Abstract
Hirschsprung disease (HSCR) is a common genetic disorder characterized by intestinal obstruction secondary to enteric aganglionosis. HSCR demonstrates a complex pattern of inheritance, with the RET proto-oncogene acting as a major gene and with several additional susceptibility loci related to the Ret-signaling pathway or to other developmental programs of neural crest cells. To test how the HSCR phenotype may be affected by the presence of genetic variants, we investigated the role of a single-nucleotide polymorphism (SNP), 2508C→T (S836S), in exon 14 of the RET gene, characterized by low frequency among patients with HSCR and overrepresentation in individuals affected by sporadic medullary thyroid carcinoma. Typing of several different markers across the RET gene demonstrated that a whole conserved haplotype displayed anomalous distribution and nonrandom segregation in families with HSCR. We provide genetic evidence about a protective role of this low-penetrant haplotype in the pathogenesis of HSCR and demonstrate a possible functional effect linked to RET messenger RNA expression.
Hirschsprung disease (HSCR [MIM 142623]) (incidence 1/5,000 live births) is a developmental disorder characterized by the absence of enteric neurons in distal segments of the gastrointestinal tract, which causes intestinal obstruction in neonates and severe constipation in adults. Several lines of evidence suggest a multigenic pattern of inheritance for the disease: a distorted sex ratio (M:F = 4:1), incomplete penetrance, variable expressivity, and the existence of syndromic forms (Badner et al. 1990; Gabriel et al. 2002). Since 1994, many studies have indicated that the RET proto-oncogene (MIM 164761), which encodes a tyrosine kinase receptor, is the major HSCR gene and have ascribed the disease to loss-of-function mutations of the gene. To date, RET mutations account for <30% of sporadic cases, and a small number of patients (5%–10%) show alterations in other genes (GDNF, NRTN, ECE1, EDN3, EDNRB, SOX10, and SIP1 [reviewed by Parisi and Kapur 2001]).
In view of the central role played by the RET proto-oncogene in HSCR development, we have investigated the existence of RET alleles that could act as susceptibility factors and modulate the expression of the disease. In our previous work, we analyzed a 2508C→T SNP in exon 14 of the RET gene (S836S), finding that the T variant was less frequent in patients with HSCR (1 [0.5%] of 184 alleles) compared with the normal population (30 [8.5%] of 352 alleles) (P=.00023 by Fisher's exact test) (Griseri et al. 2000b). We also demonstrated that the anomalous allelic distribution was due to nonrandom segregation of the T and C alleles in Italian families with HSCR (transmission/disequilibrium test P=.008). In the present study, we have attempted to determine whether the observed segregation distortion is due to an increased risk of developing HSCR conferred by the C allele or to a protective effect of the T allele.
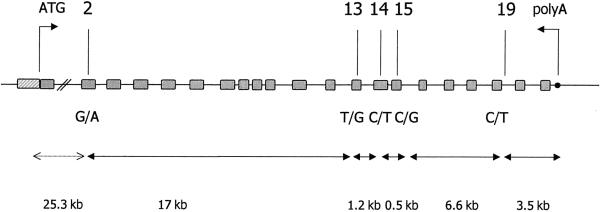
Using four SNPs distributed along the whole gene, we reconstructed RET haplotypes in seven Italian families informative for SNP14. The SNPs selected are the closest and the most distant known on each side of the 2508C→T locus, covering a total region of 25.3 kb (fig. 1). Among these markers, the 135G→A in exon 2 (A45A), the 2307T→G in exon 13 (L769L), and the 2712 C→G in exon 15 (S905S) represent known synonymous polymorphisms (Ceccherini et al. 1994), whereas the IVS19+47C→T is a new polymorphic intronic variant and is described here for the first time (see legend to fig. 1). Reconstruction of RET haplotypes in the seven selected families with HSCR showed that the T allele of SNP14 (which is never transmitted from the informative C/T parents to the affected children) lies on a single haplotype, defined by the wild-type G of SNP2, the variant G of SNP13, and the two most-common C alleles of SNP15 and SNP19 (GGTCC) (table 1). On the other hand, the C allele of SNP14 (which is always transmitted from heterozygous individuals to their affected offspring) was found to be associated with three different combinations of polymorphisms (table 1).
