Abstract
ATP binding cassette (ABC) transmembrane efflux pumps such as P-glycoprotein (ABCB1), multidrug resistance protein 1 (ABCC1), and breast cancer resistance protein (ABCG2) play an important role in anti-cancer drug resistance. A large number of structurally and functionally diverse compounds act as substrates or modulators of these pumps. In vitro assessment of the affinity of drug candidates for multidrug resistance proteins is central to predict in vivo pharmacokinetics and drug–drug interactions. The objective of this study was to identify and characterize new substrates for these transporters. As part of a collaborative project with Life Technologies, 102 fluorescent probes were investigated in a flow cytometric screen of ABC transporters. The primary screen compared substrate efflux activity in parental cell lines with their corresponding highly expressing resistant counterparts. The fluorescent compound library included a range of excitation/emission profiles and required dual laser excitation as well as multiple fluorescence detection channels. A total of 31 substrates with active efflux in one or more pumps and practical fluorescence response ranges were identified and tested for interaction with eight known inhibitors. This screening approach provides an efficient tool for identification and characterization of new fluorescent substrates for ABCB1, ABCC1, and ABCG2.
Keywords: Efflux inhibition, ABCB1, ABCC1, ABCG2, Fluorescent substrate, Flow cytometry
The transmembrane ATP binding cassette (ABC)2 efflux pumps ABCB1 (P-glycoprotein, P-gp), ABCC1 (multidrug resistance protein 1, MRP1), and ABCG2 (breast cancer resistance protein, BCRP) play an important role in the development of resistance against anticancer drugs [1,2]. To date, more than a dozen ABC transporter pumps have been observed to efflux chemotherapeutic agents in vitro [3]. ABCB1, ABCC1, and ABCG2, in particular, are highly expressed in the gut, liver, and kidneys, and they may restrict the oral bioavailability of administered drugs. ABCB1 and ABCG2 are also expressed in the epithelia of the brain and placenta as well as in stem cells, where they perform a barrier function [4]. The role played by ABC transporter pumps in protecting tissues from xenobiotics is now widely recognized, but their interplay, their relationship with other enzymes, and how they affect the disposition, distribution, and effect of individual drugs remain an active area of investigation.
Structural information for mammalian ABC transporter family members is relatively sparse, with ABCB1 being the most extensively studied. Recent investigations indicate that at least four distinct drug binding sites exist on ABCB1, which can be classified as both transport and modulation sites. At 3.8 Å resolution, the X-ray structure of mouse apo ABCB1a displays a 6000-Å3 cavity and two ATP-binding domains separated by approximately 30 Å. The apo and drug-bound ABCB1a structures show portals open to the cytoplasm and the inner leaflet of the lipid bilayer for drug entry as well as the ability to accommodate large and small substrates or even two substrates simultaneously [5]. Together, these facts can account for broad or even poly-specificity for unrelated chemical structures.
In addition, the substrate binding cavity can be formally partitioned into an upper portion with mostly hydrophobic and aromatic interactions, and a lower space containing polar interactions (with overlap in the middle). Binding of a substrate to one of the sites may induce conformational changes to adjacent binding site(s), which in turn alters experimental affinities [6]. The drug binding pocket of ABCG2 may function in a similar manner to that of ABCB1, with radioligand binding studies suggesting two or more symmetric substrate binding sites with overlapping specificity [7]. Drug–drug interactions resulting from transporter inhibition present a clinical concern [8,9]. The presence of multiple binding sites and interactions between them may account for diverse specificity of structurally and functionally unrelated modulators and substrates. Multiple binding site interactions also raise questions as to which substrate should be used to demonstrate inhibitory potential of a new chemical probe.
To understand the mechanism of action and to design more effective modulators, efforts have been made to study the interaction of substrates and modulators with these transporters [10]. For example, most ABCB1 inhibitors are also substrates of the efflux pump [11]. It is valuable not only to assess inhibitor potency for a given transporter but also to profile its activity with respect to other transporters as well as its interrelationship with substrate drugs. For instance, strong inhibition of ABCB1 by drugs such as cyclosporine and verapamil in vitro was of limited value in vivo due to toxic pharmacological effects of the inhibitors [1].
We previously reported a new platform for identification of substrates and inhibitors for three human ABC transporters using two fluorescent probes: J-aggregate-forming lipophilic cation 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1, T3168) and calcein acetoxymethyl ester (CaAM, C1430) as substrates [12]. We demonstrated differential activity of inhibitors for ABCB1, ABCC1, and ABCG2 transporters, and we noted cross-reactivity of both these substrates across the three transporters, which could help to explain such severe toxicity effects. Recently, we used a similar duplex system with T3168 to identify an ABCG2 inhibitor with high selectivity as compared with ABCB1 [13]. In the current study, we applied this high-throughput flow cytometric assay system to evaluate 102 fluorescent compounds for their substrate properties and their interactions with several known inhibitors for these transporters.
Materials and methods
Reagents and instrumentation
The collection of potential fluorescent substrates was obtained from Life Technologies (Eugene, OR, USA). The efflux pump inhibitors loxapine succinate, nicardipine hydrochloride, niclosamide, novobiocin sodium, pimozide, and verapamil hydrochloride were purchased from Sigma–Aldrich (St. Louis, MO, USA). Lasalocid sodium was purchased from Prestwick Chemical (Illkirch, France), and mometasone furoate was purchased from U.S. Pharmacopeia (Rockville, MD, USA). Unless otherwise indicated, all compound solutions were maintained and diluted in dimethyl sulfoxide (DMSO) prior to addition to assay wells. Final DMSO concentrations were no more than 1% (v/v). A Biomek NX Multichannel (Beckman Coulter, Fullerton, CA, USA) was used for all cell and compound solution transfers for volumes greater than 1 μl. Low-volume transfers (100 nl) were performed via pintool (V&P Scientific, San Diego, CA, USA). Compound dose–response plates were generated with the Biomek NX Span-8 (Beckman Coulter, Fullerton) or the Eppendorf epMotion 5070 (Westbury, NY, USA). The HyperCyt high-throughput flow cytometry platform (IntelliCyt, Albuquerque, NM, USA) was used to sequentially sample cells from 384-well microplates (2 μl/sample) for delivery to the flow cytometer at a rate of 40 samples per minute [14,15]. Flow cytometric analysis was performed with a CyAn flow cytometer (Beckman Coulter, Fort Collins, CO, USA). To cover the fluorescence range of all substrates, each sample was (i) excited at 488 nm and detected with 530/40 (FL 1), 575/25 (FL 2), 613/20 (FL 4), and 680/30 (FL 5) optical bandpass filters and (ii) excited at 635 nm and detected with 665/20 (FL 8) and 750 LP (FL 9) optical bandpass filters. The resulting time-separated data files were analyzed with HyperView software (IntelliCyt) to determine compound activity in each well. Inhibition–response curves were fitted by Prism software (Graph-Pad Software, San Diego, CA, USA) using nonlinear least-squares regression in a sigmoidal dose–response model with variable slope, also known as the four-parameter logistic equation. Analysis of time-separated flow cytometric data was detailed in a previous ABC transporter screen from our group [12].
ABCB1, ABCC1, and ABCG2 transporter-expressing cell lines
The ABCB1-overexpressing drug-resistant cell line, CCRF-ADR 5000, and its parental CCRF-CEM cells were kindly provided by T. Efferth (Pharmaceutical Biology, German Cancer Research Center, Heidelberg, Germany). We have developed and previously characterized a SupT1-vincristine (Vin) drug-resistant cell line that selectively overexpresses ABCC1 [16]. Ovarian Ig-MXP3 (ABCG2) and its parental Igrov1-sensitive cells were kindly provided by D. Ross (Department of Medicine, University of Maryland Greenebaum Cancer Center, Baltimore, MD, USA). Cells were grown in RPMI-1640 medium supplemented with 10% fetal bovine serum (Hy-clone, Logan, UT, USA), 2 mM L-glutamine, 10 mM Hepes, 10 U/ml penicillin, 10 lg/ml streptomycin, and 4 μg/ml ciprofloxacin. To ensure ABCB1 up-regulation, CCRF-ADR 5000 cell lines were grown in 20 nM daunorubicin. To up-regulate ABCC1 expression, the SupT1-Vin cell line was grown in the presence of 150 nM vincristine. Further population enrichment of the CCRF-ADR 5000 and SupT1-Vin cell lines was achieved via fluorescence cell sorting with 250 nM CaAM and 1 μM JC-1, respectively, on a MoFlo cell sorter (Beckman Coulter, Fort Collins). Up-regulation of ABCG2 pump expression in Ig-MXP3 cells was achieved via the addition of 340 nM mitoxantrone to the cell media 1 h prior to cell harvest. Flow cytometric screening was done in calcium- and magnesium-free phosphate-buffered saline (PBS, Mediatech, Manassas, VA, USA) containing 10% fetal bovine serum.
Uptake and efflux assay
To identify potential fluorescent substrates in ABCB1-, ABCC1-, and ABCG2-overexpressing cell lines, substrates were serially diluted 1:3 eight times, affording in-well dose–response ranges from 15 pM to 100 nM. Table 1 shows the catalog number and name of each substrate along with the plating protocol type and the observed fluorescence channel. Depending on the amount of each substrate available, mother plates of the substrate stocks were generated and stored at 10, 1, and 0.1 mM. Regardless of starting stock concentration, daughter dose–response plates were generated with a top final concentration of 10 μM. The screen was done in 384-well plates at 100 μl per well with a maximum of 16 substrates tested per plate, 1 per row. To avoid potential carryover from highly fluorescent substrates, the last 12 wells of each row were filled with buffer only, providing multiple wash wells between the end of one dose–response series and the beginning of the next one. Substrate (1 μl) was added to plates containing 99 μl of the transporter-expressing cell lines and separately to their parental counterparts (1.5 × 106 cells/ml). After compound addition, the plates were vortexed briefly to mix the compounds with the cell suspension and then rotated end over end at room temperature to avoid cell settling. After a 20-min incubation, cellular fluorescence representing substrate retention was measured on the flow cytometer. Median channel fluorescence (MCF) was plotted for each substrate for each cell line for direct comparison between parental and transporter-expressing values. Response was calculated for each substrate for each transporter-expressing/parental cell line pair as
Table 1.
