Abstract
The aggregation of Aβ-peptide (Aβ) is widely considered to be the critical step in the pathology of Alzheimer's disease. Small, soluble Aβ oligomers have been shown to be more neurotoxic than large, insoluble aggregates and fibrils. Recent studies suggest that biometal ions, including Zn(II), may play an important role in the aggregation process. Experimentally determining the details of the binding process is complicated by the kinetic lability of zinc. To study the dynamic nature of the zinc-bound Aβ complexes and the potential mechanisms by which Zn(II) affects Aβ oligomerization we have performed atomistic molecular dynamics (MD) simulations of Zn(Aβ) and Zn(Aβ)2. The models were based on NMR data and predicted coordination environments from previous density functional theory calculations. When modeled as 4-coordinate covalently bound Zn(Aβ)n complexes (where n = 1 or 2), zinc imposes conformational changes in the surrounding Aβ residues. Moreover, zinc reduces the helix content and increases the random coil content of the full peptide. Although zinc binds at the N-terminus of Aβ, β-sheet formation is observed exclusively at the C-terminus in the Zn(Aβ) and most of the Zn(Aβ)2 complexes. Furthermore, initial binding to zinc promotes the formation of intra-chain salt-bridges, while subsequent dissociation promotes the formation of inter-chain salt-bridges. These results suggest that Zn-binding to Aβ accelerates the aggregation of Aβ by unfolding the helical structure in Aβ peptide and stabilizing the formation of vital salt-bridges within and between Aβ peptides.
Introduction
Alzheimer's disease is the most common neurodegenerative disorder, impacting more than 35 million people worldwide [1]. The aggregation of amyloid β-peptide (Aβ) is the major pathological event that takes place in Alzheimer's disease [2]. The pathologically relevant Aβ fragments are Aβ(1-40) and Aβ(1-42) with 40 or 42 residues respectively: DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIA. Both can form soluble monomers, oligomers, insoluble tangles and amyloid plaques. Aβ(1-42) is more prone to aggregation than Aβ(1-40) [3]. Although Aβ(1-40) is the predominant form of Aβ produced, Aβ(1-42) is the major component of amyloid deposits in senile plaques [4]. Also Aβ(1-42) promotes amyloid deposition but Aβ(1-40) inhibits the process [5]. The soluble forms of Aβ(1-42) are the most toxic species that cause neuronal damage in the brains of Alzheimer's disease patients [6]. Although the detailed mechanism of Aβ aggregation is not known, the amyloid cascade hypothesis suggests that the pathology of Alzheimer's disease arises due to an imbalance between the production and clearance of Aβ [7]. Moreover, the small soluble oligomers of Aβ are allegedly the toxic species and not the amyloid fibrils [8].
Physiological examinations have revealed that disrupted biometal homeostasis is an indication of Alzheimer's disease progression [9]. Tissue analysis showed that the concentrations of transition metal ions in neuropil (normal vs. Alzheimer's disease) are: Zn2+ (346 µM vs. 786 µM), Cu2+ (69 µM vs. 304 µM) and Fe3+ (338 µM vs. 695 µM) [10]. Also, the most recent quantitative analysis in the brain tissue of Alzheimer's disease patients revealed that, compared with the surrounding tissue, concentrations of Fe3+, Cu2+ and Zn2+ in the amyloid deposits are greater by factors of 2, 2.6 and 2.5 respectively [11]. Moreover, in vitro experiments have shown that Aβ binds to these metals with relatively high affinity and metal ions generally promote the aggregation of Aβ [12], [13]. Furthermore, metal chelators have been shown to dissolve the proteinaceous deposits from the postmortem brain tissues of people who suffered from Alzheimer's disease [14], [15].
Of the three transition metals that have increased concentrations in Alzheimer's disease, zinc has a much higher extracellular concentration (up to 200 µM) [11] and it has been found at up to 1 mM concentration in amyloid plaques. Zn(II) is labile with a rapid ligand exchange rate in general. Time-resolved spectroscopic and structural experiments suggest that Zn(II) promotes aggregation more strongly through intermolecular bridges forming Zn(Aβ)2 species [16]. Studies have revealed that the residues in Aβ that coordinate to Zn(II) are found at the N-terminus, Aβ(1-16), including Asp1, His6, Glu11, His13 and His14 [17]–[35]. However, the β-sheet components of Aβ fibrils were found on the C-terminus, Aβ(17-42) [36]. To date, there is no available X-ray crystal structure for zinc-bound oligomers. However, there is an NMR solution structure (PDB: 1ZE9) [20] for the monomeric Zn-Aβ(1-16) complex, in which the peptide coordinates were obtained from NMR measurements and the zinc coordinates were found from molecular modeling [20]. In addition, NMR structures for monomeric Aβ (PDB: 1Z0Q) [37] and Aβ fibril (PDB: 2BEG) [38] are available.
In this work, molecular dynamics simulations were performed on structures of both Zn(Aβ) and Zn(Aβ)2 with different combinations of coordinating residues including the three N-terminal histidines (His6, 13, 14) and the glutamic acid (Glu11) generated from previous quantum mechanics calculations and NMR results (PDB: 1Z0Q) [37]. Our molecular dynamics simulations show how zinc binding affects the secondary structure of Aβ in the N-terminus and influences the overall conformation of the peptide. Also, our simulations show how peptide-peptide interactions promote changes in secondary structure in the presence of zinc and after the loss of zinc.
Methods
QM calculations on Zn(Aβ)n (where n = 1 or 2) complexes
For Zn(Aβ) complexes there is one available NMR solution structure (PDB: 1ZE9) [20], to which we can compare our geometric parameters. In this structure, Zn2+ binds to four ligands including three histidines (His6, 13 and 14) and one glutamic acid (Glu11). In molecular dynamics simulations using classical force-fields, metal ions are often treated as free ions that are only “bound” via electrostatic interactions. This method gives all coordination complexes octahedral geometry, which does not match zinc's coordination sphere found from the NMR result for Zn(Aβ). Statistics have shown that though other coordination numbers and geometries have been observed, zinc's coordination environment is most often a slightly distorted tetrahedral four-coordinate complex when bound to proteins [39]–[41]. In our study, the force field parameters for the Zn-peptide bonds were calculated from density functional theory (DFT) [42] calculations on model complexes including [Zn(Im)3(Ac)]+, Zn(Im)2(Ac)2, and Zn(Im)4 (where Im = imidazole and Ac = acetate). Geometry optimizations were performed using the popular B3LYP [43], [44] density functional with the 6-31+G* [45], [46] basis set. All DFT calculations were performed with Gaussian 09 [47]. Force constants for the Zn coordination sphere in each structure were generated from the Hessian matrix obtained from vibrational frequency analysis and were added to the CHARMM 22 force field with CMAP corrections (CHARMM 22/CMAP) [48] (Table 1) for the molecular dynamics simulations.
Table 1. Percentage of Time the Asp23-Lys28 Salt Bridge Is Intact.
| Structures | Inter-chain | Intra-chain |
| Zn(Aβ) | N/A | 48.1 |
| Control (Aβ) | N/A | 0.5 |
| Zn(Aβ)2 at His6 Glu11 | 0.0 | 46.7 |
| Control (Aβ)2 | 0.0 | 33.3 |
| Zn(Aβ)2 at Glu11 His13 | 0.0 | 44.8 |
| Control (Aβ)2 | 24.5 | 0 |
| Zn(Aβ)2 at Glu11 His14 | 0 | 33.7 |
| Control (Aβ)2 | 31.8 | 5.8 |
| Zn(Aβ)2 at His13 His14 | 0.0 | 0.0 |
| Control (Aβ)2 | 0.0 | 26.6 |
The data show the percentage of time after equilibration that the Asp23-Lys28 salt bridge is intact in each set of simulations. The salt bridge is defined as intact when the O(Asp23)-N(Lys28) distance is ≤4 Å. All results are the average values of three runs for each simulation. Inter- and intra-chain salt bridge percentages were measured separately.
