Abstract
Two fluorescent amino acids, including the novel fluorescent species biphenyl-phenylalanine, have been incorporated into positions 17 and 115 of dihydrofolate reductase to enable a study of conformational changes associated with inhibitor binding. Unlike most studies involving fluorescently labeled proteins, the fluorophores were incorporated into the amino acid side chains, and both probes (biphenyl-phenylalanine (1) and L-(7-hydroxycoumarin-4-yl)ethylglycine (2)) were smaller than fluorophores typically used for such studies. The DHFR positions were chosen as potentially useful for FRET measurements based on their estimated separation (~17–18 Å), and expected change in distance along the reaction coordinate. Also of interest was the steric accessibility of the two sites: Glu17 is on the surface of DHFR, while Ile115 is within a folded region of the protein. Modified DHFR I (1 at position 17; 2 at position 115) and DHFR II (2 at position 17; 1 at position 115) were both catalytically competent. However, DHFR II, containing the potentially rotatable biphenyl-phenylalanine moiety at sterically encumbered position 115 was significantly more active than DHFR I. Irradiation of the modified DHFRs at 280 nm effected excitation of biphenyl-phenylalanine (1), energy transfer to coumarin 2 and emission at 450 nm. However, energy transfer was substantially more efficient for DHFR II. The effect of inhibitor binding was also measured. Trimethoprim mediated concentration dependent diminution of the emission observed at 450 nm for DHFR II, but not for DHFR I. These findings demonstrate that amino acids containing small fluorophores can be introduced into DHFR with minimal disruption of function, and in a fashion that enables sensitive monitoring of changes in DHFR conformation.
Föster resonance energy transfer (FRET) has been utilized extensively to monitor conformational changes in macromolecules such as nucleic acids and proteins,1 and also the intermolecular association between macromolecules by measuring changes in the efficiency of energy transfer between a donor and acceptor.2 The fluorophore donor is excited by irradiation; the absorbed energy is transferred to the (fluorescent or quencher) acceptor in a nonradiative process. Distance measurements made by FRET typically range from 41 to 73 Å, although the use of dye–quencher pairs has enabled the measured distance to be ~23 Å.3 The fluorophores are typically large polycyclic aromatic molecules, and are generally attached to the macromolecules by mean of flexible tethers, such that they have conformational freedom independent of conformational changes in the macromolecules to which they are attached.
