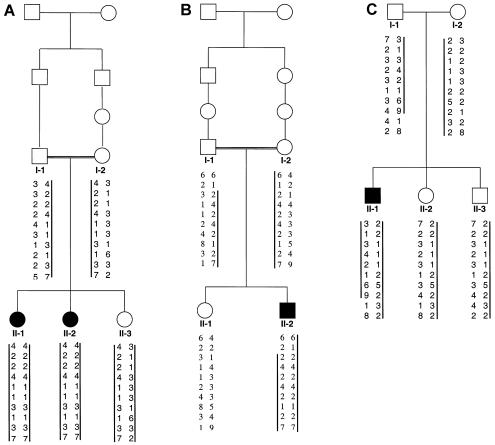
To evaluate the hypothesis that SMC and DMC are allelic phenotypes, we determined genotypes for markers at loci linked to SMC in three small nuclear families with DMC (fig. 2). The affected individuals in both families exhibited typical radiographic and clinical features of DMC, including significant developmental delay. The parents in family DMC1 (ISDR number R02-106) and DMC2 (ISDR number R94-187) are consanguineous and are of Pakistani and Dominican origin, respectively. The parents in family DMC3 (ISDR number R96-219) were not related. An inheritance pattern consistent with linkage was observed in all three families. The highest combined two-point LOD score was 1.6 at θ=0.00, which was obtained for the marker at locus D18S1118. The affected children in families DMC1 and DMC2 were homozygous across the entire 10.7-cM SMC region defined by recombination mapping. However, given the small size of the families, the data constitute only suggestive evidence that DMC is linked to the SMC region on chromosome 18.
Figure 2.
Pedigrees of families with Dyggve-Melchior-Clausen syndrome (DMC). A, Family DMC1, of Pakistani origin. B, Family DMC2, of Dominican origin. C, Family DMC3. Blackened symbols indicate affected individuals, and vertical bars indicate the haplotypes from the linked region on chromosome 18q that segregate with the disease.
The MADH2 gene is located in the 1.5-cM interval on chromosome 18q defined by homozygosity mapping. Since SMAD2, the MADH2 gene product, plays a role in chondrocyte development (Fujii et al. 1999; Ferguson et al. 2000), it was a reasonable candidate disease gene for SMC/DMC. We used published primer sequences (Takenoshita et al. 1998) for the 10 coding exons of MADH2 to amplify each fragment and determine its sequence. One patient from each family was used for the sequence analysis, but no mutations were observed in any of the individuals. These data rule out structural mutations in MADH2 as the cause of SMC or DMC in these families, but we cannot exclude reduced expression as the cause of these disorders. Apart from MADH2, there are 12 known or hypothetical genes in the 1.5-cM region of homozygosity, none of which are known to be expressed in chondrocytes. Expression studies on each of these genes will be used to prioritize analysis of the remaining candidate genes.
Our data provide evidence that the disease gene for SMC is within a 1.5-cM region on the long arm of chromosome 18, and they also provide evidence consistent with DMC being an allelic disorder. Identification of the disease gene is expected to define a new gene that is important for skeletal development and to provide insight into the function of the gene product in chondrocytes and other tissues, including the brain.
Acknowledgments
We thank the families for their participation in this research. Fiona Field was invaluable in helping to obtain the family material. We are grateful to Susan Napier for assistance with the linkage calculations. This work was supported, in part, by National Institutes of Health grant HD22657.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics (for genetic maps)
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/genome/guide/human/(for physical maps and marker information)
- University of California at Santa Cruz, http://genome.ucsc.edu/ (for physical maps and marker information)
- Stanford Human Genome Center, http://www.shgc-stanford.edu/ (for STS information)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
References
- Beighton P (1990) Dyggve-Melchior-Clausen syndrome. J Med Genet 27:512–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C, Powell BR, Hsia YE, Reinker K. Dyggve-Melchior-Clausen syndrome: report of 7 patients and review of the literature. J Pediatr Orthop (in press) [PubMed] [Google Scholar]
- Dyggve HV, Melchior JC, Clausen J (1962) Morquio-Ullrich's disease: an inborn error of metabolism? Arch Dis Child 37:525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engfeldt B, Bui T-H, Eklof O, Hjerpe A, Reinholt FP, Ritzen EM, Wilkstrom B (1983) Dyggve-Melchior-Clausen dysplasia: morphological and biochemical findings in cartilage growth zones. Acta Paediatr Scand 72:269–274 [DOI] [PubMed] [Google Scholar]
- Ferguson CM, Schwarz EM, Reynolds PR, Puzas JE, Rosier RN, O'Keefe RJ (2000) Smad2 and 3 mediate transforming growth factor-β1-induced inhibition of chondrocyte maturation. Endocrinology 141:4728–4735 [DOI] [PubMed] [Google Scholar]
- Fujii M, Takeda K, Imamura T, Aoki H, Sampath TK, Enomoto S, Kawabata M, Kato M, Ichijo H, Miyazono K (1999) Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol Biol Cell 10:3801–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton WA, Scott CI (1982) Dyggve-Melchior-Clausen syndrome: a histochemical study of the growth plate. J Bone Joint Surg Am 64:408–415 [PubMed] [Google Scholar]
- International Working Group on Constitutional Diseases of Bone (1998) International nomenclature and classification of the osteochondrodysplasias (1997). Am J Med Genet 79:376–382 [DOI] [PubMed] [Google Scholar]
- Kaufman RL, Rimoin DL, McAlister WH (1971) The Dyggve-Melchior-Clausen syndrome. Birth Defects Orig Artic Ser 7:144–149 [PubMed] [Google Scholar]
- Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E (2001) Mendel version 4.0: a complete package for the exact analysis of discrete traits in pedigree and population data sets. Am J Hum Genet 69:A504 [Google Scholar]
- Nakamura K, Kurokawa T, Nagano A, Nakamura S, Taniguchi K, Hamazaki M (1997) Dyggve-Melchior-Clausen syndrome without mental retardation (Smith-McCort dysplasia): morphological findings in the growth plate of the iliac crest. Am J Med Genet 72:11–17 [PubMed] [Google Scholar]
- Smith R, McCort JJ (1958) Osteochondrodystrophy (Morquio-Brailsford type): occurrence in three siblings. Calif Med 88:55–59 [PMC free article] [PubMed] [Google Scholar]
- Spranger JW, Bierbaum B, Herrmann J (1976) Heterogeneity of Dyggve-Melchior-Clausen dwarfism. Hum Genet 33:279–287 [DOI] [PubMed] [Google Scholar]
- Takenoshita S, Tani M, Mogi A, Nagashima M, Nagamachi Y, Bennett WP, Hagiwara K, Harris CC, Yokota J (1998) Mutation analysis of the Smad2 gene in human colon cancers using genomic DNA and intron primers. Carcinogenesis 19:803–807 [DOI] [PubMed] [Google Scholar]