MD simulations on Zn(Aβ)n (where n = 1 or 2) complexes
Aβ(1-42) (PDB: 1Z0Q) [37] models were created for all molecular dynamics simulations. In general, the initial structures for Zn(Aβ)n (where n = 1 or 2) complexes were created by using the experimental data and our aforementioned DFT calculations. The initial structure for Zn(Aβ) was created by using the NMR structure for Zn(Aβ(1-16)) (PDB: 1ZE9) [20] as the N-terminus and combining it with residues 17-42 from Aβ(1-42) (PDB: 1Z0Q) [37]. The Zn(II) was covalently bound to His6, 13, 14 and Glu11 as found by NMR, using the force-constants obtained from our DFT calculations. For the Zn(Aβ)2 complexes, quantum mechanics calculations revealed that the ligand combinations (2 Glu and 2 His) and 4 His are relatively stable dimeric cross linkages (at pH 7.0). Thus, there are six different possible dimeric structures (Glu11/His6, Glu11/His13, Glu11/His14, His6/His13, His6/His14 and His13/His14). To model the His6/His13 and His6/His14 combinations would have required changing the backbone from what was observed in the NMR. Therefore, the His6/His13 and His6/His14 combinations were not included in this study. Control models were built by removing the Zn(II) from the Zn-bound complexes, thereby keeping the same initial structures (prior to minimization) for the peptides. In this manner, it is possible to show how zinc's effect on the peptide can persist even after dissociation. Ten models were built and simulated. We used all-atom simulations with explicit water solvent (TIP3P) for the systems with our augmented CHARMM 22/CMAP force field (Table S1). We performed conjugate gradient energy minimization on the solvated system, followed by 1.5 ns simulation during which thermal energy was added to the systems gradually, which increased the temperature from 0 to 300 K. Next, we performed 200 ns simulations in triplicate for each complex at constant temperature and pressure (NPT, 1 atm, 300 K) using the program NAMD 2.7 [49]. All data analysis was done using VMD 1.9 [50].
Root mean square deviations (RMSD) of the Cα atoms were measured (Figure S1) for all simulations to evaluate the extent of equilibration. To compare the flexibilities of different regions of Aβ, root mean square fluctuations (RMSF) were measured (Figure S2) for the Cα atoms after the simulations equilibrated. Secondary structure changes over time were measured (using the STRIDE plugin as implemented in VMD [50]) for all peptide species in terms of total percentage secondary structure composition within each peptide chain. Also, we measured the percentage of time each residue exhibited either helix or β-sheet structures after equilibration. The radius of gyration was measured over time in each simulation to determine how binding to zinc affects how compact the peptides are. Salt bridges (Table 1) over time were measured to reveal certain intra-chain and inter-chain interactions.
Results
The trajectories of all the species over the 200 ns of simulation time are shown in Figures 1, 2, 3, 4, and 5. For Zn(Aβ) (Figure 1), Zn binding disturbed the helix in the N-terminus, and those areas remained random coil throughout the simulation, whereas the controls, which model the behavior of Aβ after Zn dissociates, regain helical structure in the N-terminus. For the Zn(Aβ)2 complexes with Zn bridging at His 6 and Glu11 (Figure 2), the zinc forced the two peptide chains to adopt a perpendicular orientation to accommodate the local Zn-binding residues. In the initial structure, the Zn binding brought the N-termini of the two peptides together, whereas the C-termini remained separated from each other. However, after equilibration, the C-termini of the peptide chains had moved and were in contact. For the control system, which started with the same initial structure as the Zn(Aβ)2 (His6/Glu11), both the N- and C-termini of the peptides moved closer to each other despite the absence of a Zn linkage. A stable intra-chain β-sheet was formed at the C-terminus of the control simulation after 50 ns. For Aβ dimers with Zn bridging at Glu11 and His13 (Figure 3), the peptide chains remained parallel due to the constraints imposed by the Zn-binding residues. Both peptides remained in contact with each other and maintained helical structures despite Zn-binding. In both the Zn-bound and the control simulations, the peptides acquired β-sheet structures at their respective C-termini after 100 ns. In the Zn-bound simulation, the peptides formed a single intra-chain β-sheet, while in the control simulation there was an inter-chain β-sheet. For the peptide dimers with Zn bridging at Glu11 and His14 (Figure 4), the peptides are forced into a 45° angle with respect to each other due to constraints imposed by binding to Zn and regional conformations from the Aβ(1-16) NMR structure. The peptides in the Zn-bound and control simulations both lost helix at their respective N-termini. β-sheet formation was observed at the C-terminus of one of the peptides in the control simulations. For Aβ dimers with Zn bridging at His13 and His14 (Figure 5), the two peptide chains are parallel. The peptides in the Zn-bound system lost helix at the N- and C-termini, while the peptides in the control system regained helix during the simulations. In general, once zinc binds and bridges two peptides, those peptides remain in contact even after zinc dissociates and leaves the binding site. Peptides that are not arranged in a parallel configuration tend to lose helical structure while parallel peptide configurations stabilize the helix. All β-sheet structures were observed at the C-terminus of Aβ, which suggests that this region initiates changes in secondary structure that lead to peptide aggregation.
Figure 1. Trajectories of Simulations of Zn(Aβ) and Control.
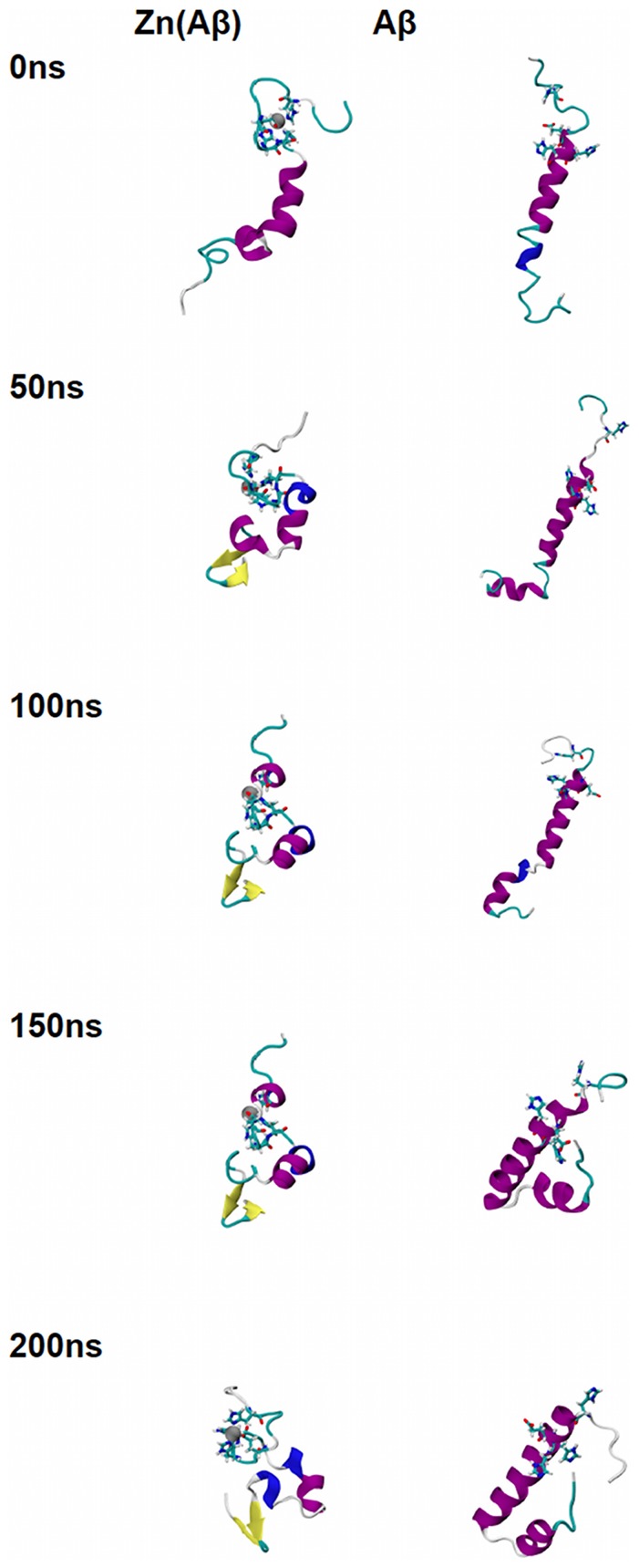
Structures of Zn(Aβ) (where Zn binds at Glu11, His6, 13 14) and the control (Aβ) at 0 ns, 50 ns, 100 ns, 150 ns and 200 ns are shown. The peptide has the same initial structure in the Zn(Aβ) complex and the control simulation. Gray spheres are zinc atoms. Zn-binding residues including Glu11, His6, 13, 14 are shown in licorice, where red, blue and cyan colors shows the atoms and bonds for oxygen, nitrogen and carbon respectively. Peptide backbones are shown in cartoon style where cyan indicates turn, white is coil, purple is alpha-helix, blue is 310 helix, and yellow is β-sheet. All simulations were performed in explicit water. Water molecules are not shown for clarity. Each is peptide oriented with the N-terminus at the top and C-terminus towards the bottom.