As part of a program to measure protein dynamics, we recently described the use of dihydrofolate reductase (DHFR) containing two pyrenylalanines to measure protein dynamics via the observation of excimer formation.4 The distance changes measured in DHFR were on the order of several Å.4,5 In an effort to develop a complementary technique enabling measurement of conformational changes in proteins over somewhat longer distances, we have explored the use of FRET. The smaller distances of interest for measurement in comparison with typical FRET measurements argued for the attachment of the dyes close to the protein backbone, and with fewer degrees of conformational freedom. This, in turn, would necessitate the use of acceptors and donors sterically similar to the side chains of proteinogenic amino acids. The use of fluorescent acceptors provides qualitative assurance of energy transfer through the longer wavelength emission from the acceptor fluorophore,6 and has been reported to have higher sensitivity than dye–quencher systems, and to enable ratiometric analysis which is generally superior to intensity based measurements.7,8
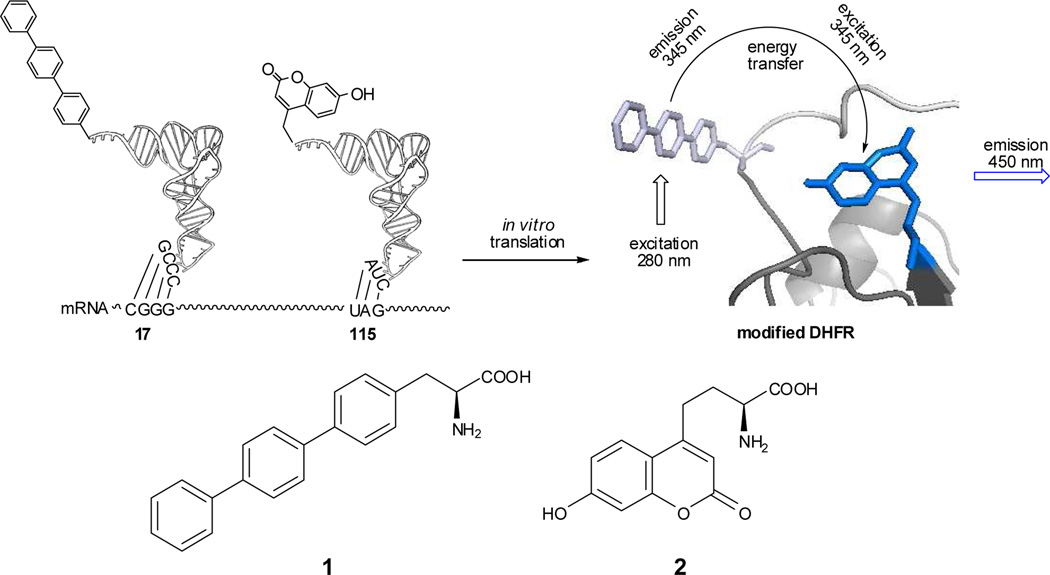
Proteins containing fluorescent amino acids have been reported,9 and a few studies have employed proteins with two such amino acids to record large changes in proximity of the fluorescent amino acids resulting from protein backbone cleavage10 or extensive reorganization of protein structure.11 Presently, the fluorescent amino acids 4-biphenyl-L-phenylalanine (1) and L-(7-hydroxycoumarin-4-yl)ethylglycine (2)12 are used to explore more subtle conformational changes in Escherichia coli DHFR. These amino acids were of interest due to the relatively small sizes and complementary shapes of their side chains. Biphenyl-L-phenylalanine is a para-substituted phenylalanine having two biphenyl linkages which should permit some conformational mobility for inclusion within folded protein structures, while L-(7-hydroxycoumarin-4-yl)ethylglycine has a bicyclic side chain whose dimensions do not differ dramatically from those of tryptophan, and which has been reported to have a high fluorescence quantum yield, and to be sensitive to pH and solvent polarity.13 Critically, the excitation and emission maxima for 1 were found to be at 280 and 345 nm, respectively, while the comparable values for 2 were 345 and 450 nm. This suggested that it should be possible to selectively excite the biphenyl-L-phenylalanine moiety in a modified DHFR containing both species by irradiation at 280 nm, and to observe fluorescence from the coumarin moiety at 450 nm if FRET had taken place (Scheme 1).
Scheme 1.
Strategy employed for incorporation of biphenyl-phenylalanine (1) and L-(7-hydroxycoumarin-4-yl)ethylgycine (2) into DHFR at positions 17 and 115, respectively.
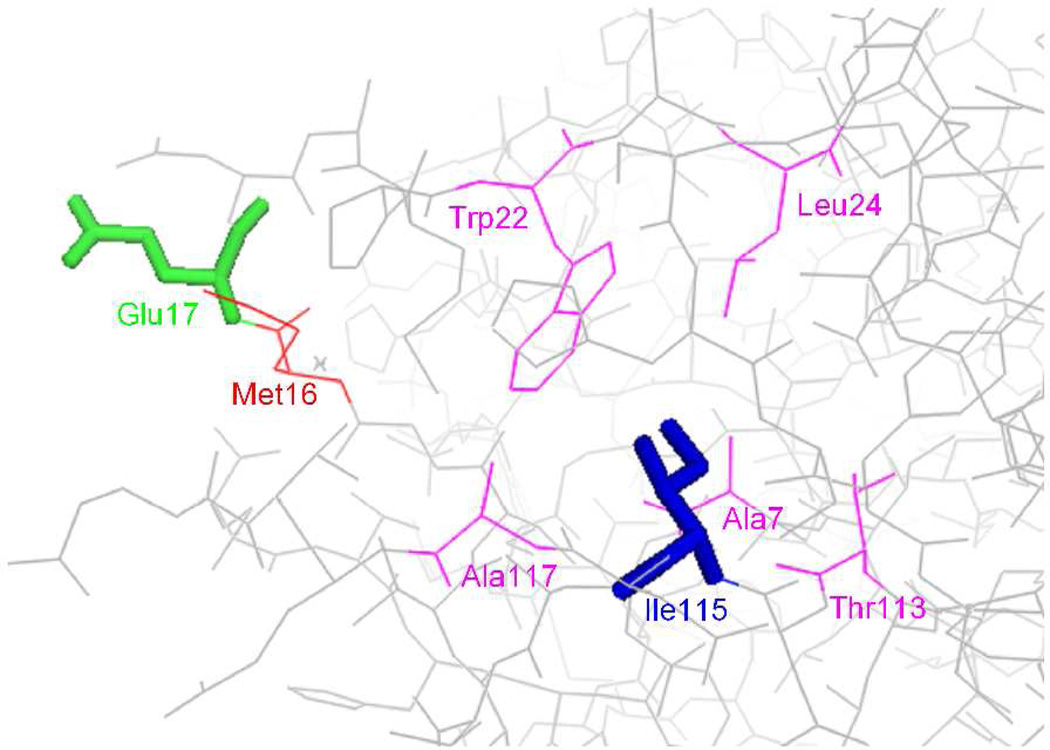
The amino acid residues chosen for substitution in DHFR were Glu17 and Ile115, which were estimated to be 17.7 Å apart, and to potentially undergo a change in relative distance of ~1.1 Å along the collective reaction coordinate.14 The calculated R0 value for 1 and 2 was 22 Å. Additionally, as may be appreciated readily from the crystal structure of DHFR shown in Figure 1, Glu17 is at the enzyme surface, and has been shown to tolerate the attachment of a fluorescence quencher,15 while Ile115 is within a folded region of the protein such that changes in side chain structure might be expected to affect protein folding to varying degrees. Nonetheless, it was anticipated that structural changes could be made without dramatically affecting DHFR structure or function.