Figure 1.
Physical map of RET markers. RET genomic region (not to scale) is represented, together with relative distances between SNP markers. Hatched rectangle indicates the promoter region. Analysis of SNPs was reported by Ceccherini et al. (1994). Analysis of the SNP IVS19+47C→T was performed using primers int19F (5′-TGG AGT GAC CGG CCA TCT CT-3′) and int19R (5′-AAG CAT CAC AGA GAG GAA GG-3′) and the MnlI restriction enzyme.
Table 1.
RET Haplotypes and Their Frequencies in Parents of Italian and Dutch Patients with HSCR
| Haplotypesa | Italian(No. [%]) | Dutch(No. [%]) |
| Nontransmitted: | ||
| GGTCC |
7 (100) | 19 (95) |
| ATCCC |
… | 1 (5) |
| Transmitted: | ||
| ATCCCb |
2 (28.6) | 8 (40) |
| AGCCTc |
3 (42.8) | 7 (35) |
| AGCGT |
… | 1 (5) |
| AGCCC |
… | 3 (15) |
| GTCCCd |
2 (28.6) | … |
| GGTCC |
… | 1 (5) |
For Italian (7) and Dutch (20) patients with HSCR, haplotypes were determined for all parents who were heterozygous for the SNP14 C→T polymorphic variant.
Associated once with the R813W RET mutation.
Associated once with the I647I RET mutation.
Associated once with the C157Y RET mutation.
To evaluate haplotype frequencies in the general population of Italy, we typed 80 Italian healthy control individuals for the same five SNPs. We calculated the frequency of the common alleles: G = 0.73 for SNP2, T = 0.79 for SNP13, C =0.93 for SNP14, C = 0.83 for SNP15, and C = 0.87 for SNP19. No significant deviation from Hardy-Weinberg equilibrium was observed for any polymorphism. The distribution of haplotypes, which was estimated using the EM algorithm as implemented in the EHPlus program (Zhao et al. 2000), demonstrated that GGTCC was the only haplotype that included the T variant of SNP14 in the Italian population, with a frequency of 7.2% in healthy control individuals.
We further tested 156 Dutch patients with HSCR for the distribution of the C/T variant of exon 14, and we found a frequency of 1.0% (3 of 312 chromosomes) of the T allele, a frequency lower than the 2.5% (2 of 80 alleles) calculated for the 40 population-based control individuals matched on the basis of geographical region (difference not significant). Further analysis of 289 parents of Dutch patients with HSCR allowed us to identify 20 C/T heterozygotes (T allele frequency 3.5%), among whom only one transmitted the T allele to the affected child, with a significant deviation from random transmission (transmission/disequilibrium test P=.000057). Haplotype determination in the same 20 Dutch informative families established that the T allele of SNP14 was always present on the same conserved haplotype, namely GGTCC, and the C variant was associated with five different haplotypes (table 1). Our data suggest that in the Italian and Dutch populations a single T haplotype (GGTCC) is present and that it is characterized by nonrandom transmission in families with HSCR.
To determine the extension of the conserved region containing the T variant of SNP14, we characterized the promoter region of the seven Italian parents with the C/T variant, using two polymorphic nucleotide substitutions: a G→A transition and a C→A transversion located at −5 and −1 bp from the transcription start site, respectively (M.S., unpublished data). Analysis of these two markers showed three different sequence combinations, namely, GA (2/7 alleles), GC (4/7 alleles), and AA (1/7 alleles), that were associated with the GGTCC haplotype. The presence of different genotypes of the promoter region restricts the extension of the conserved region and defines the 5′ boundary of the haplotype. These data suggest that the mutation that generated the T allele occurred once on a unique haplotype. Different recombinations in the 23.5-kb region from start codon to exon 2 of the RET gene have subsequently broken the allelic association with the promoter region. Both the large dimension of intron 1 and its high content in sequences of short interspersed elements and long interspersed elements could explain the high degree of recombination between the promoter and exon 2, suggesting the existence of hotspots of genomic rearrangements in the RET locus.