Figure 2. Trajectories of Simulations of Zn(Aβ)2 (Glu11 and His6) and Control.
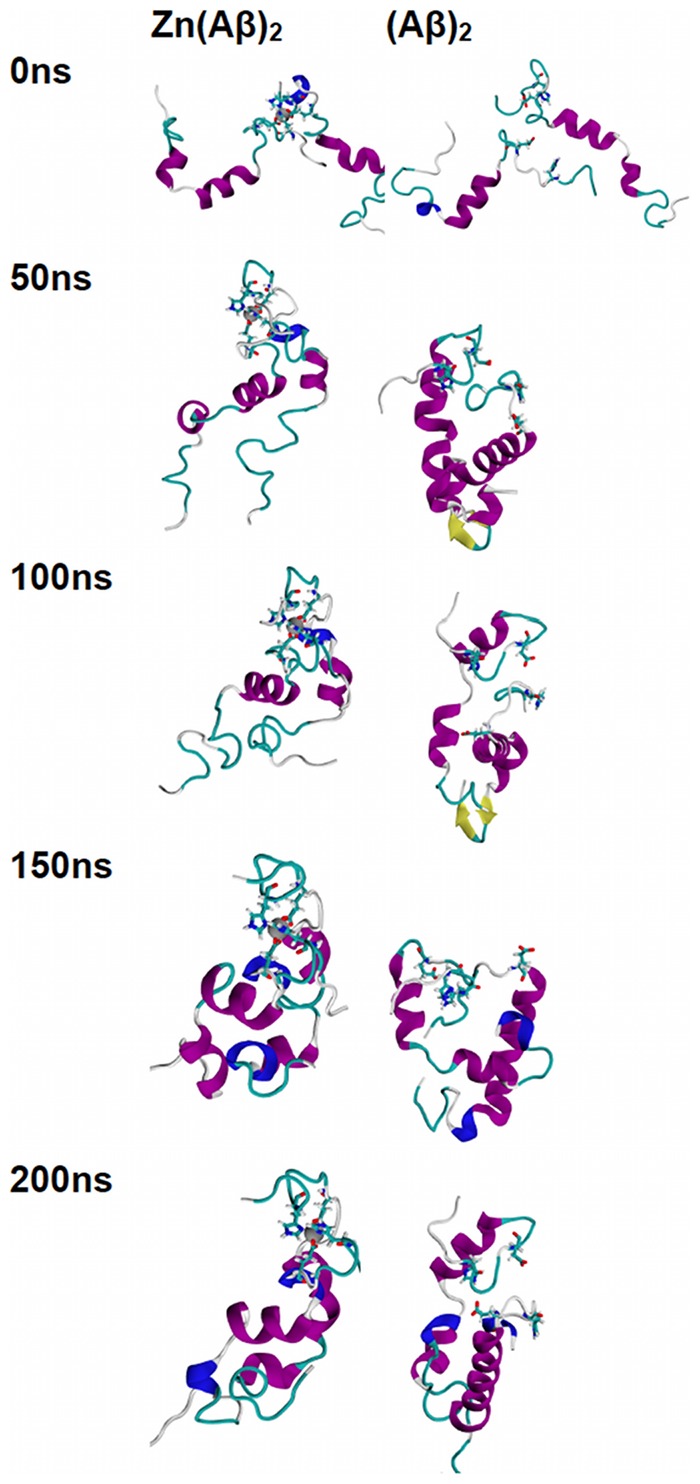
Structures of Zn(Aβ)2 (where Zn bridges at Glu11 and His6), and the control (Aβ)2 at 0 ns, 50 ns, 100 ns, 150 ns and 200 ns are shown. The peptides have the same initial structure in the Zn(Aβ)2 complex and the control simulation. Gray spheres are zinc atoms. Zn-binding residues including Glu11 and His6 are shown in licorice, where red, blue and cyan colors shows the atoms and bonds for oxygen, nitrogen and carbon respectively. Peptide backbones are shown in cartoon style where cyan indicates turn, white is coil, purple is alpha-helix, blue is 310 helix, and yellow is β-sheet. All simulations were performed in explicit water. Water molecules are not shown for clarity. Each is peptide oriented with the N-terminus at the top and C-terminus towards the bottom.
Figure 3. Trajectories of Simulations of Zn(Aβ)2 (Glu11 & His13) and Control.
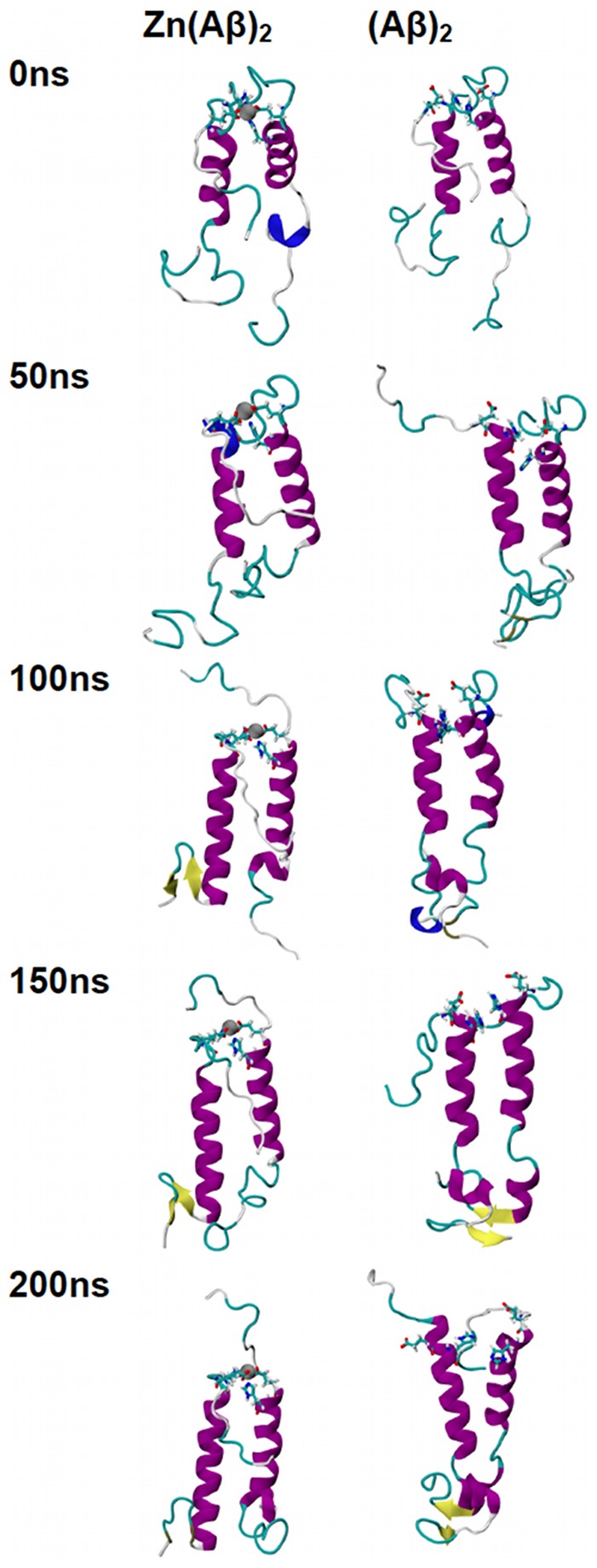
Structures of Zn(Aβ)2 (where Zn bridges at Glu11 and His13), and the control (Aβ)2 at 0 ns, 50 ns, 100 ns, 150 ns and 200 ns are shown. The peptides have the same initial structure in the Zn(Aβ)2 complex and the control simulation. Gray spheres are zinc atoms. Zn-binding residues including Glu11 and His13 are shown in licorice, where red, blue and cyan colors shows the atoms and bonds for oxygen, nitrogen and carbon respectively. Peptide backbones are shown in cartoon style where cyan indicates turn, white is coil, purple is alpha-helix, blue is 310 helix, and yellow is β-sheet. All simulations were performed in explicit water. Water molecules are not shown for clarity. Each is peptide oriented with the N-terminus at the top and C-terminus towards the bottom.