Figure 1.
Structure of wild-type E. coli DHFR structure (PBD 1RA1), showing Glu17 in green and Ile115 in blue. The amino acid residue (Met16) which is close spatially to the side chain of Glu17 (≤ 4 Å) is shown in red. The amino acid residues (Ala7, Trp22, Leu24, Thr113 and Ala117) which are close to the side chain of Ile115 (≤ 4 Å) are shown in magenta.
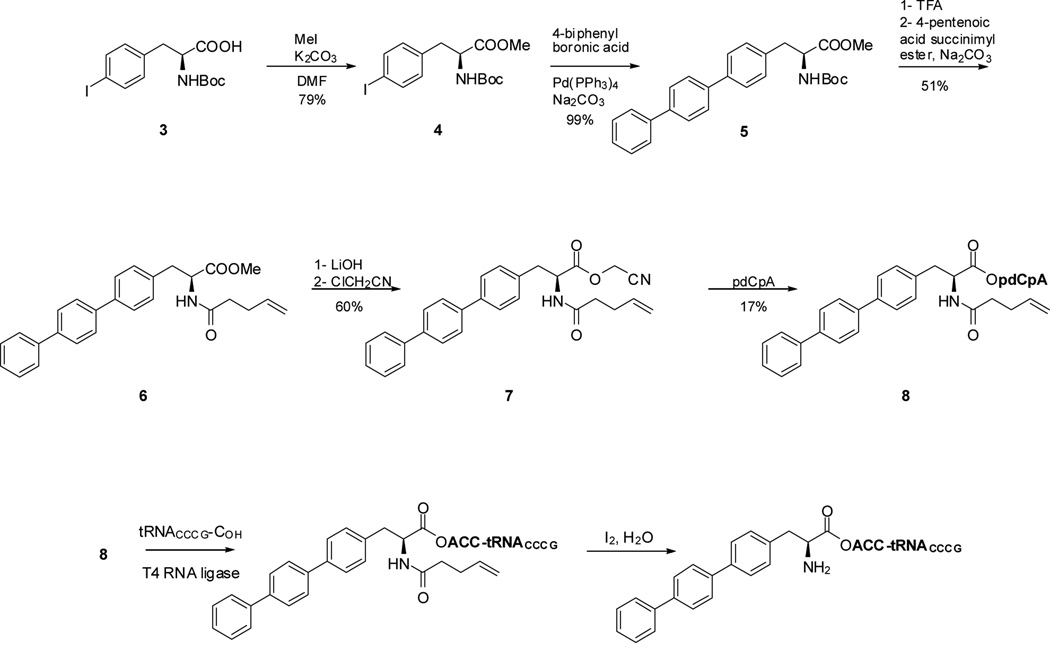
The strategy employed for the preparation of the modified DHFRs is illustrated in Scheme 1 for DHFR I. Plasmids expressing the mRNA for DHFR were modified to contain the four-base codon CGGG16 in lieu of the Glu17 codon GAA, and nonsense codon UAG in place of the Ile115 codon AUC. Biphenyl-phenylalanyl-tRNACCCG was used for suppression of the CGGG codon, while L-(7-hydroxycoumarin-4-yl)ethylgycinyl-tRNACUA was employed for UAG codon suppression. Biphenyl-L-phenylalanine was synthesized as shown in Scheme 2. The key step involved a Suzuki coupling of 4-biphenylboronic acid and protected p-iodophenylalanine (4) in the presence of Pd(PPh3)4, which afforded Boc-4-biphenyl-L-phenylalanine methyl ester (5) in 99% yield. Subsequent replacement of the Boc protecting group with an N-pentenoyl group (51% yield) afforded 6 and replacement of the methyl ester with a cyanomethyl ester gave N-(4-pentenoyl)-4[(4’;1’,1”)biphenyl]-L-phenylalanine cyanomethyl ester (7) in 60% yield. Cyanomethyl ester 7 was then treated with 5′-phosphoro-2′-deoxycytidylyl[3′→5′]adenosine (pdCpA),17 affording aminoacylated dinucleotide 8. Incubation of this dinucleotide with an abbreviated tRNACCCG-COH transcript lacking the 3'-terminal nucleotides C and A in the presence of T4 RNA ligase18 afforded biphenyl-phenylalanyl-tRNACCCG (Scheme 2); the course of the ligation reaction was monitored by polyacrylamide gel electrophoresis under acidic conditions.19 The preparation of L-(7-hydroxycoumarin-4-yl)ethylglycinyl-tRNACUA employed the same overall strategy; synthesis of the requisite aminoacyl-pdCpA intermediate 11 was carried out as shown in Scheme S1 by modification of a reported12 procedure.
Scheme 2.