Summary of potential fluorescent substrate efflux probes.
| Catalog number | Product | Formata | FLb |
|---|---|---|---|
| A10254 | Alexa Fluor 488 C5-maleimide | B | 1 |
| A10255 | Alexa Fluor 532 C5-maleimide | A | 2 |
| A10256 | Alexa Fluor 594 C5-maleimide | A | 2 |
| A10258 | Alexa Fluor 546 C5-maleimide | B | 2 |
| A12410 | BODIPY FL ATP | B | 1 |
| A1318 | Tetramethylrhodamine cadaverine | A | 2 |
| A20341 | Alexa Fluor 568 C5-maleimide | A | 2 |
| A20501MP | Alexa Fluor 555 hydrazide | B* | 2 |
| A20502 | Alexa Fluor 647 hydrazide | B* | 8 |
| A22184 | BODIPY FL ATP-γ-S, thioester | B* | 1 |
| B10250 | BODIPY FL N-(2-aminoethyl) maleimide | A | 1 |
| B20340 | BODIPY FL l-cystine | A | 1 |
| B2103 | BODIPY 493/503 methyl bromide | A | 1 |
| B22356 | BODIPY FL AMPPNP | B* | 1 |
| B22461 | BODIPY FL histamine | A | 1 |
| B23461 | BODIPY FL ouabain | B | 1 |
| B6905 | BODIPY FL amiloride | C | 1 |
| B7431 | BODIPY FL verapamil, HCl | A | 1 |
| B7433 | BODIPY FL prazosin | B | 1 |
| B7436 | BODIPY FL pirenzepine, HCl | B | 1 |
| B7447 | BODIPY FL brefeldin A | C | 1 |
| B7469 | BODIPY FL forskolin | B | 1 |
| B7487 | BODIPY FL thapsigargin | B | 1 |
| B7505 | BODIPY FL-X ryanodine | C | 1 |
| C1430 | Calcein, AM | A | 1 |
| C2102 | CellTracker Green BODIPY | A | 1 |
| C2925 | CellTracker Green CMFDA | A | 1 |
| C2927 | CellTracker Orange CMTMR | A | 2 |
| C300 | 5(6)-TAMRA (mixed isomers) | A | 1 |
| C6121 | 5-TAMRA (single isomer) | A | 2 |
| C6122 | 6-TAMRA (single isomer) | A | 2 |
| C6124 | 5-ROX (single isomer) | B | 1 |
| C6156 | 6-ROX (single isomer) | A | 2 |
| C7606MP | ChromaTide tetramethylrhodamine-6-dUTP | C* | 2 |
| C7614 | ChromaTide BODIPY FL-14-dUTP | C | 1 |
| C7629 | ChromaTide Rhodamine Green-5-dUTP | C* | 1 |
| D14730 | DiOC2(3) | A | 1 |
| D20350 | BODIPY 499/508 maleimide | A | 1 |
| D2183 | BODIPY FL propionic acid | A | 1 |
| D22421 | DiNOC1(3) (JC-9) | A | 1 |
| D2390 | BODIPY FL EDA | A | 1 |
| D272 | DiOC5(3) | A | 1 |
| D273 | DiOC6(3) | A | 1 |
| D306 | DiSC3(5) | A | 8 |
| D3238 | BODIPY 492/515 disulfonate | A | 1 |
| D378 | DiOC7(3) | B | 1 |
| D383 | DiIC12(3) | A | 2 |
| D3834 | BODIPY FL C5 | A | 1 |
| D3921 | BODIPY 505/515 | A | 1 |
| D3922 | BODIPY 493/503 | A | 1 |
| D632 | Dihydrorhodamine 123 | A | 1 |
| D633 | Dihydrorhodamine 6G | A | 2 |
| D7443 | DM-BODIPY (-)-dihydropyridine | C | 1 |
| D7776 | DiIC18(3)-DS | B | 1 |
| E34251 | ER-Tracker Green | B | 1 |
| G12411 | BODIPY FL GTP | B | 1 |
| G22183 | BODIPY FL GTP-γ-S, thioester | B* | 1 |
| G22360 | BODIPY FL GDP | B* | 1 |
| G35778 | BODIPY FL GTP-γ-NH, amide | B* | 1 |
| H14700 | DiIC1(5) | A | 8 |
| L2424 | Lissamine rhodamine B EDA | A | 2 |
| L7526 | LysoTracker Green DND-26 | B | 1 |
| L7595 | LDS 751 | A | 4 |
| M1198MP | Fluorescein methotrexate | A | 1 |
| M23271 | Alexa Fluor 488 methotrexate | B | 1 |
| M23272 | BODIPY FL methotrexate | B | 1 |
| M23273 | Texas Red-X methotrexate | B | 2 |
| M7510 | MitoTracker Orange CMTMRos | B | 2 |
| M7511 | MitoTracker Orange CM-H2TMRos | B | 2 |
| M7512 | MitoTracker Red CMXRos | B | 3 |
| M7513 | MitoTracker Red CM-H2XRos | B | 3 |
| M7514 | MitoTracker Green FM | B | 1 |
| N1142 | Nile red | A | 2 |
| O246 | Octadecyl rhodamine B chloride (R18) | A | 2 |
| P7500 | BODIPY FL taxol | C | 1 |
| P7581 | Quant-iT PicoGreen | B | 1 |
| R302 | Rhodamine 123 | A | 1 |
| R6029 | Rhodamine Red C2 maleimide | A | 2 |
| R634 | Rhodamine 6G chloride | A | 2 |
| R6479 | Rhodamine 110 (R110) | A | 1 |
| R648MP | Rhodamine B, hexyl ester, perchlorate (R6) | A | 2 |
| S1307 | Sulforhodamine B | A | 2 |
| S34854 | SYTO 9 | B | 1 |
| S359 | Sulforhodamine 101 | A | 2 |
| S6957 | Sulforhodamine G | A | 2 |
| S7575 | SYTO 13 | B | 1 |
| S7578 | SYTO 16 | B | 1 |
| T12921 | Biocytin TMR | A | 2 |
| T2425 | Texas Red cadaverine | A | 2 |
| T30453 | TS-Link BODIPY FL C2-thiosulfate, sodium salt | A | 1 |
| T3168 | CBIC2(3) (JC-1) | A | 1 |
| T3602 | TO-PRO-1 iodide (515/531) | B | 1 |
| T6027 | Tetramethylrhodamine-5-maleimide | A | 2 |
| T6028 | Tetramethylrhodamine-6-maleimide | A | 2 |
| T6256 | Texas Red hydrazide | A | 2 |
| T639 | tetramethylrosamine chloride | A | 2 |
| T668 | TMRM | A | 2 |
| T669 | TMRE | A | 2 |
| V12390 | BODIPY FL vinblastine | B | 1 |
| V34850 | BODIPY FL vancomycin | C | 1 |
| Y3601 | YOYO-1 iodide (491/509) | B | 1 |
| Y3603 | YO-PRO-1 iodide (491/509) | B | 1 |
Plate formatting protocol. Despite varied starting concentrations, all mother stocks were diluted into 10-μM daughter plates for 1:100 transfers to assay wells. A: mother stocks maintained in 10 mM DMSO; B: mother stocks maintained in 1 mM DMSO; C: mother stocks maintained in 100 μM DMSO. An asterisk (*) denotes aqueous sample handling rather than DMSO.
Fluorescence channel: 488-nm excitation–optical bandpass filters: FL 1, 530/40; FL 2, 575/25; FL 3, 613/20; FL 4, 680/30; FL 5, 750 LP; 635-nm excitation–optical bandpass filters: FL 8, 665/20; FL 9, 750 LP.
where MCF_Test, MCF_HC-P, and MCF_NC represent the MCF of wells containing transporter-expressing cells and substrate, MCF of parental cells containing 100 nM substrate, and MCF of wells containing cells with no substrate, respectively. Although responses were calculated for each substrate concentration, only the 100-nM substrate data were used for comparison across cell lines. JC-1 (T3168) and CaAM (C1430) dose responses were used as efflux controls for each run.