Figure 4. Trajectories of Simulations of Zn(Aβ)2 (Glu11 & His14) and Control.
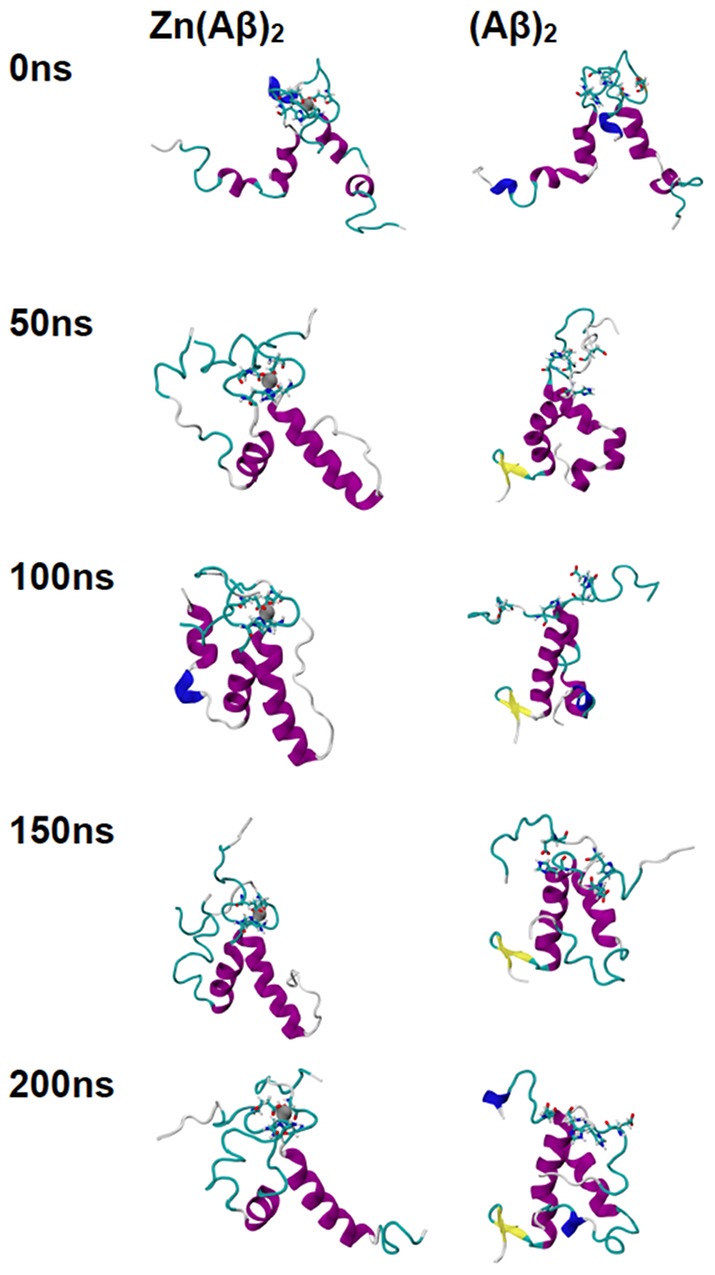
Structures of Zn(Aβ)2 (where Zn bridges at Glu11 and His14), and the control (Aβ)2 at 0 ns, 50 ns, 100 ns, 150 ns and 200 ns are shown. The peptides have the same initial structure in the Zn(Aβ)2 complex and the control simulation. Gray spheres are zinc atoms. Zn-binding residues including Glu11 and His14 are shown in licorice, where red, blue and cyan colors shows the atoms and bonds for oxygen, nitrogen and carbon respectively. Peptide backbones are shown in cartoon style where cyan indicates turn, white is coil, purple is alpha-helix, blue is 310 helix, and yellow is β-sheet. All simulations were performed in explicit water. Water molecules are not shown for clarity. Each is peptide oriented with the N-terminus at the top and C-terminus towards the bottom.
Figure 5. Trajectories of Simulations of Zn(Aβ)2 (His13 & His14) and Control.
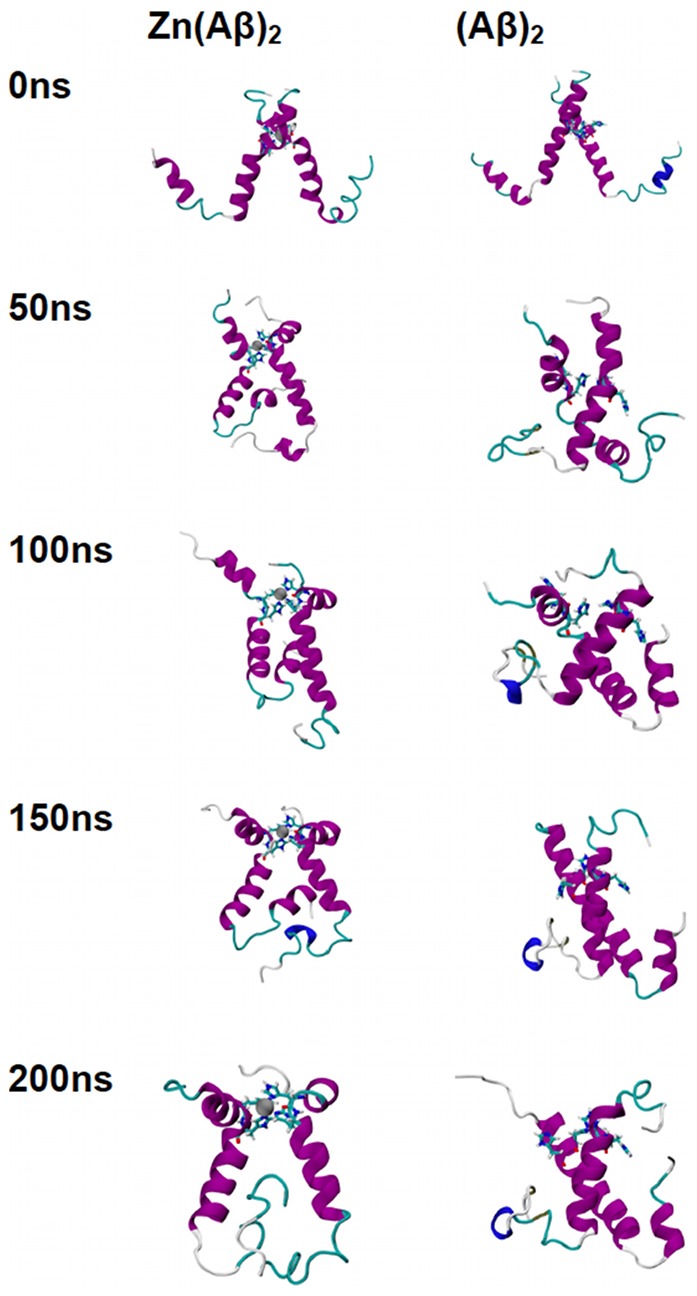
Structures of Zn(Aβ)2 (where Zn bridges at His13 and His14), and the control (Aβ)2 at 0 ns, 50 ns, 100 ns, 150 ns and 200 ns are shown. The peptides have the same initial structure in the Zn(Aβ)2 complex and the control simulation. Gray spheres are zinc atoms. Zn-binding residues including His13 and His14 are shown in licorice, where red, blue and cyan colors shows the atoms and bonds for oxygen, nitrogen and carbon respectively. Peptide backbones are shown in cartoon style where cyan indicates turn, white is coil, purple is alpha-helix, blue is 310 helix, and yellow is β-sheet. All simulations were performed in explicit water. Water molecules are not shown for clarity. Each is peptide oriented with the N-terminus at the top and C-terminus towards the bottom.