The first construct to be synthesized was the modified DHFR (DHFR I) containing biphenyl-L-phenylalanine (1) at position 17 and L-(7-hydroxycoumarin-4-yl)ethylglycine (2) at position 115 (Figure S1). Also prepared were the two DHFRs containing only one of the two modified amino acids at the same position. The-modified DHFRs were purified by successive chromatography on Ni-NTA and then DEAE-Sepharose columns. The ability of these modified enzymes to effect the reduction of dihydrofolate to tetrahydrofolate was studied to assure that the isolated proteins were properly folded. As shown in Table S1 and Figure S2, substitution of position 17 with 1 was well tolerated, reducing the relative activity of the enzyme to 76% that of wild type. In contrast, introduction of amino acid 2 into position 115 of DHFR reduced the consumption of NADPH to 23%, relative to wild type. The doubly modified DHFR was 20% as active as the wild-type enzyme.
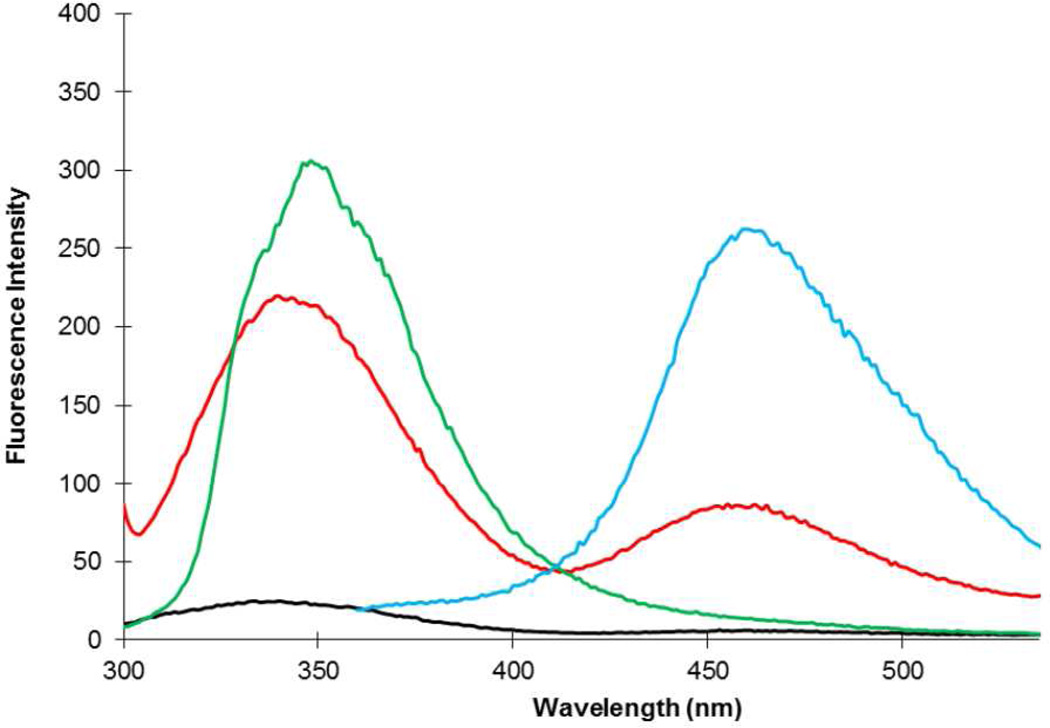
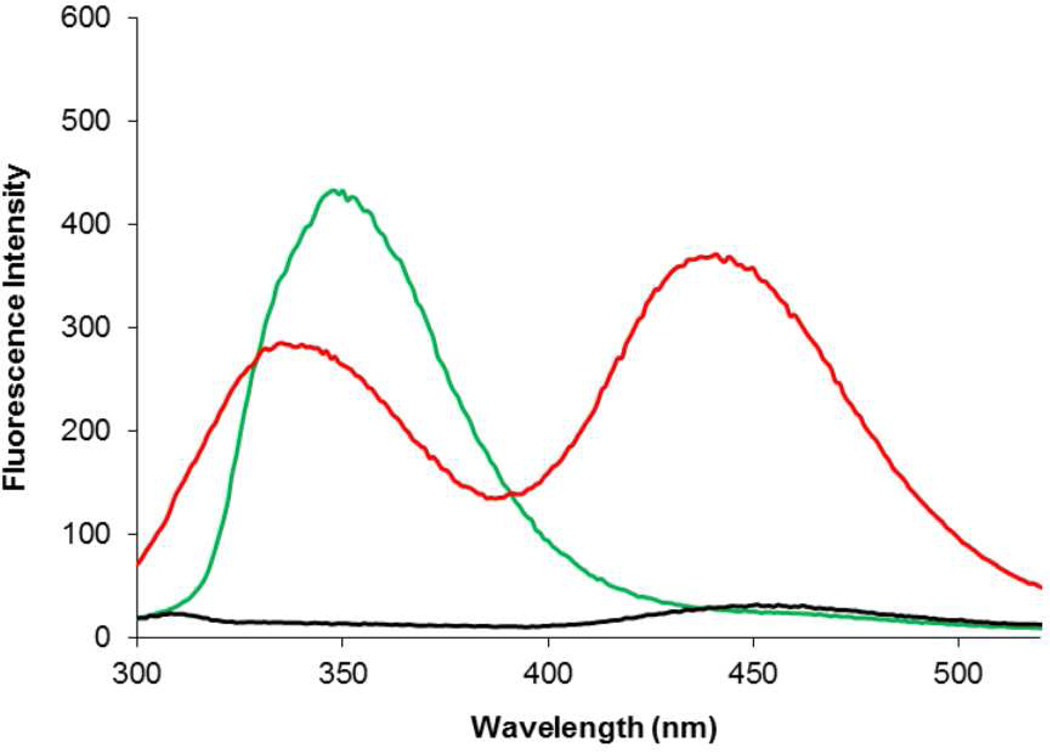
The singly and doubly-modified DHFRs were then employed to determine whether energy transfer between the two fluorescent amino acids could be observed. As shown in Figure 2 (green curve), when irradiated at 280 nm the singly modified DHFR containing 1 at position 17 exhibited an emission spectrum centered at 345 nm. In contrast, the singly modified DHFR containing 2 at position 115 failed to emit significantly at any wavelength when irradiated at 280 nm (black curve), but gave a strong emission at 450 nm when irradiated at 340 nm (cyan curve). This demonstrated that irradiation at 280 nm in the doubly substituted DHFR should effect excitation only of the biphenyl-phenylalanine moiety. When irradiated at 280 nm, the doubly modified DHFR I produced emission peaks at 345 and 450 nm (red curve), with the longer wavelength emission being of lesser intensity. This established the successful transfer of energy from excited biphenyl-phenylalanine (1) to coumarin amino acid 2.
Figure 2.
Fluorescence of modified DHFRs (0.5 µM), measured at pH 8.0. Green line is the modified DHFR containing biphenyl-phenylalanine (1) at position 17 (excitation at 280 nm); red line is the modified DHFR I containing biphenyl-phenylalanine (1) at position 17 and L-(7-hydroxycoumarin-4-yl)ethylglycine (2) at position 115 (excitation at 280 nm); black line is the modified DHFR containing L-(7-hydroxycoumarin-4-yl)ethylglycine (2) at position 115 (excitation at 280 nm); cyan line is the modified DHFR containing L-(7-hydroxycoumarin-4-yl)ethylglycine (2) at position 115 (excitation at 340 nm).