Inhibition assay
Numerous ABC transporter efflux inhibitors have been described previously [17], eight of which were chosen for their specific efflux pump inhibition profiles here: ABCB1 (mometasone), ABCC1 (loxapine and pimozide), ABCG2 (niclosamide and novobiocin), ABCB1/ABCC1 (verapamil), ABCB1/ABCG2 (lasalocid), and ABCB1/ABCC1/ABCG2 (nicardipine) [12,18]. To investigate the inhibition profile of each potential substrate, a 9-point dilution series of each inhibitor (assay well concentrations ranging from 7.6 nM to 50 μM) was added in combination with an optimal concentration of each substrate to ABCB1, ABCC1, and ABCG2 transporter-expressing cells. The optimal concentration of each fluorescent substrate was based on the maximal efflux activity in the transporter-expressing cells. The assay protocol and order of addition were essentially the same as described in the efflux assay; however, the screening volume was reduced to 20 μl per well (19.8 μl of cells with 100 nl of substrate and inhibitor). Time-separated MCF values were plotted for each substrate/inhibitor dose response, resulting in eight inhibition curves per fluorescent sub-strate per cell line. IC50 values were calculated with the four-parameter logistic equation via Prism:
where Log[Inhib] is the log of the inhibitor concentration, MCF_Test is the MCF value indicating substrate retention in the transporter-expressing cell line, and Bottom and Top define the range of the fitted variable slope sigmoidal curve. Subsequently, each curve was subjected to a set of validation cutoff criteria (Fig. 1). Due to imperfect automated curve fitting, curves were excluded where the standard error for the logIC50 was greater than 15% of the value. Curves with negative Hill slope values were also excluded. Several fluorescence limits were established to exclude low-level or inconsistent response levels. Of the fitted curves, a minimum change from bottom to top of 50 MCF units was required. This helped to eliminate the background fluorescence for several inhibitors, including mometasone, verapamil, and pimozide. For substrates with lower fluorescence ranges (50–500 MCF units), a 2.5-fold change was required from bottom to top of the fitted curve. For fluorescent responses greater than 500 MCF units, a relaxed 2-fold change allowed inclusion of substrates with high baselines (i.e., 1500–3000 MCF inhibition levels). A total of four dose–response curves were analyzed for each inhibitor with each substrate. The IC50 values were averaged, and standard deviations were calculated.
Fig.1.
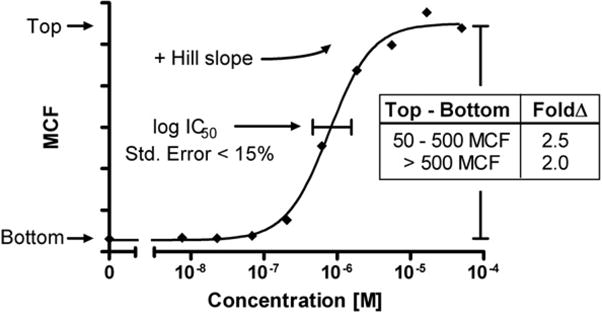
Illustration of curve validation cutoff criteria for efflux inhibition. The curve fit was first validated by excluding curves in which the standard error of the log IC50 was greater than 15% of the value. A positive Hill slope was also required. A baseline fluorescence change also needed to be maintained where bottom to top needed to be at least 50 MCF units. For substrates with lower fluorescence (top between 50 and 500 MCF units), a fold change in fluorescence of 2.5 from top to bottom was required. For substrates with high fluorescence (top >500 MCF units), the fold change was reduced to a 2-fold change to offset possible exclusion of curves with high baselines that still maintained workable fluorescence ranges.
Results
Uptake and efflux of fluorescent dyes in parental and transporter-expressing cell lines
The 102-member fluorescent library of potential efflux substrates included labeling agents and tracer dyes as well as fluorescently labeled small molecules. The fluorophores included fluorescein, rhodamine, rosamine, Alexa Fluor, BODIPY, cyanine, and several others. Each compound was incubated with both parental and transporter-expressing ABCB1, ABCC1, and ABCG2 cell lines using two lasers for excitation and seven fluorescence emission bands for analysis as specified in Materials and Methods. Because each dye had unique fluorescence and cell penetration qualities, it was challenging to find the optimal concentration range. An initial screen was conducted at a higher concentration range of 3 pM to 1 μM, and in some cases the fluorescence levels of parental cell lines were off-scale (data not shown). Therefore, we rescreened all samples at concentrations ranging from 0.3 pM to 100 nM. Although each fluorescent compound was tested in dose response, the 100-nM concentration response was used to compare across cell lines and between substrates. Although we were unable to characterize the full retention curve, optimal probe concentrations could be inferred for use in subsequent testing.
Fig. 2 illustrates dose–response data for two carbocyanine dyes, JC-1 (T3168) and DiOC2(3) (D14730) in the green fluorescence channel (FL 1, 530 nm) for each dye.3 Efficient T3168 efflux was observed for all three overexpressing cell lines, whereas selective efflux was noted with D14730 by ABCB1 and ABCC1. Fig. 2 also shows that normalized response data provided a sensitive index for comparing transporter-expressing cell efflux to parental retention. The single-point 100-nM MCF was extracted from each curve and plotted for the entire compound set (Fig. 3). A majority of tested compounds (61 of 102) failed the primary efflux assay based on low observed cellular fluorescence (as opposed to uptake without efflux). This may have been due to poor permeability of the dye or possibly to compound instability (i.e., hydrolysis). For example, several compounds such as the Alexa Fluor hydrazides A20501MP and A20502 were noted as membrane-impermeant dyes and acted as negative controls. In fact, all tested Alexa Fluor-containing fluorescent probes, including the Alexa Fluor maleimide labeling reagents A10254, A10255, A10256, A10258, and A20341, were determined to be membrane impermeant in the primary efflux screen, and none of them was considered for screening in the inhibitor assay. In addition, redox-sensitive dyes such as MitoTracker M7511 and M7513 and the dihydrorhodamines D632 and D633 were apparently not oxidized to their fluorescent counterparts, resulting in no observed cellular fluorescence.
Fig.2.
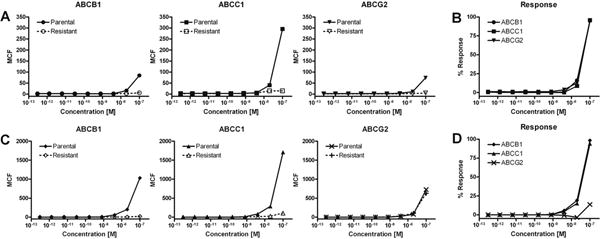
Representative examples of dose–response uptake and efflux analysis in transporter-expressing cells and their parental counterparts via the carbocyanine dyes JC-1 (T3168) and DiOC2(3) (D14730). (A) Comparison of JC-1 efflux in ABCB1, ABCC1, and ABCG2 transporter-expressing cells (dashed line) as compared with parental cellular fluorescence retention (solid line). (B) Normalized percentage response of each transporter-expressing cell line using parental fluorescence to establish the activity range. Each cell line effectively pumped out all of the T3168 substrate. (C) Comparison of D14730 ABCB1, ABCC1, and ABCG2 transporter-expressing cell line efflux (dashed line) as compared with parental cellular fluorescence retention (solid line). (D) The normalized percentage response readily demonstrates the ABCB1/ABCC1 (solid circles/solid triangles) efflux specificity of this cyanine substrate.
Fig.3.
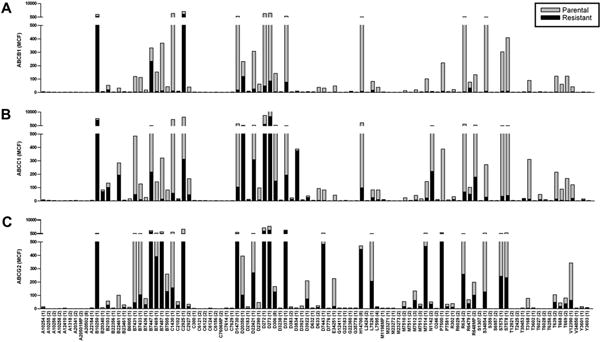
The fluorescence retention of each substrate at 100 nM is plotted for ABCB1, ABCC1, and ABCG2 transporter-expressing cells (black bars, overlay) and their parental counterparts (gray bars). (A) High ABCB1 expression in the CCRF-ADR 5000 cell line facilitated efficient efflux of most membrane-permeable substrates, indicated by the low-level fluorescence in comparison with the parental CCRF-CEM cells. (B) Greater substrate efflux specificity was observed for ABCC1-overexpressing SupT1-Vin cells as compared with ABCB1. (C) ABCG2 overexpression in Ig-MXP3 cells demonstrated the most specific substrate–pump interactions. Many of the responses were at overall lower levels compared with the other two pumps.
Although some selectivity data have been compiled for commonly used dyes in flow cytometric transporter analysis as well as recently described detection kits [19,20], here we report a direct comparison of efflux activity in ABCB1, ABCC1, and ABCG2 within one screening protocol. For the full compound set, ABCB1 and ABCC1 shared similar uptake and efflux profiles. Although substrates of ABCC1 were also substrates of ABCB1 (i.e., the cyanines S7575 and S7578), the reverse was not necessarily true (i.e., the cyanines D22421, D273, and D378). The only exception was the ABCC1/ABCG2 efflux substrate CellTracker Green CMFDA (C2925), which was pumped poorly by ABCB1. ABCG2 pumped the fewest overlapping substrates with ABCB1 and ABCC1 and tended to have higher fluorescence baselines. Cellular fluorescence was not observed with any of the labeled methotrexate analogs regardless of fluorophore, including Alexa Fluor (M23271), fluorescein (M1198MP), rhodamine (M23273), and BODIPY (M23272). Six probes (B10250, B7447, D20350, L7526, R6479, and N1142) with membrane permeability were not substrates for any of the transporters.
Fluorescent dyes that were actively effluxed by one or more of the transporter-expressing cell lines (response >75%) and displayed appropriate fluorescence levels (∼100 MCF units or more) were chosen for the inhibition screening protocol. These criteria were based primarily on the reference substrate T3168, which has previously been used in high-throughput screening (HTS) and was effluxed by ABCB1, ABCC1, and ABCG2 (Fig. 2A and B). Fig. 4 shows the 31 selected substrates displayed on the basis of the efflux response in transporter-expressing cells along with the fluorescence retention in the parental cell line. Retention in the parental cell line was used to gauge the potential fluorescence response. For simplicity, we assume that full inhibition of efflux in the transporter-expressing cells affords similar cellular fluorescence retention levels as in the cells with low pump expression. This data subset demonstrates that high responses were observed for the majority of active pump substrates in ABCB1, with the only exceptions being C2925 and D20350. Fig. 4 also shows that the efflux activity of ABCC1 closely mirrors that of ABCB1, with the exception of substrate C2925 as noted above. The ABCG2 profile of effluxed substrates was significantly narrower than the other two transporters, indicating higher specificity in pump–substrate interactions. Fewer than a dozen substrates showed responses above 75%, including substrate D20350, which appeared to be ABCG2 selective at approximately 65% response.