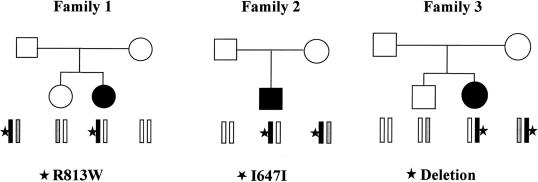
The low frequency of the GGTCC haplotype in patients with HSCR suggests that it could have a role as a low-penetrance “gain of function” RET allele and that, for this reason, it may exert a protective effect against development of megacolon. To test this hypothesis, we analyzed the genotype-phenotype correlation in families with HSCR that showed segregation of this haplotype. First, we focused on the only three families who had co-occurrence of both a RET mutation and the GGTCC haplotype and were characterized by nontransmission of this haplotype to the affected child (fig. 2). In family 1, the father was found to be a compound heterozygote for the R813W mutation in exon 14 and the T variant of SNP14 (on the GGTCC haplotype), and he appeared healthy; the daughter, who inherited only the mutated allele, was affected by HSCR. In family 2, a synonymous substitution (1941C→T [I647I], which was previously demonstrated to interfere with RET splicing but was not sufficient to determine the HSCR phenotype by itself) was transmitted to the patient with HSCR from an unaffected mother who carried the GGTCC haplotype on the other allele (Auricchio et al. 1999). Finally, in family 3, which was characterized by recurrence of a whole RET gene deletion, the hemizygous mother, with GGTCC as the only RET allele, unexpectedly did not show any symptom of the disease; however, the daughter, who was hemizygous for a different allele, was affected with HSCR.
Figure 2.
Families with HSCR with co-occurrence of RET mutations and the GGTCC haplotype. RET-mutated alleles are shown in black, and the GGTCC haplotype is shown in gray. Segregating RET mutations are indicated below each pedigree.
We have also analyzed the phenotype of all the available patients with HSCR who were carriers of the T allele, irrespective of nationality and type of mutation (familial or sporadic). The DGGE screening of 188 patients with HSCR showed only four C/T heterozygotes. Two patients with sporadic disease were affected by Down syndrome; the association between HSCR and Down syndrome has been frequently observed and has led other investigators to hypothesize the presence of a HSCR susceptibility gene on chromosome 21 (Puffenberg et al. 1994). In the third individual, we detected a de novo RET substitution (IVS12+47C→T), which was demonstrated to interfere with RET splicing; this mutation was in a cis position with the SNP14 T allele and may have counteracted its protective effect (Griseri et al. 2000a). Finally, in an Italian family with several instances of HSCR among the father’s relatives, the two affected sibs shared the same paternal haplotype (GTCCC) with no RET mutations in the coding region. Interestingly, one sib had inherited the maternal GGTCC allele and was affected with a short form of HSCR. Her sister, who inherited the alternative maternal haplotype (GTCCC), showed a severe phenotype of total colonic aganglionosis.
Two hypotheses can be formulated about the protective role of the GGTCC haplotype: (1) the effect is related to the particular combinations of polymorphic variants (combinatory hypothesis); (2) the effect is due to a genetic variant, as yet unidentified, in linkage disequilibrium with this haplotype (linkage disequilibrium hypothesis).
The hypothesis of an additive/combinatorial effect of multiple polymorphisms is intriguing, considering that single polymorphic variants were found to be differentially associated with HSCR (Borrego et al. 2000; Fitze et al. 2002). In agreement with this hypothesis, we noticed that the GGTCC haplotype does not contain the A allele of SNP2, which was suggested to be predisposing for HSCR disease; in contrast, the A allele of SNP2 is overrepresented among the Italian and Dutch transmitted haplotypes. As an alternative hypothesis, it is possible that the effect of the GGTCC haplotype does not derive from any of the single variants or their combinations. In this case, our observations may be explained by the existence of an additional variant, as yet unidentified, in linkage disequilibrium with the GGTCC haplotype.