Analysis of root mean square deviations of each simulation (Figure S1), shows that all systems achieve at least 80 ns of equilibrated dynamics. For the monomers, both the control and the Zn-bound Aβ simulations reach equilibrium after 120 ns. For the dimers, the four Zn-bound simulations and their respective control simulations all reach equilibrium before 100 ns.
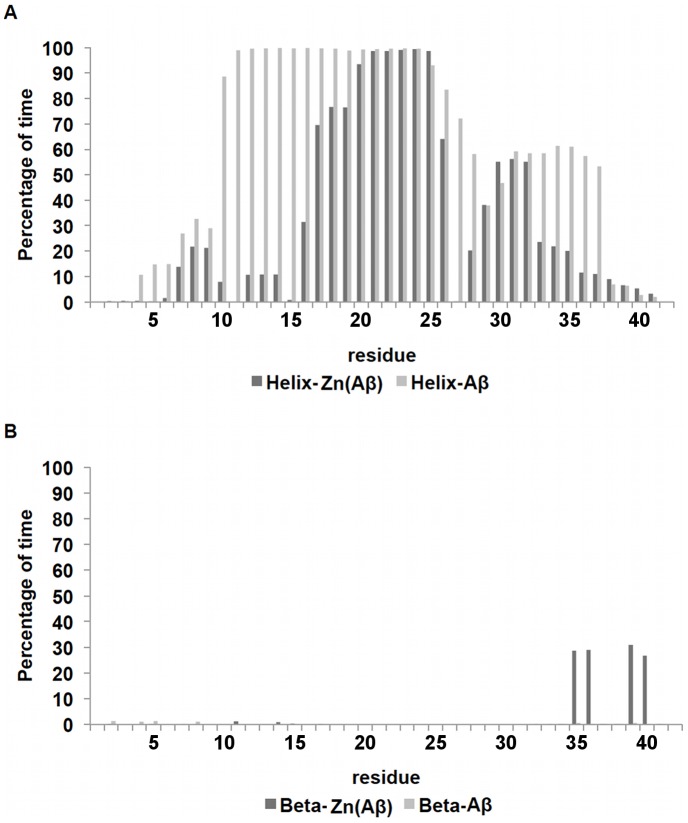
The total percentage of secondary structure composition (Figure 6) shows that Zn-binding results in a lower total helix percentage after equilibration, but increased β-sheet percentage at the C-termini of various species. Analysis of the per-residue-helicity for each peptide (Figure 7 and Figure S3), shows that for the monomeric species, Zn binding caused the loss of helix structure from residue 6 to residue 14, which extended the random coil structure from residue 1 to residue 20. On the other hand, the control peptide remains mostly helical from residue 6 to the C-terminus, with the exception of residues N27KGA30 and the last three residues. Both systems possess the loop region N27KGA30 which is the same loop found in the Aβ fibril structure [44]. For the dimeric complexes (Figure S3), the N- and C-termini show a decrease of helical structures in general. However, the total percentage of helix at the C-terminus only showed a modest decrease. Interestingly, for all simulations of Zn(Aβ)2, the decrease in helix occurred near residue Asn27 which is in the loop that is vital to the formation of β-sheet structure in the Aβ fibrils. Similarly, the formation of β-sheet among all the dimer simulations occurred in the same region, G33LMVGGVV40, regardless of whether it was inter-chain or intra-chain β-sheet, which suggests a common mechanism for the secondary structure transition.
Figure 6. Total Percentage of Secondary Structure.
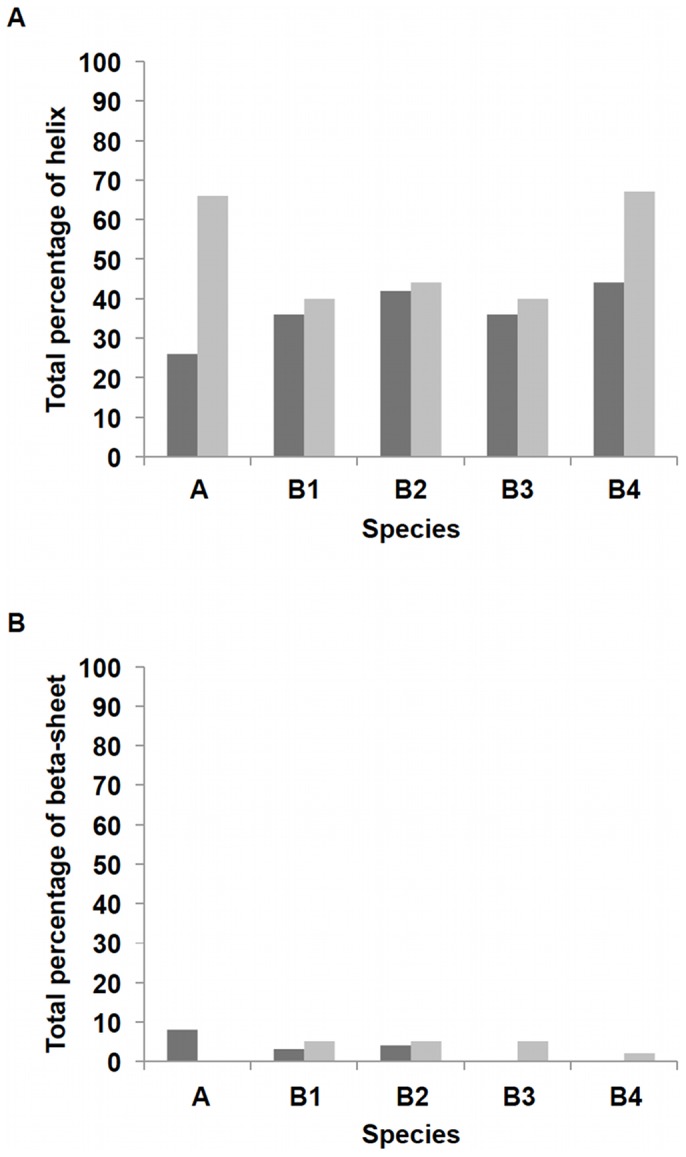
Percentages of helix and β-sheet for each chain after equilibrium are shown. The peptides in each Zn-bound complex and their corresponding controls have the same initial structures. A shows Zn(Aβ) (where Zn binds at Glu11 His6, 13 14) and control. B1 shows Zn(Aβ)2 (where Zn bridges at Glu11 and His6) and control. B2 shows Zn(Aβ)2 (where Zn bridges at Glu11 and His13) and control. B3 shows Zn(Aβ)2 (where Zn bridges at His13 and His 14) and control. B4 shows Zn(Aβ)2 (where Zn bridges at Glu11 and His14) and control. Dark gray lines are for Zn-bound complexes. Gray lines are for controls.
Figure 7. Per-Residue Secondary Structure of Zn(Aβ) and Control.
The percentage of time (after equilibration) peptide residues adopt helix or β-sheet structure is shown. (A) shows the per-residue helix. (B) shows the per-residue β-sheet. Dark gray lines are Zn(Aβ) where Zn binds at Glu11 His6, 13 14. Gray lines are the control. The peptide has the same initial structure in the Zn(Aβ) complex and the control simulation.