Fig.4.
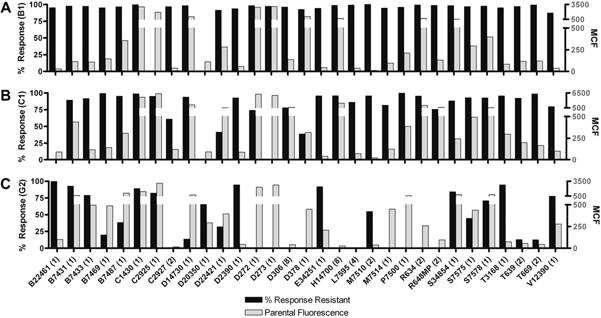
The 31 substrates (100 nM) chosen for further study were plotted as percentage response of transporter-expressing cells (black bars) as compared with the parental MCF cellular retention values (gray bars) to determine substrates with both high efflux responses and reasonable fluorescence ranges. (A) High efflux responses were observed for the majority of the substrates in ABCB1 with the exceptions of C2925 and D20350. (B) ABCC1 closely mirrors the efflux activity of ABCB1 with some selectivity observed, such as substrate C2925, which appears not to be an ABCB1 substrate. (C) The efflux profile of ABCG2 is significantly more substrate specific than either of the other two pumps. Fewer than a dozen substrates have responses above 75%. Substrate D20350 appears to be ABCG2 selective at approximately 65% response.
Substrate efflux inhibition
The 31 fluorescence substrates from Fig. 4 were tested against a panel of eight inhibitors known for their activity versus the three transporters. Nicardipine is a strong cross-reactive inhibitor for all three transporters and was used as the reference standard [12]. MCF was used to plot the dose–response data for the eight inhibitors with each fluorescent substrate and to generate IC50 values. After curve fitting, Hill slopes ranged from 0.3 to 10.9 with a mean of 2.1 ± 1.3 (median = 2.1), with 42% between 0.5 and 1.5 and 82% between 0 and 3. Fig. 5 illustrates the results for the fully cross-reactive efflux substrate, BODIPY histamine (B22461), inhibited by both mometasone and nicardipine for ABCB1, ABCC1, and ABCG2.
Fig.5.
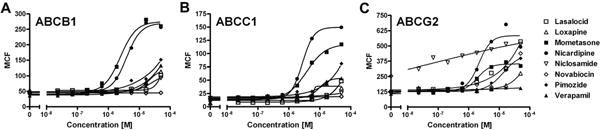
Representative dose–response curves of the efflux inhibition of BODIPY FL histamine (B22461). The concentration of each inhibitor ranged from 7.6 nM to 50 μM. All IC50 values are given as the average of multiple runs (n ≥ 2; subsequent curves not shown). (A) B22461 efflux in ABCB1-overexpressing CCRF-ADR 5000 cells was inhibited by both mometasone (closed squares) and nicardipine (closed circles), with IC50 values of 9.1 ± 8.4 and 3.2 ± 1.0 μM, respectively. Weak nonquantitative inhibition was also noted for pimozide (closed diamonds) and verapamil (closed upward-pointing triangles). (B) The ABCC1 (SupT1-Vin) efflux inhibition profile of B22461 closely mirrored that of ABCB1; however, pimozide and verapamil inhibition levels were within the measured concentration range, affording IC50 values of 11.1 ± 6.4 and 12.4 ± 4.9 μM, respectively. The IC50 value for mometasone was 3.7 ± 1.0 μM, and that for nicardipine was 1.2 ± 0.9 μM. (C) Inhibition of B22461 efflux in ABCG2 (Ig-MXP3) was seen with mometasone, nicardipine, and pimozide, with IC50 values of 4.4 ± 3.4, 4.3 ± 2.3, and 14.2 ± 4.3 μM, respectively. Over multiple runs, a weak inhibition was also noticed with niclosamide (open downward-pointing triangles), albeit with lower than average Hill slopes and relatively high baseline fluorescence.
Table 2 shows the IC50 values calculated for the active inhibitor/substrate pairs. Novobiocin was inactive and excluded from further consideration. Although efflux was noted for CellTracker Green CMFDA (C2925), D20350, DiOC7(3) (D378), and BODIPY taxol (P7500), no notable inhibition was observed and these substrates are also excluded from Table 2. For visualization, substrate/inhibitor pairs, clustered by fluorophore, are also depicted as a heat map in Fig. 6, showing the activity of each substrate in each cell line. It should be noted that for fluorescently labeled small molecules (i.e., the majority of active BODIPY probes), this clustering method only roughly indicates structure–activity relationships due to the variation in the nonfluorophore moiety. A distinction also needs to be made between efflux in the first round of screening and efflux inhibition by one or more inhibitors for each ABC transporter-expressing cell line. Although not depicted, substrates with noninhibited efflux activity are discussed by fluorophore. Of the 27 inhibitable substrates, 19 contained BODIPY or cyanine fluorophore, with 7 of the remaining 8 containing rhodamine, rosamine, or fluorescein.
Table 2.
Summary of known efflux inhibitor profile with potential fluorescent probes.
| Fluorescent substrate | Lasalocid | Loxapine | Mometasone | Nicardipine | Pimozide | Verapamil | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|||||||||||||
| B1 | C1 | G2 | B1 | C1 | G2 | B1 | C1 | G2 | B1 | C1 | G2 | B1 | C1 | G2 | B1 | C1 | G2 | |
| B22461c | – | – | w | – | – | – | 9.1 ± 8.4 | 3.7 ± 1.0 | 4.4 ± 3.4 | 3.2 ± 1.0 | 1.2 ± 0.9 | 4.3 ± 2.3 | w | 11.1 ± 6.4 | 14.2 ± 4.3 | w | 12.4 ± 4.9 | – |
| B7431a | – | – | – | 15.6 ± 10.9 | 10.1 ± 4.4 | – | 1.9 ± 1.1 | 1.6 ± 1.0 | – | 6.8 ± 4.8 | 2.6 ± 1.1 | – | 5.2 ± 5.2 | 9.7 ± 8.2 | – | 9.9 ± 6.6 | 12.3 ± 2.5 | – |
| B7433a | – | – | w | 15.9 ± 6.3 | 8.0 ± 1.9 | w | 2.6 ± 1.1 | 1.0 ± 0.5 | 2.1 ± 0.8 | 0.6 ± 0.3 | 2.6 ± 2.2 | 1.0 ± 0.8 | 9.4 ± 4.6 | w | W | 12.3 ± 7.4 | 7.2 ± 7.3 | – |
| B7469a | – | – | w | 4.3 ± 1.4 | w | w | 0.5 ± 0.1 | w | – | 2.0 ± 2.0 | 6.2 ± 6.0 | 0.7 ± 0.4 | 7.0 ± 1.9 | w | – | w | w | w |
| B7487a | – | – | – | 6.1 ± 2.3 | 15.5 ± 5.8 | – | 1.2 ± 1.3 | 1.0 ± 0.7 | – | 1.9 ± 2.3 | 0.6 ± 0.0 | – | w | w | – | 7.1 ± 6.2 | w | – |
| C1430b | – | – | – | w | w | – | 1.9 ± 1.6 | 5.4 ± 5.6 | – | 5.8 ± 2.8 | 4.3 ± 5.0 | – | 13.0 ± 3.0 | 7.7 ± 3.2 | – | w | w | – |
| C2927c | – | – | – | w | w | – | 0.8 ± 0.2 | w | – | 1.1 ± 0.8 | 0.6 ± 0.4 | – | 6.1 ± 1.3 | w | W | 17.9 ± 19.9 | 15.5 ± 6.7 | – |
| D14730a | – | – | – | 8.3 ± 6.5 | 12.9 ± 8.0 | – | 2.4 ± 1.6 | 1.4 ± 0.3 | – | 4.0 ± 2.1 | 5.3 ± 2.9 | – | 2.4 ± 1.3 | 1.9 ± 0.7 | – | 12.8 ± 1.9 | 5.8 ± 2.7 | – |
| D22421a | – | – | – | 1.7 ± 0.9 | 2.5 ± 1.7 | – | 0.9 ± 0.6 | 1.9 ± 1.4 | – | 0.6 ± 0.5 | 2.7 ± 4.1 | – | 3.5 ± 2.6 | 9.4 ± 3.6 | – | 1.5 ± 0.4 | 1.1 ± 0.3 | – |
| D2390c | 9.0 ± 6.2 | 13.5 ± 2.1 | 4.2 ± 2.4 | 9.8 ± 6.0 | w | w | 1.5 ± 0.6 | 2.5 ± 1.8 | w | 1.2 ± 0.9 | 1.0 ± 0.4 | w | 5.9 ± 1.9 | 3.7 ± 2.8 | – | 6.2 ± 5.3 | 9.6 ± 4.4 | – |
| D272a | – | – | – | w | w | – | 0.7 ± 0.2 | 2.1 ± 2.5 | – | 2.6 ± 3.8 | w | – | 3.8 ± 1.5 | 2.9 ± 0.7 | W | w | 22.1 ± 11.7 | – |
| D273a | – | – | – | w | w | – | 0.3 ± 0.1 | w | – | 0.8 ± 0.9 | w | – | 1.7 ± 0.9 | 1.4 ± 0.7 | W | 6.3 ± 7.3 | 7.1 ± 3.2 | – |
| D306c | – | – | – | 20.1 ± 19.4 | – | – | 3.2 ± 4.6 | 10.2 ± 9.3 | – | 1.4 ± 0.6 | – | – | 3.9 ± 3.2 | w | – | 8.0 ± 8.8 | w | – |
| E34251c | – | – | w | w | w | – | 2.7 ± 1.9 | w | – | 5.0 ± 1.5 | 4.0 ± 3.2 | – | w | 12.9 ± 9.1 | – | w | w | – |
| H14700a | w | w | – | w | w | – | 4.1 ± 2.5 | 3.9 ± 2.2 | – | 3.3 ± 2.5 | w | – | 10.8 ± 2.9 | 6.0 ± 2.1 | – | w | w | – |