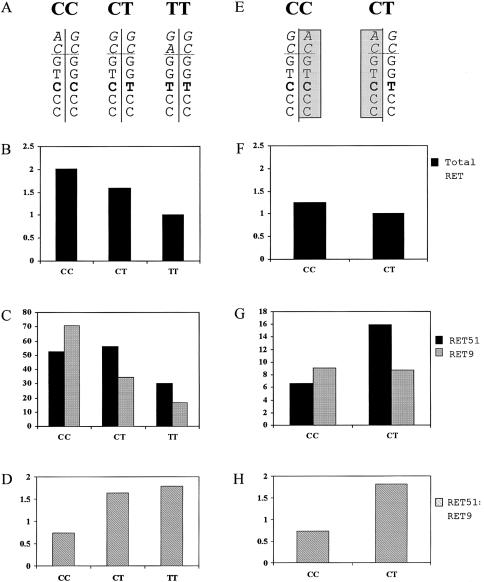
In the attempt to explain the protective effect of the GGTCC haplotype, we performed a semiquantitative RT-PCR, using RNA from lymphoblastoid cells treated with sodium butyrate. The histone deacetylase inhibitor sodium butyrate up-regulates both the levels of RET mRNA and the nuclear transcription rate in RET-negative cells, such as lymphoblasts (Puppo et al. 2002). We evaluated RET mRNA levels in lymphoblastoid cells obtained from three normal control individuals with different genotypes of SNP14; one homozygote C/C, one heterozygote C/T, and one homozygote T/T (fig. 3). We found the highest level of RET expression in the C/C homozygote, the second highest level in the C/T heterozygote, and the lowest level in the T/T homozygote (50% of the C/C control) (fig. 3B). To verify that the observed effect could be exclusively ascribed to the GGTCC haplotype and not to other differences in the genetic background of the three individuals, we performed the same experiments in two additional healthy control individuals, a father and his son, who shared an identical-by-descent C allele (see family 3 in fig. 2). In this case, the difference in the level of RET transcript could be attributed to a single different haplotype harboring the C (father) or the T (son) variant of SNP14 (fig. 3E). This test confirmed that a decreased expression of the RET gene was related to the presence of the GGTCC haplotype (fig. 3F).
Figure 3.
Results of semiquantitative RT-PCR. Cell growth conditions were reported elsewhere (Auricchio al. 1999). Sodium butyrate was dissolved in water (1 M stock solution) and was delivered to cells at a final concentration of 5 mM. Treatment was performed overnight. The fluorescence-based semiquantitative PCR approach has been described elsewhere (Patrone et al. 2000). Regression lines correlating PCR yield to cDNA concentration were determined for each sample by pooling three dilutions of three independent PCRs. Relative transcript abundance was evaluated as the ratio between the RET and the G3PDH line slopes. A and E, Haplotypes of three unrelated control individuals and two unaffected members of a family, sharing an identical-by-descent allele (shaded boxes). From top to bottom, the two alleles of promoter region (in italics) SNP2, SNP13, SNP14 (bold), SNP15, and SNP19 are indicated. B and F, Semiquantitative analysis of total RET mRNA (relative values). C and G, the expression profiles of Ret isoform 51 (black) and isoform 9 (gray) are reported (absolute values). D and H, Graphical representation of RET51:RET9 ratio.
To investigate further the expression profile of the GGTCC haplotype (and considering that the conserved region is maintained at least down to intron 19), we analyzed the presence of 3′ alternatively spliced RET isoforms. There are two major isoforms, Ret9 and Ret51, which differ in the amino acid sequence of the C-terminal tail, as a result of alternative splicing and different use of terminal codons (Myers et al. 1995). To date, compelling pieces of evidence indicate that RET9 and RET51 present distinct biochemical and biological properties, playing different roles in tumorigenesis and development (Ivanchuck et al. 1998; Le Hir et al. 2000). Analysis of the same five control individuals mentioned above showed that, in the presence of the GGTCC haplotype, a clear inversion in the proportion of the two transcripts occurred—resulting from a relative increase of RET51 expression with respect to RET9 (fig. 3C and 3G)—as is also revealed by isoform histograms and graphical representation of RET51:RET9 ratio (fig. 3D and 3H).