Also prepared using an analogous strategy was isomeric DHFR II having L-(7-hydroxycoumarin-4-yl)ethylglycine (2) at position 17 and biphenyl-L-phenylalanine (1) at position 115. The singly modified DHFRs having one or the other of these amino acids were also prepared. Experiments illustrating activation of the suppressor tRNAs, as well as the synthesis and purification of the proteins are shown in Figures S3, S4 and S5, respectively. Interestingly, the single modification of DHFR with amino acid 2 at position 17, or amino acid 1 at position 115, had essentially no effect on the activity of DHFR (Table S1 and Figure S2). Doubly substituted DHFR II retained 50% of the activity of wild type.
The use of DHFR II in a FRET experiment is shown in Figure 3, and is qualitatively similar to the results obtained with DHFR I. Thus irradiation at 280 nm, effecting excitation of the DHFR containing 1 at position 115, gave a single emission centered at 345 nm. Similar excitation of the DHFR containing 2 at position 17 gave minimal emission at 450 nm. Excitation of doubly modified DHFR II gave two emission peaks centered at 345 and 450 nm. However, in this case the emission at 450 nm was the stronger of the two. This established both the greater efficiency of energy transfer in the case of DHFR II as compared with DHFR I, and also the value of ratiometric analysis in establishing the relative efficiencies of energy transfer.8 Repetition of the FRET experiment at pH 7.0 gave essentially the same result, but fluorescence emission was dramatically diminished at pH 6.0 (Figure S6).
Figure 3.
Fluorescence of modified DHFRs (0.5 µM), measured at pH 8.0. Fluorescence emission of modified DHFRs following excitation at 280 nm. Green line is the singly modified DHFR containing biphenyl-phenylalanine (1) at position 115; red line is the doubly modified DHFR II containing L-(7-hydroxycoumarin-4-yl)ethylglycine (2) at position 17 and biphenyl-phenylalanine (1) at position 115; black line is the singly modified DHFR containing L-(7-hydroxycoumarin-4-yl)ethylglycine (2) at position 17.