| L7595c | – | – | – | 15.2 ± 7.5 | – | – | 0.9 ± 0.2 | w | – | 1.7 ± 1.0 | w | – | 7.1 ± 3.0 | – | – | 8.9 ± 5.7 | w | – |
| M7510c | – | – | – | w | w | – | 2.2 ± 1.6 | w | – | 2.0 ± 0.7 | w | – | 5.8 ± 3.4 | w | – | w | w | – |
| M7514c | – | – | – | 6.7 ± 0.1 | w | – | 6.6 ± 8.7 | 3.6 ± 0.9 | – | 7.8 ± 1.9 | 11.9 ± 10.8 | – | 7.8 ± 9.3 | w | – | 7.1 ± 1.1 | 11.1 ± 1.5 | – |
| R634a | – | – | – | w | – | – | 3.6 ± 2.2 | w | – | 10.1 ± 5.6 | w | – | w | w | – | w | – | – |
| R648MPc | w | – | – | w | – | – | 2.9 ± 2.0 | 2.2 ± 1.1 | – | 0.4 ± 0.2 | w | – | 5.3 ± 4.9 | 2.6 ± 2.3 | – | w | – | – |
| S34854a | – | – | – | w | w | – | 3.1 ± 2.5 | 1.2 ± 1.0 | – | 2.0 ± 0.6 | 5.6 ± 6.7 | – | 6.9 ± 1.3 | w | W | 23.1 ± 0.3 | 9.9 ± 0.2 | – |
| S7575a | – | – | – | 9.8 ± 3.1 | 16.2 ± 4.8 | – | 1.6 ± 1.6 | 1.0 ± 0.8 | – | 2.3 ± 1.1 | 1.4 ± 1.1 | – | 7.1 ± 4.5 | 7.2 ± 2.6 | – | 7.1 ± 1.9 | 15.3 ± 7.5 | – |
| S7578a | – | – | – | w | w | – | 0.6 ± 0.3 | w | – | 1.8 ± 2.6 | – | – | 8.3 ± 7.0 | 3.3 ± 0.4 | W | w | w | – |
| T3168c | – | – | 5.1 ± 3.9 | w | w | w | 2.6 ± 1.9 | 7.0 ± 3.6 | 0.9 ± 0.1 | 3.7 ± 0.8 | 2.6 ± 1.5 | 0.5 ± 0.1 | 15.6 ± 7.6 | 5.3 ± 2.6 | 5.7 ± 4.2 | w | w | – |
| T639c | – | – | – | w | w | – | 1.1 ± 0.4 | 3.6 ± 2.3 | – | 1.1 ± 0.5 | w | – | 4.6 ± 0.4 | 7.5 ± 2.7 | – | w | 16.7 ± 4.0 | – |
| T669c | – | – | – | – | – | – | 7.1 ± 8.2 | w | – | 9.4 ± 3.6 | w | – | w | – | – | – | – | – |
| V12390c | – | – | – | 22.3 ± 2.5 | w | – | 3.0 ± 2.6 | 1.8 ± 1.1 | – | 0.6 ± 0.4 | 1.8 ± 1.4 | – | w | w | – | 6.2 ± 0.2 | 9.2 ± 6.1 | – |
Note. B1, ABCB1-overexpressing CCRF-ADR 5000 cells; C1, ABCB1-overexpressing SupT1-Vin cells; G2, ABCG2-overexpressing Ig-MXP3 cells. Values are mean IC50 values ± standard deviations (n ≥ 2). The fluorescent substrate concentrations used were one of the following: a100 nM; b250 nM; c500 nM. A dash (–) indicates a lack of response, and a “w” indicates an apparent inhibitory response that fell outside of the tested concentration range or the established cutoff parameters.
Fig.6.
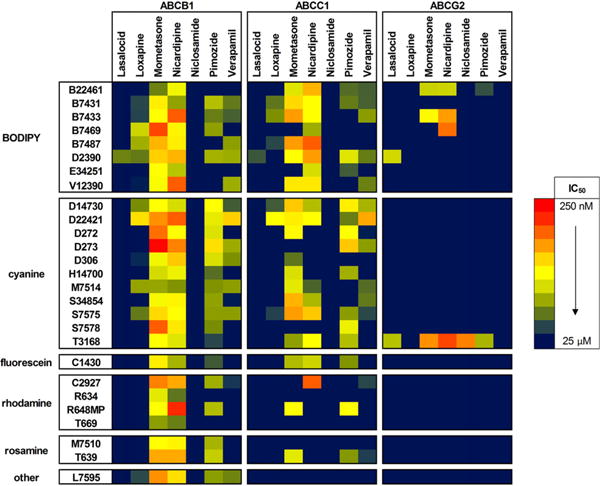
Heat map of inhibition responses for seven inhibitors against fluorescent substrates in each of the ABC transporter-overexpressing cell lines. Values are represented as IC50 values without error values for the sake of clarity. ABCB1 (CCRF-ADR 5000) demonstrated the highest number of effluxed fluorescent substrates of the three tested transporter pumps. Mometasone and nicardipine inhibition was observed for all of the shown substrates, which included some of the lowest IC50 values. For example, sub-micromolar inhibition of the cyanine DiOC6(3) (D273) was observed with both mometasone (0.3 ± 0.1 μM) and nicardipine (0.8 ± 0.9 μM). Pimozide and verapamil were also shown to be inhibitors of a majority of the substrates, albeit with higher IC50 values. No activity was seen with niclosamide in ABCB1, and only BODIPY FL EDA (D2390) was inhibited by lasalocid (IC50 = 9.0 ± 6.2 μM). Similar inhibitory activity was seen in ABCC1 (SupT1-Vin) as compared with ABCB1 with the BODIPY, cyanine, and single fluorescein substrates. Of the rhodamine/rosamine substrates shown, only CellTracker Orange CMTMR (C2927), rhodamine B, hexyl ester, perchlorate (R6, R648MP), and tetramethylrosamine chloride (T639) were shown to have inhibitable efflux activity. As in ABCB1, mometasone, nicardipine, pimozide, and verapamil were the common inhibitors. Again, no activity was seen with niclosamide in ABCC1, and only D2390 was inhibited by lasalocid (IC50 = 13.5 ± 2.1 μM). Much less activity was seen in ABCG2 (Ig-MXP3), and except for the three-pump cross-substrate JC-1 (T3168), the inhibition profiles seemed to be more selective. T3168 efflux was inhibited by lasalocid, mometasone,nicardipine, and pimozide in a range of IC50 values. Interestingly, T3168 in ABCG2 was the only substrate efflux inhibited by niclosamide (IC50 = 0.8 ± 0.6 μM). The remaining active substrates were all members of the BODIPY fluorophore subset and included the nicardipine-inhibited three-pump cross-substrates BODIPY FL histamine (B22461), BODIPY FL prazosin (B7433), and BODIPY FL forskolin (B7469). Once again, D2390 was inhibited by lasalocid (IC50 = 4.2 ± 2.4 μM).
Aside from the previously mentioned efflux-inactive M1198MP, the fluorescein probes CaAM (C1430) and C2925 demonstrated efflux activity in the primary screen for ABCB1/ABCC1/ABCG2 and ABCC1/ABCG2, respectively. Only C1430 was taken forward into the inhibition screen, where inhibition of ABCB1/ABCC1 responses with mometasone, nicardipine, and pimozide was observed. No significant selectivity was seen between ABCB1 and ABCC1, with all IC50 values being in the low micromolar (μM) range for mometasone (1.9 ± 1.6 and 5.4 ± 5.6 μM, respectively) and nicardipine (5.8 ± 2.8 and 4.3 ± 5.0 μM, respectively). In a flow cytometric fluorescence retention analysis, Wang and coworkers reported ABCB1 efflux inhibition of CaAM with nicardipine at an IC50 of 6.6 ± 0.4 μM [21], which correlated well with the IC50 value reported here.
A total of 34 rhodamine/rosamine-based compounds were represented in the collection. Unconjugated alkyl amine-substituted rhodamine probes tended to be active in ABCB1 or ABCB1/ABCC1 efflux and inhibitor assays provided that the carboxylic acid was ester protected (R634, R648MP, and T669). The exception was the membrane probe R18 (O246), with its octadecyl ester demonstrating no cellular fluorescence in the efflux assay. An exception to the ester-based activity rule was the free carboxylate-containing CellTracker Orange CMTMR (C2927), where the aryl amide substitution appears to maintain adequate lipophilicity to facilitate membrane permeability. All four of the rhodamine substrates tested in the inhibitor assay (R634, R648MP, T669, and C2927) showed quantifiable ABCB1 efflux inhibition with both mometasone and nicardipine. Although not fully illustrated in Fig. 6, each of these substrates was at least weakly inhibited by mometasone and nicardipine in ABCC1 as well (Table 2). However, the potential for high selectivity of ABCB1 over ABCC1 can be seen in the sub-μM ABCB1 efflux inhibition example of R648MP with nicardipine. Inhibitor-based substrate efflux variation can also be seen with R648MP, which was observed to have an ABCB1/ABCC1 cross-pump interaction with pimozide. C2927 efflux was inhibited by pimozide as well as verapamil in ABCB1 and ABCC1. No significant inhibition was seen for these four rhodamine substrates with lasa-locid, loxapine, or niclosamide. Despite a long history of use in transporter efflux assays [22], rhodamine 123 (R302) was observed to have comparatively low fluorescence levels at the available wavelengths and was not explored further in the inhibition protocol.