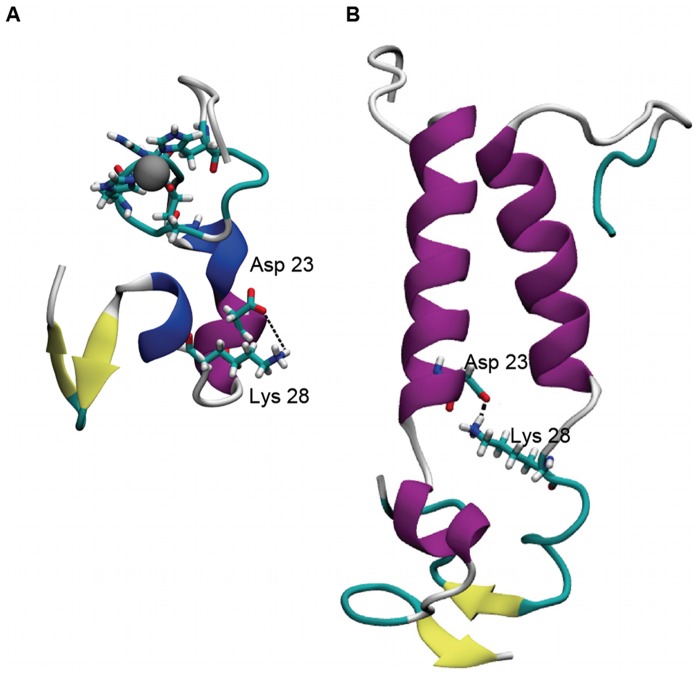
The disruption of the helix structures in the N27KGA30 region facilitates the formation of the Asp23-Lys28 salt bridge (Figure 8) that stabilizes the Aβ fibrils. Hence, we analyzed this salt bridge over time and the results are shown in Table 1. For the monomeric species, the Zn-bound Aβ showed a constant intra-chain salt bridge Asp23-Lys28 formation over 48.1% of time whereas in the control species the salt bridge did not form. Zn binding promoted the formation of the salt bridge. For the Zn(Aβ)2 complexes, various intra-chain and inter-chain Asp23-Lys28 salt bridges were observed for both Zn(Aβ)2 and (Aβ)2 species (Table 1). In general, the intra-chain salt bridge helps maintain the loop N27KGA30. The inter-chain salt bridges enhance the peptide-peptide interactions and keep both peptides in contact even without the Zn linkage.
Figure 8. Intra- and Inter-chain Salt Bridge (Asp23-Lys28).
Structure A shows an intra-chain salt bridge between Asp23 and Lys28 in Zn(Aβ) at 200 ns. Structure B shows an inter-chain salt bridge between Asp23 and Lys28 in Zn(Aβ)2 with Zn bridging at Glu11 and His13 at 200 ns. Gray spheres are Zn. Zn-binding residues (A: Glu11, His6, 13, 14; B:Glu11 and His13) are represented in licorice where red, blue and cyan represent oxygen, nitrogen and carbon respectively. Peptide backbones are shown in cartoon style where cyan indicates turn, white is coil, purple is alpha-helix, blue is 310 helix, and yellow is β-sheet.
The radius of gyration analysis reveals the general compactness of the peptide complexes. Comparison between Zn-bound and control species (Figure 9) showed that Zn binding results in a slightly more compact conformation during equilibrium. For proteins with the same number of residues, those with α helix structures have the largest radii of gyration [51]. In general, decreases in the radius of gyration reflect the loss of helix.
Figure 9. Average Equilibrium Radius of Gyration (Rygr) (Å).
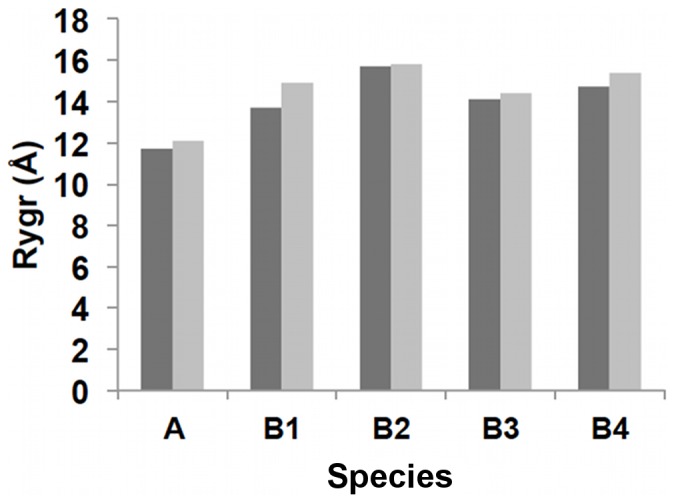
Average equilibrium radii of gyration are shown. A shows Zn(Aβ) (where Zn binds at Glu11 His6, 13 14) and control. B1 shows Zn(Aβ)2 (where Zn bridges at Glu11 and His6) and control. B2 shows Zn(Aβ)2 (where Zn bridges at Glu11 and His13) and control. B3 shows Zn(Aβ)2 (where Zn bridges at His13 and His 14) and control. B4 shows Zn(Aβ)2 (where Zn bridges at Glu11 and His14) and control. Dark gray lines are for Zn-bound complexes. Gray lines are for controls.
Discussion
Aβ peptide is one of the intrinsically disordered proteins that have no native tertiary structure. Therefore, its conformation largely depends on the complex and often transient interactions between multiple environmental factors. For example, the free soluble Aβ peptide in water/hexafluoroisopropanol (HFIP) 4∶1 solution [37] is mostly helical whereas the fibril Aβ is dominated by β-sheet [38]. The transformation of helix to β-sheet needs to go through the unfolding of helical turns (to coil) and refolding (to β-sheet). Previous experiments showed that the ratio of zinc: Aβ in senile plaques ranges from 1∶1 to 1∶200 [52]. The Zn(Aβ) simulation results suggests that if zinc: Aβ has a 1∶1 ratio, zinc binding to Aβ monomer can disrupt the helix structure not only on the zinc binding regions but also in both the N- and C-terminus. Kinetically, the ligand exchange rate of zinc is greater than that of most other first row transition metals [53]. Therefore, zinc is constantly associating to and dissociating from Aβ. The results of the Zn-bound and control Aβ dimer models demonstrate the influence of Zn binding and its extended effects on the peptide conformation after dissociation. In this work, Zn was covalently bound to Aβ peptides to simulate a sufficient concentration of Zn such that Aβ peptides would be effectively coordinated for enough time to form Aβ-Aβ interactions. Once these interactions form, even if Zn(II) subsequently dissociates, conformational changes in the C-terminus can still occur. Although the peptides in the Zn-bound and the control Aβ dimer simulations have similar behavior, they do differ in the nature of the salt-bridges that they form. In particular, the Zn-bound dimer simulations show that formation of the critical D23/K28 intra-chain salt-bridge occurs and persists with the exception of the Zn(Aβ)2 (His13/His14) case. The formation and stabilization of the intra-chain D23/K28 salt-bridge has been shown to be important in accelerating the fibrillation of Aβ(1-40) [54]. Nevertheless, none of the Zn-bound dimer simulations shows the formation of inter-chain D23/K28 salt-bridges. Interestingly, inter-chain D23/K28 salt-bridges do form in the simulations for controls for the Zn(Aβ)2 (Glu11/His13) and (Glu11/His14) simulations. It worth noting that inter-chain D23/K28 salt-bridges have been observed by NMR for the Aβ(1-40) fibrils [55]. This suggests that zinc accelerates fibrillation of Aβ at least in part as a result of its lability. By binding and dissociating from Aβ, Zn alternately promotes the formation of intra- and inter-chain D23/K28 salt-bridges, respectively.
In all of the simulations, β-sheet formation is observed exclusively at the C-terminus, G31LMVGGVVI42, which suggests that the peptide's secondary structure transition starts in this region. This result is in agreement with a recent study on the assembly processes of Aβ(1-42) and Aβ(1-40) [56], which showed that β-sheet structures were observed in the G31LMVGGVVI42 region for Aβ(1-42). In addition, it was shown that C-terminal β-sheet formation is positively correlated to the increased toxicity of Aβ peptide species.
Our results suggests that a sufficient concentration Zn(II) ions can promote the oligomerization of Aβ by disrupting the helical structures in the N-terminus, inducing β-sheet formation in the C-terminus and promoting the formation of both intra- and inter-chain salt-bridges. These results agree with the observation that zinc promotes rapid aggregation with both parallel and non-parallel assembly and form non-fibril aggregated species [41]. The simulations in this work shows how Zn binding to Aβ can promote and accelerate the helix-to-β-sheet transition in this region, which might explain how zinc enhances the toxicity of Aβ.