FRET efficiency E depends both upon the distance between the fluorophores and a dipole orientation factor κ2. In the present case, the FRET donor and acceptor are not freely rotating, so dipole orientation can be important to E. We suggest that amino acid 2 can be accommodated less well at position 115 than 1, which has rotatable biphenyl linkages, and that the resulting distortion in DHFR structure results in the lesser FRET and enzymatic efficiency of DHFR I as compared with DHFR II.20
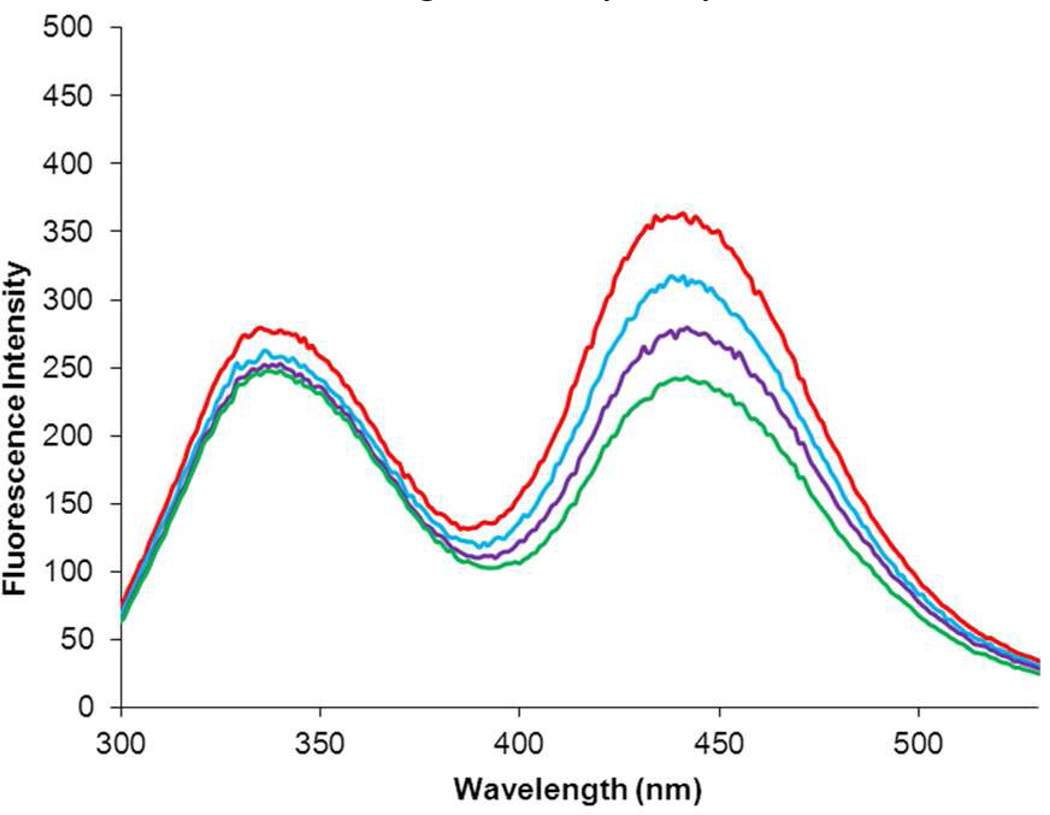
The ability of the modified DHFRs to monitor changes in protein conformation was studied by adding the DHFR inhibitor trimethoprim. When 0.5 µM DHFR I was treated with excess trimethoprim, the intensity of the emission at both 345 and 450 nm decreased in a concentration dependent fashion (Figure S9). In contrast, when 0.5 µM DHFR II was treated with trimethoprim, the intensity of the emission at 345 nm remained constant while the emission at 450 nm decreased in a concentration dependent fashion, reflecting a lessening in the efficiency of energy transfer (Figure 4) as a consequence of trimethoprim addition.20 Thus, for DHFR II the change in protein structure occasioned by the addition of trimethoprim can be monitored conveniently by FRET. The binding of trimethoprim and numerous other DHFR inhibitors is known to produce only modest changes in the structure of DHFR,21,22 far smaller than what is ordinarily studied by FRET. Thus the strategy employed here involving the careful placement of multiple small fluorescent amino acids within a protein holds promise for the monitoring of small conformational changes, such as those which occur during the catalytic cycle.
Figure 4.
Effect of the DHFR inhibitor trimethoprim on the fluorescence emission spectrum of 0.5 µM modified DHFR II containing L-(7-hydroxycoumarin-4-yl)ethylglycine (2) at position 17 and biphenyl-phenylalanine (1) at position 115 following excitation at 280 nm. The red line is the emission spectrum of modified DHFR II alone; the blue, purple and green lines reflect the effect of 2, 4 and 6 µM trimethoprim (TMP), respectively, on the emission spectrum of the modified DHFR.