Rosamine-based tetramethylrosamine chloride (T639) and the MitoTracker dyes M7510 and M7512 showed ABCB1/ABCC1 efflux potential, albeit at lower than optimal fluorescence levels. Low-μM efflux inhibition of M7510 and T639 was observed in ABCB1 with mometasone, nicardipine, and pimozide. T639 also demonstrated similar ABCC1 efflux inhibition with mometasone, pimozide, and (to a lesser degree) verapamil. The ABCB1 T639 efflux inhibition result also correlated with low-μM nicardipine inhibition (IC50 = 11.7 μM) previously reported by Wang and coworkers [21].
A total of 37 BODIPY-based probes were tested in the primary efflux screen, with 8 going forward into the inhibition assay. Aqueous solubility of BODIPY analogs is often of concern and likely affected those compounds without polar functional groups, resulting in low cellular fluorescence in the efflux assay. Although efflux by ABCB1/ABCC1/ABCG2 was noted for the acidic compartment tracer LysoTracker Green DND-26 (L7526), the low-level efflux response coupled with less than optimal fluorescence ranges excluded it from further investigation. BODIPY EDA (D2390) demonstrated a low fluorescence but high response efflux activity across all three pumps, which translated into an interesting inhibition profile in the subsequent screen. D2390 showed full cross-pump inhibition in ABCB1, ABCB2, and ABCG2 with lasalocid and was one of only two probes inhibited by lasalocid. The efflux inhibition of D2390 by loxapine, mometasone, nicardipine, pimozide, and verapamil was noted only for ABCB1 and ABCC1 at low-μM IC50 values.
Although little activity was seen with the majority of BODIPY labeling agents, BODIPY-labeled small molecules demonstrated activities apparently related to the labeled molecule rather than the fluorophore. In the primary efflux screen, pan-ABCB1, -ABCC1, and -ABCG2 activity was observed for BODIPY prazosin (B7433), glibenclamide (E34251, ER-Tracker Green), verapamil HCl (B7431), and vinblastine (V12390). The inhibition profile of these four substrates was more varied. B7433 has previously been described as an ABCG2 substrate and used as a fluorescent probe for inhibitors in ABCG2-transfected HEK293 cells [23]. Here, B7433 demonstrated ABCB1, ABCC1, and ABCG2 inhibition with mometasone and nicardipine and showed moderate ABCB1 and ABCC1 efflux inhibition with loxapine, pimozide, and verapamil. Unlabeled glibenclamide is a competitive ABCB1 inhibitor [24]. Here, labeled glibenclamide (E34251) showed both ABCB1 and ABCC1 efflux inhibition with nicardipine. Verapamil is well known in multidrug resistance (MDR) reversal [25] and the BODIPY-labeled analog (B7431) has been described as a substrate for both ABCB1 and ABCC1 [26,27]. We observed low to moderate ABCB1 and ABCC1 efflux inhibition by B7431 with loxapine, mometasone, nicardipine, pimozide, and verapamil. Vinca alkaloids are ABCB1 and ABCC1 pump substrates [28], and low-μM ABCB1 and ABCC1 efflux inhibition was noted here for BODIPY vinblastine (V12390) with mometasone, nicardipine, verapamil, and (to a lesser degree) loxapine. Although active efflux of BODIPY histamine (B22461) was noted only for ABCB1 and ABCG2, inhibition was observed with mometasone and nicardipine across all three pumps, indicating that it was indeed an efflux substrate for ABCC1. Reduced inhibition was also noted for B22461 with pimozide (ABCC1 and ABCG2) and verapamil (ABCC1). ABCB1 and ABCC1 efflux activity was observed for BODIPY forskolin (B7469), BODIPY thapsigargin (B7487), and BODIPY taxol (P7500). B7469 efflux was inhibited by nicardipine across all three pumps along with ABCB1 inhibition by loxapine, mometasone, and pimozide. Efflux of B7487 was inhibited to varying degrees for ABCB1 and ABCC1 by loxapine, mometasone, and nicardipine. The cost and availability of P7500 made it less attractive, and it was not carried forward into the inhibition screen.
Of the 17 cyanine dyes tested in the efflux assay, 12 were taken into the inhibition screen. The monomeric cyanine dyes, the dead cell indicator TO-PRO-1 iodide (T3602), the apoptotic cell stain YO-PRO-1 iodide (Y3603), and the cell-impermeant nucleic acid stain YOYO-1 iodide (Y3601), not surprisingly, showed no parental cell uptake in the efflux screening protocol. The asymmetric cyanine dyes (M7514, 34854, S7575, and S7578) showed varying degrees of inhibition of ABCB1 and ABCC1 by loxapine, mometasone, nicardipine, pimozide, and verapamil. Limited cellular fluorescence was noted for the lipophilic tracer indocarbocya-nine dyes D383 and D7776. However, the potentiometric probes indodicarbocyanine (DiIC1(5), H14700) and thiadicarbocyanine (DiSC3(5), D306) both demonstrated inhibitable efflux responses. H14700 efflux was inhibited by mometasone and pimozide for ABCB1 and ABCC1 and by nicardipine for ABCB1 alone. ABCB1 efflux of D306 was inhibited by loxapine, nicardipine, pimozide, and verapamil along with ABCB1 and ABCC1 inhibition with mometasone. The oxacarbocyanine probes (D272, D273, D378, D14730, and D22421) exhibited ABCB1 or ABCB1/ABCC1 efflux activity, which was inhibited to varying degrees by mometasone, nicardipine, pimozide, and verapamil. The benzimidazolylcarbocy-anine JC-1 (T3168) [12,29] was shown to be an efflux substrate for all three ABC transporter pumps, and at an optimal 500-nM concentration efflux was inhibited by mometasone, nicardipine, and pimozide for ABCB1, ABCC1, and ABCG2. The nicardipine ABCB1 efflux response for T3168 matches earlier data (IC50 values of 5.8 ± 2.8 vs. our 7.1 ± 0.7 μM) [21]. T3168 efflux inhibition was also reported in ABCG2 alone for lasalocid (5.1 ± 3.9 μM) and niclosamide (0.8 ± 0.6 μM), demonstrating unique substrate/inhibitor specificity.
Of three probes (L7595, N1142, and P7581) not placed into fluorophore categories, the efflux of the ABCB1 substrate LDS 751 (L7595) [30,31] was inhibited with loxapine, mometasone, nicardipine, pimozide, and verapamil. Reported IC50 values for nicardipine (5.6 ± 0.5 μM) and verapamil (4.7 ± 1.3 μM) [21] compare with ours (1.7 ± 1.0 and 8.9 ± 5.7 μM, respectively).
Discussion
Taken together, our results reveal a general correspondence of efflux substrates between ABCB1 and ABCC1, with ABCB1 exhibiting somewhat more diversity. In contrast, the list of efflux substrates for ABCG2 was more restricted, indicating higher selectivity in pump–substrate interactions. The majority of tested efflux substrates have BODIPY or cyanine fluorophores and, interestingly, BODIPY is analogous to a rigid monomethine cyanine dye. Moreover, the selectivity of efflux activities by BODIPY-labeled small molecules correlated more with the labeled molecule than the fluorophore. Evaluations of inhibitors and efflux substrate pairs yielded mometasone and nicardipine as pan-inhibitors for multiple efflux substrates across ABCB1 and ABCC1. Our control inhibitor nicardipine blocked efflux from all three transporters (ABCB1, ABCC1, and ABCG2) for four substrates: carbocyanine JC-1 (T3168) and BODIPY-labeled small molecules BODIPY prazosin (B7433), BODIPY forskolin (B7469), and BODIPY histamine (B22461). In an orthogonal view, the cyanine compound T3168 was a pan-substrate for ABCG2 across a majority of the inhibitors and was the sole ABCG2 substrate inhibited by niclosamide. The combination of BODIPY-labeled substrate D2390 with lasalocid was active for all three pumps. D2390 was the only substrate besides T3168 inhibited by lasalocid, suggesting related binding sites for this substrate/inhibitor pair.
Based on the response of the control inhibitor nicardipine, 12 substrates demonstrated ABCB1/ABCC1 efflux inhibition; however, no ABCC1, ABCG2, ABCB1/ABCG2, or ABCC1/ABCG2 selectivity was observed. Mometasone, pimozide, and (to a lesser degree) verapamil displayed similar selectivity profiles as nicardipine across the substrate set. Lasalocid was originally chosen as a specific inhibitor of T3168 efflux for ABCG2, and this interaction was replicated here with an IC50 value of 5.1 ± 3.9 μM, with limited activity across the rest of the substrates. Niclosamide, the other ABCG2-specific T3168 efflux inhibitor, was found to inhibit only T3168 efflux for ABCG2 (IC50 = 0.8 ± 0.6 μM). The two ABCC1 inhibitors, loxapine and verapamil, produced similar ABCB1 and ABCC1 inhibition profiles over the substrate set.