Although these experiments, performed on sodium butyrate–treated lymphoblasts, do not reflect tissue-specific RET expression, our results indicate a decrease in RET mRNA and an unbalance in RET isoforms associated with the GGTCC haplotype.
The lower RET transcription rate that we have observed in association with the GGTCC haplotype seems to be a contrast with the putative protective effect of the latter allele, since the pathogenesis of HSCR is generally ascribed to loss of function or haploinsufficiency of the RET gene. This reduced expression could be explained by a selective underproduction of one of the two isoforms, RET9, and the resulting enrichment of the other, RET51. The identification of a missense RET mutation within exon 20 (iso51-specific M1064T) in a family with HSCR suggests that RET51 (long isoform) is essential to normal enteric development (Attiè et al. 1995), and the relative increase of RET51 that we observed may represent a protective factor. Nevertheless, experiments with recently developed transgenic mice support the idea that RET9 plays a central role with respect to RET51 in vertebrate development (de Graaf et al. 2001). On the basis of these apparently discordant studies, we suggest that there may be a critical role, not only for the absolute amount of the two isoforms but also for the RET51:RET9 ratio; this would agree with a possible cross-talk between the two proteins: in this respect, formation of homo- and heterodimers between long and short isoforms has already been demonstrated (Alberti et al. 1998).
In conclusion, we have demonstrated the existence of a rare RET haplotype that acts as a risk-modifier allele in HSCR and related neurocristopathies, and we have associated this effect with the haplotype's peculiar expression profile (a decrease in the total amount of RET, with enrichment of the RET51 isoform). We are now investigating the characterization of the genetic/functional variant responsible for this effect.
Acknowledgments
G.P. and M.S. are supported by a fellowship awarded by the Fondazione Italiana per la Ricerca sul Cancro. The financial support of Italian Telethon grant E791, European Community contract N. QLG1-2001-01646), and the CEBR-Center of Excellence at the University of Genoa are gratefully acknowledged.
Electronic-Database Information
Accession numbers and the URL for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HSCR [MIM 142623] and RET [164761])
References
- Alberti L, Borrello MG, Ghizzoni S, Torriti F, Pizzetti MG, Pierotti M (1998) Grb2 binding to the different isoforms of RET tyrosine-kinase. Oncogene 17:1079–1087 [DOI] [PubMed] [Google Scholar]
- Attiè T, Pelet A, Edery P, Eng C, Mulligan LM, Amiel J, Boutrand L, Beldjor C, Nihoul-Feketè C, Munnich A, Ponder BAJ, Lyonnet S (1995) Diversity of RET proto-oncogene mutations in familial and sporadic Hirschsprung disease. Hum Mol Genet 4:1381–1386 [DOI] [PubMed] [Google Scholar]
- Auricchio A, Griseri P, Carpentieri L, Betsos N, Staiano A, Tozzi A, Priolo M, Thompson H, Bocciardi R, Romeo G, Ballabio A, Ceccherini I (1999) Double heterozygosity for a RET substitution interfering with splicing and a EDNRB missense mutation in Hirschsprung disease. Am J Hum Genet 64:1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badner JA, Sieber WK, Garver KL, Chakravarti A (1990) A genetic study of Hirschsprung disease. Am J Hum Genet 46:568–590 [PMC free article] [PubMed] [Google Scholar]