It may be noted that the use of suppressor tRNAs activated with fluorescent amino acids for the introduction of fluorophores into proteins has some important advantages relative to other techniques for producing fluorescently labeled proteins. For example, post-translational derivatization of cysteine residues is not position-selective, such that mixtures of products can result, as well as unmodified cysteine residues that have failed to undergo derivaization with the fluorophore. The latter is also a potential problem with proteins prepared using unnatural amino acids having unique functional groups that can undergo chemoselective modification with fluorophores following translation.23 In comparison, the use of amino acids with fluorescent side chains small enough to permit efficient utilization by the ribosome enables uniform, site-specific labeling to be verified.24
In conclusion, we have demonstrated the specific incorporation of two fluorescent amino acids into E. coli dihydrofolate reductase, at sites anticipated to undergo a limited change in distance from each other along the reaction coordinate. One of the sites (Glu17) was known from earlier work to tolerate the introduction of a fluorescence quencher without significant loss of enzyme function, while the other (Ile115) was known from crystallographic studies to be more constrained sterically. DHFR I, containing fluorescent amino acids 1 at position 17 and 2 at position 115 retained catalytic competence but exhibited an 80% reduction in the rate of NADPH oxidation. In contrast, the introduction of amino acid 2 into position 17 had no effect on the rate of NADPH oxidation, nor did the substitution of amino acid 1 at position 115. Doubly modified DHFR II, having 2 at position 17 and 1 at position 115 retained 50% of the activity of the wild-type DHFR. This verified our hypothesis concerning the need to choose the appropriate amino acid for introduction into position 115 to achieve an optimal fluorescently labeled DHFR. While both doubly modified DHFRs emitted light at 450 nm when the biphenyl-phenylalanine moiety was excited at 280 nm, DHFR II exhibited much more efficient energy transfer than DHFR I. Additionally, only DHFR II exhibited a measurable, dose dependent change in FRET when treated with the inhibitor trimethoprim.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Research Grant GM 092946 from the National Institutes of Health.
Footnotes
Supporting Information
Methods employed for the synthesis of amino acids, activated suppressor tRNAs and modified DHFRs. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Dietrich A, Buschmann V, Muller C, Sauer M. Rev. Mol. Biotech. 2002;82:211. doi: 10.1016/s1389-0352(01)00039-3. [DOI] [PubMed] [Google Scholar]; (b) Jares-Erijman EA, Jovin TM. Nat. Biotech. 2003;21:1387. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]; (c) Dickenson NE, Picking WD. Int. J. Mol. Sci. 2012;13:15137. doi: 10.3390/ijms131115137. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Okumoto S, Jones A, Frommer WB. Annu. Rev. Plant Biol. 2012;63:663. doi: 10.1146/annurev-arplant-042110-103745. [DOI] [PubMed] [Google Scholar]
- 2.(a) Periasamy A. J. Biomed. Opt. 2001;6:287. doi: 10.1117/1.1383063. [DOI] [PubMed] [Google Scholar]; (b) Grohmann D, Klose D, Klare JP, Kay CWM, Steinhoff H-J, Werner F. J. Am. Chem. Soc. 2010;132:5954. doi: 10.1021/ja101663d. [DOI] [PubMed] [Google Scholar]
- 3. http://search.barnesandnoble.com/Principles-of-Fluorescence-Spectroscopy-Joseph-R-Lakowicz/e/9780387312781?r=1&cm_mmc=GooglePLA-_-Textbook-_-Q000000633-_-9780387312781&cm_mmca2=pla.
- 4.Chen S, Wang L, Fahmi NE, Benkovic SJ, Hecht SM. J. Am. Chem. Soc. 2012;134:18883. doi: 10.1021/ja307179q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conibear PB, Bagshaw CR, Fajer PG, Kovacs M, Malnasi-Csizmadia A. Nat. Struct. Biol. 2003;10:831. doi: 10.1038/nsb986. [DOI] [PubMed] [Google Scholar]
- 6.Jockusch S, Marti AA, Turro NJ, Li Z, Li X, Ju J, Stevens N, Akins DL. Photochem. Photobiol. Sci. 2006;5:493. doi: 10.1039/b600213g. [DOI] [PubMed] [Google Scholar]
- 7.While dye–quencher pairs have been employed over distances of potential utility for studying protein dynamics, observed changes in fluorescence intensity can also be due to changes in the local environment of the donor, or to adventitious quenching induced by species commonly encountered in experimental settings, such as oxygen, iodine and acrylamide ( Phillips SR, Wilson LJ, Borkman RF. Curr. Eye Res. 1986;5:611. doi: 10.3109/02713688609015126.