The three transporters (ABCB1, ABCC1, and ABCG2) reported on here are known to significantly influence the ADME-Tox (absorption, distribution, metabolism, excretion, and toxicity) properties of drugs [32]. Although a large number of compounds possess ABC transporter inhibitory properties, only a few of these agents are appropriate candidates for clinical use as MDR reversal agents [33]. Characterization of the transporter/substrate/inhibitor interactions might provide further clues about their structure and mechanism of action as well as aid in predicting potential drug interactions among different therapeutic agents. Ongoing clinical trials with third-generation modulators (e.g., biricodar, zosuquidar, laniquidar) [34] have not yet defined an ideal MDR reversal agent [35]. Main liabilities from cross-reactivity of these inhibitors with major ABC transporters involved in the body's physiological protection from xenobiotics and endogenous metabolites result in high toxicity and mortality in patients. Acquired mutations in transporter genes introduce more complexity, altering the pattern of resistance and improving the ability of the mutants to efflux new drugs [36]. Moreover, drug-resistant human tumor cell lines express different ABCG2 variants, suggested to be gain-of-function mutations acquired during the course of drug exposure [37]. Single amino acid changes alter the drug resistance profile and substrate specificity compared with wild-type ABCG2 [38]. Thus, daunorubicin, used to identify ABCB1 inhibitors, is a substrate for ABCG2 mutants [39].
A diverse set of compounds are substrates for efflux pumps, with many showing cross-pump activity. It stands to reason that, for each pump, more than one combination of substrate and inhibitor is required to properly characterize that particular pump's activity. For example, rhodamine 123 has been used in combination with Hoechst 33342 to describe two functional transport sites in ABCB1 with complex allosteric interactions [40]. In combination with LDS 751, it was also shown that the rhodamine 123 may bind to a different or overlapping region within the same large flexible binding site as LDS 751 [41]. It has also been shown that ABCB1 possesses two allosterically coupled drug acceptor sites where one binds vinblastine, doxorubucin, etoposide, and cyclosporin A and the other binds dexniguldipine-HCl and other 1,4-dihydropyri-dines [42]. Fluorescent substrates in combination with high-throughput flow cytometry can be a powerful tool for transporter interaction studies. The single-transporter fluorescent substrate, pheophorbide A, was shown to be ABCG2 specific. Its transport correlated with ABCG2 expression, and an HTS campaign identified ABCG2 inhibitors [43,44]. Further development of high-throughput assay systems to screen for potential transporter-interacting partners may be of particular interest to help elucidate structure and function within a given transporter.
The current study indicates that each substrate has not only diverse activity for multiple transporters but also unique interactions with different inhibitors, suggesting the involvement of different binding sites in substrate recognition and transport inhibition processes. Such widely reported promiscuity (or polyspecificity) does not allow a definitive correlation between substrate specificity and structural characteristics of the fluorophores investigated here. However, several trends in transporter/inhibitor/substrate interactions were identified. We found striking similarity in substrate specificity profiles between ABCB1 and ABCC1 transporters. All ABCC1 efflux compounds in our test system demonstrated variable levels of interaction with ABCB1. Moreover, mometasone and nicardipine, two structurally diverse compounds, inhibited most of the ABCB1/ABCC1-specific compounds at low concentration. On the other hand, only 10 of 102 tested fluorescent dyes were effluxed by ABCG2, and only 5 of them could be inhibited by the tested inhibitors. Interestingly, T3168 efflux by ABCG2 was inhibited by six of seven tested inhibitors. The other four ABCG2-specific dyes belong to the BODIPY family.
One of the major aims of the current research was to provide novel fluorescent tools for understanding drug resistance. Well-characterized fluorescent probes with variable profiles of selectivity can be used to identify new inhibitors as well as to characterize functional expression of ABCB1, ABCC1, and ABCG2 in tissue samples. Pan-efflux substrate probes such as T3168 and the BODIPY-FL-labeled small molecules histamine (B22461), prazosin (B7433), and forskolin (B7469) could help to define the expression of several transporters in combination with inhibitors such as nicardipine and mometasone. By varying the inhibitor, in principle, one could evaluate pump expression such as T3168 combined with lasalocid (IC50 = 5.1 μM) or niclosamide (IC50 = 0.8 μM) for ABCG2 expression or ABCB1 expression probed with LDS 751 (L7595) and inhibitors such as loxapine, mometasone, nicardipine, pimozide, and verapamil. Depending on the system of interest, combinations of such substrate/inhibitor pairs might describe the ABC transporter phenotype. These data could also be useful for predicting common drug inhibition/drug binding patterns of ABC transporters and contribute to a better understanding of the pharmacological mechanisms of transporter–reversal agent interactions. Further refinement of the data presented here could also lead to multicolor probe sets that identify unique binding motifs in a single pump or across pumps, allowing for concurrent elucidation of the mode of action for existing or new ABC transporter modulators and substrates.
Acknowledgments
This work was supported by grant U54MH084690 from the National Institutes of Health, the University of New Mexico Center for Molecular Discovery, the University of New Mexico Shared Flow Cytometry Resource, and the University of New Mexico Cancer Research and Treatment Center. We thank M. Ignatius (formerly of Life Technologies) for helpful discussions about fluorescent substrates, D. D. Ross and T. Nakanishi (University of Maryland School of Medicine) for providing Igrov1 and Ig-MXP3 cell lines, and T. Efferth (German Cancer Research Center) for providing the CCRF-ADR 5000 drug-resistant cell line. We honor Susan M. Young for her extraordinary flow cytometry skills and infectious laughter, sorely missed by all of us.
Footnotes
Abbreviations used: ABC, ATP binding cassette; P-gp, P-glycoprotein (ABCB1); MRP1, multidrug resistance protein 1 (ABCC1); BCRP, breast cancer resistance protein (ABCG2); JC-1, 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide; CaAM, calcein acetoxymethyl ester; DMSO, dimethyl sulfoxide; Vin, vincristine; PBS, phosphate-buffered saline; MCF, median channel fluorescence; HTS, high-throughput screening; MDR, multidrug resistance.
T3168 cellular retention can be followed via its green monomeric emission or its red J-aggregate emission. The filter set of our cytometer was not optimal for the approximately 590-nm red emission that is more often used for the fluorescent readout.
References
- 1.Krishna R, Mayer LD. Multidrug resistance (MDR) in cancer: mechanisms, reversal using modulators of MDR, and the role of MDR modulators in influencing the pharmacokinetics of anticancer drugs. Eur J Pharm Sci. 2000;11:265–283. doi: 10.1016/s0928-0987(00)00114-7. [DOI] [PubMed] [Google Scholar]
- 2.Goldman B. Multidrug resistance: can new drugs help chemotherapy score against cancer? J Natl Cancer Inst. 2003;95:255–257. doi: 10.1093/jnci/95.4.255. [DOI] [PubMed] [Google Scholar]
- 3.Schinkel AH, Jonker JW. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv Drug Deliv Rev. 2003;55:3–29. doi: 10.1016/s0169-409x(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 4.Sarkadi B, Homolya L, Szakacs G, Varadi A. Human multidrug resistance ABCB and ABCG transporters: Participation in a chemoimmunity defense system. Physiol Rev. 2006;86:1179–1236. doi: 10.1152/physrev.00037.2005. [DOI] [PubMed] [Google Scholar]
- 5.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin C, Berridge G, Higgins CF, Mistry P, Charlton P, Callaghan R. Communication between multiple drug binding sites on P-glycoprotein. Mol Pharmacol. 2000;58:624–632. doi: 10.1124/mol.58.3.624. [DOI] [PubMed] [Google Scholar]
- 7.Clark R, Kerr ID, Callaghan R. Multiple drug-binding sites on the R482G isoform of the ABCG2 transporter. Br J Pharmacol. 2006;149:506–515. doi: 10.1038/sj.bjp.0706904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin JH. Drug–drug interaction mediated by inhibition and induction of P-glycoprotein. Adv Drug Deliv Rev. 2003;55:53–81. doi: 10.1016/s0169-409x(02)00171-0. [DOI] [PubMed] [Google Scholar]
- 9.Kruijtzer CMF, Beijnen JH, Rosing H, ten Bokkel Huinink WW, Schot M, Jewell RC, Paul EM, Schellens JHM. Increased oral bioavailability of topotecan in combination with the breast cancer resistance protein and P-glycoprotein inhibitor GF120918. J Clin Oncol. 2002;20:2943–2950. doi: 10.1200/JCO.2002.12.116. [DOI] [PubMed] [Google Scholar]