Supporting Information
Root Mean Square Deviations (Cα only) for each Simulation. For each simulation, root mean square deviation (RMSD) is calculated for the entire trajectory using the first trajectory as the reference. Gray lines are Zn-bound complexes. Dark gray lines are controls.
(DOCX)
Root Mean Square Fluctuation (Cα only) of each Simulation. For each simulation, root mean square fluctuation (RMSF) is calculated for the equilibrated portion of the simulation (last 80 ns). Gray lines are Zn-bound complexes. Dark gray lines are controls.
(DOCX)
Per-Residue Helix and Beta-sheet Content for each Zn(Aβ)2. All per-residue secondary structure results are average values of all dimeric species over three runs for each simulation. Gray lines are Zn-bound complexes. Dark gray lines are controls.
(DOCX)
Force Constants for Zn and Coordinating Residues. List of force constants between zinc and the coordinating atoms obtained from DFT calculations at the B3LYP/6-31+G* level that were added to the CHARMM 22/CMAP force field parameters. The force-field potential energy as found in CHARMM22 is as follows: BONDS: V(bond) = Kb(b−b0)2, Kb: kcal/mole/A2, b0; ANGLES: V(angle) = Kθ(θ−θ0)2, Kθ: kcal/mole/rad2, θ0: degrees; DIHEDRALS: V(dihedral) = Kχ(1+cos(n(χ)−γ)), Kχ: kcal/mole, n: multiplicity, γ: degrees.
(DOCX)
Funding Statement
The work was supported by Department of Chemistry, University of Alabama at Birmingham. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Alzheimer's Association (2009) Alzheimer's Disease Facts and Figures Alzheimer's & Dementia 5: 3. [DOI] [PubMed] [Google Scholar]
- 2. Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, et al. (1985) Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A 82: 4245–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jarrett JT, Berger EP, Lansbury PT (1993) The C-terminus of the beta protein is critical in amyloidogenesis. Ann N Y Acad Sci 695: 144–148. [DOI] [PubMed] [Google Scholar]
- 4. Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, et al. (1994) Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43). Neuron 13: 45–53. [DOI] [PubMed] [Google Scholar]
- 5. Kim J, Onstead L, Randle S, Price R, Smithson L, et al. (2007) Abeta40 inhibits amyloid deposition in vivo. J Neurosci 27: 627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iijima K, Liu HP, Chiang AS, Hearn SA, Konsolaki M, et al. (2004) Dissecting the pathological effects of human Abeta40 and Abeta42 in Drosophila: a potential model for Alzheimer's disease. Proc Natl Acad Sci U S A 101: 6623–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hardy J, Selkoe DJ (2002) The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 297: 353–356. [DOI] [PubMed] [Google Scholar]
- 8. Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, et al. (2005) Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat Neurosci 8: 79–84. [DOI] [PubMed] [Google Scholar]
- 9. Becker JS, Zoriy MV, Pickhardt C, Palomero-Gallagher N, Zilles K (2005) Imaging of copper, zinc, and other elements in thin section of human brain samples (hippocampus) by laser ablation inductively coupled plasma mass spectrometry. Anal Chem 77: 3208–3216. [DOI] [PubMed] [Google Scholar]
- 10. Chen WT, Liao YH, Yu HM, Cheng IH, Chen YR (2011) Distinct effects of Zn2+, Cu2+, Fe3+, and Al3+ on amyloid-beta stability, oligomerization, and aggregation: amyloid-beta destabilization promotes annular protofibril formation. J Biol Chem 286: 9646–9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rajendran R, Minqin R, Ynsa MD, Casadesus G, Smith MA, et al. (2009) A novel approach to the identification and quantitative elemental analysis of amyloid deposits–insights into the pathology of Alzheimer's disease. Biochem Biophys Res Commun 382: 91–95. [DOI] [PubMed] [Google Scholar]
- 12. Atwood CS, Moir RD, Huang X, Scarpa RC, Bacarra NM, et al. (1998) Dramatic aggregation of Alzheimer abeta by Cu(II) is induced by conditions representing physiological acidosis. J Biol Chem 273: 12817–12826. [DOI] [PubMed] [Google Scholar]
- 13. Esler WP, Stimson ER, Jennings JM, Ghilardi JR, Mantyh PW, et al. (1996) Zinc-induced aggregation of human and rat beta-amyloid peptides in vitro. J Neurochem 66: 723–732. [DOI] [PubMed] [Google Scholar]
- 14. Cherny RA, Atwood CS, Xilinas ME, Gray DN, Jones WD, et al. (2001) Treatment with a copper-zinc chelator markedly and rapidly inhibits beta-amyloid accumulation in Alzheimer's disease transgenic mice. Neuron 30: 665–676. [DOI] [PubMed] [Google Scholar]
- 15. Cherny RA, Legg JT, McLean CA, Fairlie DP, Huang X, et al. (1999) Aqueous dissolution of Alzheimer's disease Abeta amyloid deposits by biometal depletion. J Biol Chem 274: 23223–23228. [DOI] [PubMed] [Google Scholar]
- 16. Noy D, Solomonov I, Sinkevich O, Arad T, Kjaer K, et al. (2008) Zinc-amyloid beta interactions on a millisecond time-scale stabilize non-fibrillar Alzheimer-related species. J Am Chem Soc 130: 1376–1383. [DOI] [PubMed] [Google Scholar]
- 17. Shin BK, Saxena S (2008) Direct evidence that all three histidine residues coordinate to Cu(II) in amyloid-beta1-16. Biochemistry 47: 9117–9123. [DOI] [PubMed] [Google Scholar]
- 18. Mekmouche Y, Coppel Y, Hochgräfe K, Guilloreau L, Talmard C, et al. (2005) Characterization of the ZnII binding to the peptide amyloid-beta1-16 linked to Alzheimer's disease. Chembiochem 6: 1663–1671. [DOI] [PubMed] [Google Scholar]
- 19. Syme CD, Viles JH (2006) Solution 1H NMR investigation of Zn2+ and Cd2+ binding to amyloid-beta peptide (Abeta) of Alzheimer's disease. Biochim Biophys Acta 1764: 246–256. [DOI] [PubMed] [Google Scholar]
- 20. Zirah S, Kozin SA, Mazur AK, Blond A, Cheminant M, et al. (2006) Structural changes of region 1-16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging. J Biol Chem 281: 2151–2161. [DOI] [PubMed] [Google Scholar]
- 21. Danielsson J, Pierattelli R, Banci L, Gräslund A (2007) High-resolution NMR studies of the zinc-binding site of the Alzheimer's amyloid beta-peptide. FEBS J 274: 46–59. [DOI] [PubMed] [Google Scholar]
- 22. Liu ST, Howlett G, Barrow CJ (1999) Histidine-13 is a crucial residue in the zinc ion-induced aggregation of the A beta peptide of Alzheimer's disease. Biochemistry 38: 9373–9378. [DOI] [PubMed] [Google Scholar]
- 23. Miura T, Suzuki K, Kohata N, Takeuchi H (2000) Metal binding modes of Alzheimer's amyloid beta-peptide in insoluble aggregates and soluble complexes. Biochemistry 39: 7024–7031. [DOI] [PubMed] [Google Scholar]
- 24. Yang DS, McLaurin J, Qin K, Westaway D, Fraser PE (2000) Examining the zinc binding site of the amyloid-beta peptide. Eur J Biochem 267: 6692–6698. [DOI] [PubMed] [Google Scholar]
- 25. Kozin SA, Zirah S, Rebuffat S, Hoa GH, Debey P (2001) Zinc binding to Alzheimer's Abeta(1-16) peptide results in stable soluble complex. Biochem Biophys Res Commun 285: 959–964. [DOI] [PubMed] [Google Scholar]
- 26. Curtain CC, Ali F, Volitakis I, Cherny RA, Norton RS, et al. (2001) Alzheimer's disease amyloid-beta binds copper and zinc to generate an allosterically ordered membrane-penetrating structure containing superoxide dismutase-like subunits. J Biol Chem 276: 20466–20473. [DOI] [PubMed] [Google Scholar]