- 8.(a) Zhang P, Beck T, Tan W. Angew. Chem. Int. Ed. 2001;40:402. [PubMed] [Google Scholar]; (b) Marti AA, Jockusch S, Li Z, Ju J, Turro NJ. Nucleic Acids Res. 2006;34:e50. doi: 10.1093/nar/gkl134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Cornish VW, Benson DR, Altenbach CA, Hideg K, Hubbell WL, Schultz PG. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2910. doi: 10.1073/pnas.91.8.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mendel D, Cornish VW, Schultz PG. Annu. Rev. Biophys. Biomol. Struct. 1995;24:435. doi: 10.1146/annurev.bb.24.060195.002251. [DOI] [PubMed] [Google Scholar]; (c) Steward LE, Collins CS, Gilmore MA, Gilmore MA, Carlson JE, Ross JBA, Chamberlin AR. J. Am. Chem. Soc. 1997;119:6. [Google Scholar]; (d) Hohsaka T, Kajihara D, Ashizuka Y, Murakami H, Sisido M. J. Am. Chem. Soc. 1999;121:34. [Google Scholar]; (e) Hohsaka T, Muranaka N, Komiyama C, Matsui K, Takaura S, Abe R, Murakami H, Sisido M. FEBS Lett. 2004;560:173. doi: 10.1016/S0014-5793(04)00099-7. [DOI] [PubMed] [Google Scholar]; (f) Hamada H, Kameshima N, Szymanska A, Wegner K, Lankiewicz L, Shinohara H, Taki M, Sisido M. Bioorg. Med. Chem. 2005;13:3379. doi: 10.1016/j.bmc.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Anderson RD, III, Zhou J, Hecht SM. J. Am. Chem. Soc. 2002;124:9674. doi: 10.1021/ja0205939. [DOI] [PubMed] [Google Scholar]
- 11.(a) Murakami H, Hohsaka T, Ashizuka Y, Hashimoto K, Sisido M. Biomacromolecules. 2000;1:118. doi: 10.1021/bm990012g. [DOI] [PubMed] [Google Scholar]; (b) Kajihara D, Abe R, Iijima I, Komiyama C, Sisido M, Hohsaka T. Nat. Methods. 2006;3:923. doi: 10.1038/nmeth945. [DOI] [PubMed] [Google Scholar]
- 12.(a) Wang J, Xie J, Schultz PG. J. Am. Chem. Soc. 2006;128:8738. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]; (b) Ugwumba IN, Ozawa K, Xu Z-Q, Ely F, Foo J-L, Herlt AJ, Coppin C, Brown S, Taylor MC, Ollis DL, Mander LN, Schenk G, Dixon NE, Otting G, Oakeshott JG, Jackson CJ. J. Am. Chem. Soc. 2011;133:326. doi: 10.1021/ja106416g. [DOI] [PubMed] [Google Scholar]
- 13.Zinsli PE. J. Photochem. 1974;3:55. [Google Scholar]
- 14.(a) Wong KF, Watney JB, Hammes-Schiffer S. J. Phys. Chem. B. 2004;108:12231. [Google Scholar]; (b) Wong KF, Selzer T, Benkovic SJ, Hammes-Schiffer S. Proc. Nat. Acad. Sci. U.S.A. 102:6807. doi: 10.1073/pnas.0408343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antikainen NM, Smiley RD, Benkovic SJ, Hammes GG. Biochemistry. 2005;44:16835. doi: 10.1021/bi051378i. [DOI] [PubMed] [Google Scholar]
- 16.(a) Hohsaka T, Ashizuka Y, Sasaki H, Murakami H, Sisido M. J. Am. Chem. Soc. 1999;121:12194. [Google Scholar]; (b) Hohsaka T, Ashizuka Y, Taira H, Murakami H, Sisido M. Biochemistry. 2001;40:11060. doi: 10.1021/bi0108204. [DOI] [PubMed] [Google Scholar]
- 17.Robertson SA, Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. Nucleic Acids Res. 1989;17:9649. doi: 10.1093/nar/17.23.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Heckler TG, Zama Y, Naka T, Hecht SM. J. Biol. Chem. 1983;258:4492. [PubMed] [Google Scholar]; (b) Heckler TG, Chang LH, Zama Y, Naka T, Hecht SM. Tetrahedron. 1984;40:87. [Google Scholar]
- 19.Varshney U, Lee CP, RajBhandary UL. J. Biol. Chem. 1991;266:24712. [PubMed] [Google Scholar]
- 20.The possible involvement of Trp22 in the FRET behavior of DHFR I was excluded by introducing Phe at this position. This modified DHFR I had the same affinity for TMP as DHFR I and exhibited essentially the same FRET behavior upon TMP addition as did DHFR I. (Figure S7). The absence of direct effects of TMP on fluorescence emission was verified by studying DHFR analogues having 1 or 2 at position 17 (Figure S8). The restricted rotational freedom of 1 and 2 at DHFR positions 17 and 115 was also verified by anisotropy emission measurements (Table S2).
- 21.Sawaya MR, Kraut J. Biochemistry. 1997;36:586. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 22.This is illustrated in Figure S10 for the effects of binding of NADPH and methotrexate to E. coli DHFR.
- 23.(a) Kim J, Seo M-H, Lee S, Cho K, Woo K, Kim H-S, Park H-S. Anal. Chem. 2013;85:1468. doi: 10.1021/ac303089v. [DOI] [PubMed] [Google Scholar]; (b) Chatterjee A, Sun SB, Furman JL, Xiao H, Schultz PG. Biochemistry. 2013;52:1828. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DHFRs putatively containing amino acids 1 or 2 at position 17 were digested with trypsin as described ( Maini R, Nguyen D, Chen S, Dedkova LM, Roy Chowdhury S, Alcana-Torano R, Hecht SM. Bioorg. Med. Chem. 2013;21:1088. doi: 10.1016/j.bmc.2013.01.002. ). The expected unique typtic fragments corresponding to DHFR amino acids 13–32 were observed at m/z 2473.9 (calc 2474) and m/z 2420.3 (calc 2420) for the DHFRs containing 1 and 2, respectively (Table S3).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.