- Borrego S, Ruiz A, Saez ME, Gimm O, Gao X, Lopez-Alonso M, Hernandez A, Wright FA, Antinolo G, Eng C (2000) RET genotypes comprising specific haplotypes of polymorphic variants predispose to isolated Hirschsprung disease. J Med Genet 37:572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccherini I, Hofstra R, Luo Y, Stulp R, Barone V, Stelwagen T, Bocciardi R, Nijveen H, Bolino A, Seri M, Ronchetto P, Pasini B, Bozzano M, Buys C, Romeo G (1994) DNA polymorphisms and conditions for SSCP analysis of the exons of the RET proto-oncogene. Oncogene 9:3025–3029 [PubMed] [Google Scholar]
- de Graaf E, Sriniva S, Kilknny C, D’Agati V, Mankoo B, Costantini F, Pachnis V (2001) Differential activities of the RET tyrosine-kinase receptor isoforms during mammalian embryogenesis. Genes Dev 15:2433–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitze G, Cramer J, Ziegler A, Schierz M, Schreiber M, Kuhlisch E, Roesner D, Schackert HK (2002) Association between c135G/A genotype and RET proto-oncogene germline mutations and phenotype of Hirschsprung's disease. Lancet 359:1200–1205 [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Salomon R, Pelet A, Angrist M, Amiel J, Fornage M, Attié-Bitach T, Olson JM, Hofstra R, Buys C, Steffann J, Munnich A, Lyonnet S, Chakravarti A (2002) Segregation at three loci explains familial and population risk in Hirschsprung disease. Nat Genet 31:89–93 [DOI] [PubMed] [Google Scholar]
- Griseri P, Mishto M, Priolo M, Pesce B, Romeo G, Ravazzolo R, Ceccherini I (2000a) An intronic nucleotide variant of the RET proto-oncogene causes Hirschsprung disease by interfering with RNA splicing. Gene Funct Dis 1:184–188 [Google Scholar]
- Griseri P, Sancandi M, Patrone G, Bocciardi R, Hofstra R, Ravazzolo R, Devoto M, Romeo G, Ceccherini I (2000b) Decreased frequency of a single nucleotide polymorphism of the RET proto-oncogene in sporadic Hirschsprung disease. Eur J Hum Genet 8:721–724 [DOI] [PubMed] [Google Scholar]
- Ivanchuck SM, Myers S, Mulligan LM (1998) Expression of RET 3′ splicing variants during human kidney development. Oncogene 16:991–996 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Charlet-Berguerand N, Gimenez-Roqueplo A, Mannelli M, Plouin P, de Franciscis V, Thermes C (2000) Relative expression of the RET9 and RET51 isoforms in human pheochromocytomas. Oncology 58:311–318 [DOI] [PubMed] [Google Scholar]
- Myers SM, Eng C, Ponder BA, Mulligan LM (1995) Characterization of RET proto-oncogene 3′ splicing variants and polyadenylation sites: a novel C-terminus for RET. Oncogene 11:2039–2045 [PubMed] [Google Scholar]
- Parisi MA, Kapur RP (2000) Genetics of Hirschsprung disease. Curr Opin Pediatr 12:610–617 [DOI] [PubMed] [Google Scholar]
- Patrone G, Puppo F, Cusano R, Scaranari M, Ceccherini I, Puliti A, Ravazzolo R (2000) Nuclear run-on assay using biotin labeling, magnetic bead capture and analysis by fluorescence-based RT-PCR. Biotechniques 29:1012–1013 [DOI] [PubMed] [Google Scholar]
- Puffenberger EG, Hosoda, SS, Washington K, Nakao D, De Wit M, Yaganisawa Y, Chakravarti A (1994) A missense mutation of endothelin-B receptor gene in multigenic Hirschsprung disease. Cell 79:1257–1266 [DOI] [PubMed] [Google Scholar]
- Puppo F, Griseri P, Fanelli M, Schena F, Romeo G, Pelicci PG, Ceccherini I, Ravazzolo R, Patrone G (2002) Cell-line specific chromatin acetylation at the Sox10-Pax3 enhancer site modulates the RET proto-oncogene expression. FEBS Lett 523:123–127 [DOI] [PubMed] [Google Scholar]
- Zhao JH, Curtis D, Sham PC (2000) Model-free analysis and permutation tests for allelic associations. Hum Hered 50:133–139 [DOI] [PubMed] [Google Scholar]