- 27. Dong J, Shokes JE, Scott RA, Lynn DG (2006) Modulating amyloid self-assembly and fibril morphology with Zn(II). J Am Chem Soc 128: 3540–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stellato F, Menestrina G, Serra MD, Potrich C, Tomazzolli R, et al. (2006) Metal binding in amyloid beta-peptides shows intra- and inter-peptide coordination modes. Eur Biophys J Biophy 35: 340–351. [DOI] [PubMed] [Google Scholar]
- 29. Talmard C, Guilloreau L, Coppel Y, Mazarguil H, Faller P (2007) Amyloid-beta peptide forms monomeric complexes with Cu-II and Zn-II prior to aggregation. Chembiochem 8: 163–165. [DOI] [PubMed] [Google Scholar]
- 30. Gaggelli E, Janicka-Klos A, Jankowska E, Kozlowski H, Migliorini C, et al. (2008) NMR studies of the Zn2+ interactions with rat and human beta-amyloid (1-28) peptides in water-micelle environment. J Phys Chem B 112: 100–109. [DOI] [PubMed] [Google Scholar]
- 31. Minicozzi V, Stellato F, Comai M, Dalla Serra M, Potrich C, et al. (2008) Identifying the minimal copper- and zinc-binding site sequence in amyloid-beta peptides. J Biol Chem 283: 10784–10792. [DOI] [PubMed] [Google Scholar]
- 32. Faller P, Hureau C (2009) Bioinorganic chemistry of copper and zinc ions coordinated to amyloid-beta peptide. Dalton T 2009: 1080–1094. [DOI] [PubMed] [Google Scholar]
- 33. Talmard C, Leuma Yona R, Faller P (2009) Mechanism of zinc(II)-promoted amyloid formation: Zinc(II) binding facilitates the transition from the partially alpha-helical conformer to aggregates of amyloid beta protein(1-28). J Biol Inorg Chem 14: 449–455. [DOI] [PubMed] [Google Scholar]
- 34. Miller Y, Ma B, Nussinov R (2010) Zinc ions promote Alzheimer Abeta aggregation via population shift of polymorphic states. Proc Natl Acad Sci U S A 107: 9490–9495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller Y, Ma B, Nussinov R (2010) Polymorphism in Alzheimer Abeta amyloid organization reflects conformational selection in rugged energy landscape. Chem Rev 110: 4820–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shewmaker F, McGlinchey RP, Wickner RB (2011) Structural insights into functional and pathological amyloid. J Biol Chem 286: 16533–16540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tomaselli S, Esposito V, Vangone P, van Nuland NA, Bonvin AM, et al. (2006) The alpha-to-beta conformational transition of Alzheimer's Abeta-(1-42) peptide in aqueous media is reversible: a step by step conformational analysis suggests the location of beta conformation seeding. ChemBioChem 7: 257–267. [DOI] [PubMed] [Google Scholar]
- 38. Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, et al. (2005) 3D structure of Alzheimer's amyloid-beta(1-42) fibrils. PNAS 102: 17342–17347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lipscomb WN, Sträter N (1996) Recent Advances in Zinc Enzymology. Chem Rev 96: 2375–2434. [DOI] [PubMed] [Google Scholar]
- 40. Parkin G (2004) Synthetic analogues relevant to the structure and function of zinc enzymes. Chem Rev 104: 699–767. [DOI] [PubMed] [Google Scholar]
- 41. Sousa SF, Lopes AB, Fernandes PA, Ramos MJ (2009) The Zinc proteome: a tale of stability and functionality. Dalton Trans 14: 7946–7956. [DOI] [PubMed] [Google Scholar]
- 42. Parr RG, Gadre SR, Bartolotti LJ (1979) Local density functional theory of atoms and molecules. Proc Natl Acad Sci U S A 76: 2522–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Becke AD, Johnson ER (2007) A unified density-functional treatment of dynamical, nondynamical, and dispersion correlations. J Chem Phys 127: 124108. [DOI] [PubMed] [Google Scholar]
- 44. Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ (1994) Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J Phys Chem 98: 11623–11627. [Google Scholar]
- 45. Petersson GA, Bennett A, Tensfeldt TG, Al-Laham MA, Shirley WA, et al. (1988) A complete basis set model chemistry. I. The total energies of closed-shell atoms and hydrides of the first-row atoms. J Chem Phys 89: 2193–218. [Google Scholar]
- 46. Clark T, Chandrasekhar J, Spitznagel GW, Schleyer PR (1983) Efficient diffuse function-augmented basis-sets for anion calculations. 3. The 3-21+G basis set for 1st-row elements, Li-F. J Comp Chem 4: 294–301. [Google Scholar]
- 47.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, et al.. (2009) Gaussian 09, Revision A.02, Gaussian, Inc., Wallingford CT.
- 48.Becker OM, MacKerell AD Jr, Roux B, Watanabe M, Editors (2001) Computational Biochemistry and Biophysics. Marcel Dekker, Inc. New York. pp 7–38.
- 49. Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, et al. (1996) Scalable molecular dynamics with NAMD. J Comput Chem 26: 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Humphrey W, Dalke A, Schulten K (1996) VMD - Visual Molecular Dynamics. J Molec Graphics 14: 33–38. [DOI] [PubMed] [Google Scholar]
- 51. Lobanov MY, Bogatyreva NS, Galzitskaya OV (2008) Radius of gyration as an indicator of protein structure compactness. Molecular Biology 42: 623–628. [PubMed] [Google Scholar]
- 52. Beauchemin D, Kisilevsky R (1998) A method based on ICP-MS for the analysis of Alzheimer's amyloid plaques. Anal Chem 70: 1026–1029. [DOI] [PubMed] [Google Scholar]
- 53. Deeth RJ, Randell K (2008) Ligand field stabilization and activation energies revisited: molecular modeling of the thermodynamic and kinetic properties of divalent, first-row aqua complexes. Inorg Chem 47: 7377–7388. [DOI] [PubMed] [Google Scholar]
- 54. Sciarretta KL, Gordon DJ, Petkova AT, Tycko R, Meredith SC (2005) Abeta40-lactam(D23/K28) models a conformation highly favorable for nucleation of amyloid. Biochemistry 44: 6003–6014. [DOI] [PubMed] [Google Scholar]
- 55. Petkova A, Yau WM, Tycko R (2006) Experimental constraints on quaternary structure in Alzheimer's beta-amyloid fibrils. Biochemistry 45: 498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roychaudhuri R, Yang M, Deshpande A, Cole GM, Frautschy S, et al. (2013) C-terminal turn stability determines assembly differences between Aβ40 and Aβ42. J Mol Biol 2013 425: 292–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Root Mean Square Deviations (Cα only) for each Simulation. For each simulation, root mean square deviation (RMSD) is calculated for the entire trajectory using the first trajectory as the reference. Gray lines are Zn-bound complexes. Dark gray lines are controls.
(DOCX)
Root Mean Square Fluctuation (Cα only) of each Simulation. For each simulation, root mean square fluctuation (RMSF) is calculated for the equilibrated portion of the simulation (last 80 ns). Gray lines are Zn-bound complexes. Dark gray lines are controls.
(DOCX)
Per-Residue Helix and Beta-sheet Content for each Zn(Aβ)2. All per-residue secondary structure results are average values of all dimeric species over three runs for each simulation. Gray lines are Zn-bound complexes. Dark gray lines are controls.
(DOCX)
Force Constants for Zn and Coordinating Residues. List of force constants between zinc and the coordinating atoms obtained from DFT calculations at the B3LYP/6-31+G* level that were added to the CHARMM 22/CMAP force field parameters. The force-field potential energy as found in CHARMM22 is as follows: BONDS: V(bond) = Kb(b−b0)2, Kb: kcal/mole/A2, b0; ANGLES: V(angle) = Kθ(θ−θ0)2, Kθ: kcal/mole/rad2, θ0: degrees; DIHEDRALS: V(dihedral) = Kχ(1+cos(n(χ)−γ)), Kχ: kcal/mole, n: multiplicity, γ: degrees.
(DOCX)