- 10.Xia CQ, Milton MN, Gan LS. Evaluation of drug–transporter interactions using in vitro and in vivo models. Curr Drug Metab. 2007;8:341–363. doi: 10.2174/138920007780655423. [DOI] [PubMed] [Google Scholar]
- 11.Sharom FJ. The P-glycoprotein efflux pump: how does it transport drugs? J Membr Biol. 1997;160:161–175. doi: 10.1007/s002329900305. [DOI] [PubMed] [Google Scholar]
- 12.Ivnitski-Steele I, Larson RS, Lovato DM, Khawaja HM, Winter SS, Oprea TI, Sklar LA, Edwards BS. High-throughput flow cytometry to detect selective inhibitors of ABCB1, ABCC1, and ABCG2 transporters. Assay Drug Dev Technol. 2008;6:263–276. doi: 10.1089/adt.2007.107. [DOI] [PubMed] [Google Scholar]
- 13.Strouse JJ, Ivnitski-Steele I, Khawaja HM, Perez D, Ricci J, Yao T, Weiner WS, Schroeder CE, Simpson DS, Maki BE, Li K, Golden JE, Foutz TD, Waller A, Evangelisti AM, Young SM, Chavez SE, Garcia MJ, Ursu O, Bologa CG, Carter MB, Salas VM, Gouveia K, Tegos GP, Oprea TI, Edwards BS, Aubé J, Larson RS, Sklar LA. A selective ATP-binding cassette subfamily G member 2 efflux inhibitor revealed via high-throughput flow cytometry. J Biomol Screen. 2013;18:26–38. doi: 10.1177/1087057112456875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuckuck FW, Edwards BS, Sklar LA. High throughput flow cytometry. Cytometry. 2001;44:83–90. [PubMed] [Google Scholar]
- 15.Ramirez S, Aiken CT, Andrzejewski B, Sklar LA, Edwards BS. High-throughput flow cytometry: validation in microvolume bioassays. Cytometry. 2003;53:55–65. doi: 10.1002/cyto.a.10035. [DOI] [PubMed] [Google Scholar]
- 16.Winter SS, Jiang Z, Khawaja HM, Griffin T, Devidas M, Asselin BL, Larson RS. Identification of genomic classifiers that distinguish induction failure in T-lineage acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood. 2007;110:1429–1438. doi: 10.1182/blood-2006-12-059790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckford PDW, Sharom FJ. ABC efflux pump-based resistance to chemotherapy drugs. Chem Rev. 2009;109:2989–3011. doi: 10.1021/cr9000226. [DOI] [PubMed] [Google Scholar]
- 18.Shiozawa K, Oka M, Soda H, Yoshikawa M, Ikegami Y, Tsurutani J, Nakatomi K, Nakamura Y, Doi S, Kitazaki T, Mizuta Y, Murase K, Yoshida H, Douglas DR, Kohno S. Reversal of breast cancer resistance protein (BCRP/ABCG2)-mediated drug resistance by novobiocin, a coumermycin antibiotic. Int J Cancer. 2004;108:146–151. doi: 10.1002/ijc.11528. [DOI] [PubMed] [Google Scholar]
- 19.Calcagno AM, Kim IW, Wu CP, Shukla S, Ambudkar SV. ABC drug transporters as molecular targets for the prevention of multidrug resistance and drug–drug interactions. Curr Drug Deliv. 2007;4:324–333. doi: 10.2174/156720107782151241. [DOI] [PubMed] [Google Scholar]
- 20.Lebedeva IV, Pande P, Patton WF. Sensitive and specific fluorescent probes for functional analysis of the three major types of mammalian ABC transporters. PLoS One. 2011;6:e22429. doi: 10.1371/journal.pone.0022429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang EJ, Casciano CN, Clement RP, Johnson WW. Active transport of fluorescent P-glycoprotein substrates: evaluation as markers and interaction with inhibitors. Biochem Biophys Res Commun. 2001;289:580–585. doi: 10.1006/bbrc.2001.6000. [DOI] [PubMed] [Google Scholar]
- 22.Efferth T, Lohrke H, Volm M. Reciprocal correlation between expression of P-glycoprotein and accumulation of rhodamine 123 in human tumors. Anticancer Res. 1989;9:1633–1637. [PubMed] [Google Scholar]
- 23.Henrich CJ, Robey RW, Takada K, Bokesch HR, Bates SE, Shukla S, Ambudkar SV, McMahon JB, Gustafson KR. Botryllamides: natural product inhibitors of ABCG2. ACS Chem Biol. 2009;4:637–647. doi: 10.1021/cb900134c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golstein PE, Boom A, van Geffel J, Jacobs P, Masereel B, Beauwens R. P-glycoprotein inhibition by glibenclamide and related compounds. Pfluegers Arch. 1999;437:652–660. doi: 10.1007/s004240050829. [DOI] [PubMed] [Google Scholar]
- 25.Ozols RF, Cunnion RE, Klecker RW, Jr, Hamilton TC, Ostchega Y, Parrillo JE, Young RC. Verapamil and adriamycin in the treatment of drug-resistant ovarian cancer patients. J Clin Oncol. 1987;5:641–647. doi: 10.1200/JCO.1987.5.4.641. [DOI] [PubMed] [Google Scholar]
- 26.Crivellato E, Candussio L, Rosati AM, Bartoli-Klugmann F, Mallardi F, Decorti G. The fluorescent probe Bodipy-FL-verapamil is a substrate for both P-glycoprotein and multidrug resistance-related protein (MRP)-1. J Histochem Cytochem. 2002;50:731–734. doi: 10.1177/002215540205000514. [DOI] [PubMed] [Google Scholar]
- 27.Rosati A, Candussio L, Crivellato E, Klugmann FB, Giraldi T, Damiani D, Michelutti A, Decorti G. Bodipy-FL-verapamil: a fluorescent probe for the study of multidrug resistance proteins. Cell Oncol. 2004;26:3–11. doi: 10.1155/2004/576173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avendano C, Menendez JC. Inhibitors of multidrug resistance to antitumor agents (MDR) Curr Med Chem. 2002;9:159–193. doi: 10.2174/0929867023371175. [DOI] [PubMed] [Google Scholar]
- 29.Kuhnel JM, Perrot JY, Faussat AM, Marie JP, Schwaller MA. Functional assay of multidrug resistant cells using JC-1, a carbocyanine fluorescent probe. Leukemia. 1997;11:1147–1155. doi: 10.1038/sj.leu.2400698. [DOI] [PubMed] [Google Scholar]
- 30.Lugo MR, Sharom FJ. Interaction of LDS-751 with P-glycoprotein and mapping of the location of the R drug binding site. Biochemistry. 2005;44:643–655. doi: 10.1021/bi0485326. [DOI] [PubMed] [Google Scholar]
- 31.Shapiro AB, Ling V. Transport of LDS-751 from the cytoplasmic leaflet of the plasma membrane by the rhodamine-123-selective site of P-glycoprotein. Eur J Biochem. 1998;254:181–188. doi: 10.1046/j.1432-1327.1998.2540181.x. [DOI] [PubMed] [Google Scholar]
- 32.Szakacs G, Varadi A, Ozvegy-Laczka C, Sarkadi B. The role of ABC transporters in drug absorption, distribution, metabolism, excretion, and toxicity (ADME-Tox) Drug Discov Today. 2008;13:379–393. doi: 10.1016/j.drudis.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 33.Mayur YC, Peters GJ, Prasad VV, Lemo C, Sathish NK. Design of new drug molecules to be used in reversing multidrug resistance in cancer cells. Curr Cancer Drug Targets. 2009;9:298–306. doi: 10.2174/156800909788166619. [DOI] [PubMed] [Google Scholar]
- 34.Seelig A, Gatlik-Landwojtowicz E. Inhibitors of multidrug efflux transporters: their membrane and protein interactions. Mini-Rev Med Chem. 2005;5:135–151. doi: 10.2174/1389557053402693. [DOI] [PubMed] [Google Scholar]
- 35.Gandhi L, Harding MW, Neubauer M, Langer CJ, Moore M, Ross HJ, Johnson BE, Lynch TJ. A phase II study of the safety and efficacy of the multidrug resistance inhibitor VX-710 combined with doxorubicin and vincristine in patients with recurrent small cell lung cancer. Cancer. 2007;109:924–932. doi: 10.1002/cncr.22492. [DOI] [PubMed] [Google Scholar]
- 36.Allen JD, Jackson SC, Schinkel AH. A mutation hot spot in the Bcrp1 (Abcg2) multidrug transporter in mouse cell lines selected for doxorubicin resistance. Cancer Res. 2002;62:2294–2299. [PubMed] [Google Scholar]
- 37.Mitomo H, Kato R, Ito A, Kasamatsu S, Ikegami Y, Kii I, Kudo A, Kobatake E, Sumino Y, Ishikawa T. A functional study on polymorphism of the ATP-binding cassette transporter ABCG2: critical role of arginine-482 in methotrexate transport. Biochem J. 2003;373:767–774. doi: 10.1042/BJ20030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozvegy-Laczka C, Koblos G, Sarkadi B, Varadi A. Single amino acid (482) variants of the ABCG2 multidrug transporter: major differences in transport capacity and substrate recognition. Biochim Biophys Acta. 2005;1668:53–63. doi: 10.1016/j.bbamem.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Coburger C, Lage H, Molnar J, Hilgeroth A. Impact of novel MDR modulators on human cancer cells: reversal activities and induction studies. Pharm Res. 2009;26:182–188. doi: 10.1007/s11095-008-9736-7. [DOI] [PubMed] [Google Scholar]
- 40.Shapiro AB, Ling V. Positively cooperative sites for drug transport by P-glycoprotein with distinct drug specificities. Eur J Biochem. 1997;250:130–137. doi: 10.1111/j.1432-1033.1997.00130.x. [DOI] [PubMed] [Google Scholar]
- 41.Lugo MR, Sharom FJ. Interaction of LDS-751 and rhodamine 123 with P-glycoprotein: evidence for simultaneous binding of both drugs. Biochemistry. 2005;44:14020–14029. doi: 10.1021/bi0511179. [DOI] [PubMed] [Google Scholar]
- 42.Malkhandi J, Ferry DR, Boer R, Gekeler V, Ise W, Kerr DJ. Dexniguldipine–HCl is a potent allosteric inhibitor of [3H]vinblastine binding to P-glycoprotein of CCRF ADR 5000 cells. Eur J Pharmacol. 1994;288:105–114. doi: 10.1016/0922-4106(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 43.Robey RW, Steadman K, Polgar O, Morisaki K, Blayney M, Mistry P, Bates SE. Pheophorbide a is a specific probe for ABCG2 function and inhibition. Cancer Res. 2004;64:1242–1246. doi: 10.1158/0008-5472.can-03-3298. [DOI] [PubMed] [Google Scholar]
- 44.Henrich CJ, Robey RW, Bokesch HR, Bates SE, Shukla S, Ambudkar SV, Dean M, McMahon JB. New inhibitors of ABCG2 identified by high-throughput screening. Mol Cancer Ther. 2007;6:3271–3278. doi: 10.1158/1535-7163.MCT-07-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]


