Abstract
Background
Congenital cytomegalovirus (cCMV) is a common congenital infection and a leading non-genetic cause of sensorineural hearing loss (SNHL). CMV exhibits extensive genetic variability and infection with multiple CMV strains (mixed infection) was shown to be common in cCMV. The role of mixed infections in disease and outcome remains to be defined.
Methods
Genotyping of envelope glycoproteins, UL55 (gB), UL73 (gN) and UL75 (gH) was performed on saliva specimens from 79 infants from the ongoing CMV and Hearing Multicenter Screening Study (CHIMES) and on blood and urine specimens from 52 infants who participated in natural history studies at the University of Alabama at Birmingham. Genotyping of UL144 and US28 was also performed in the CHIMES cohort. The association of individual genotypes and mixed infection with clinical findings at birth and SNHL was examined.
Results
Thirty seven of 131 infants (28%) were symptomatic at birth and 26 (20%) had SNHL at birth. All known genotypes of UL55, UL75, UL73, and US28 were represented and no particular genotype was associated with symptomatic infection or SNHL. UL144 subtype C was more common in symptomatic babies but not associated with SNHL. Mixed infection was observed in 59 infants (45%) and not associated with symptoms (p = 0.43) or SNHL at birth (p = 0.82). In the cohort of 52 infants with long-term hearing outcome, mixed infection at birth was not predictive of SNHL.
Conclusions
Mixed infection is common in infants with cCMV but is neither associated with symptomatic infection nor with SNHL.
Keywords: Cytomegalovirus, congenital infection, genotypes, mixed infection, hearing loss
Introduction
Congenital cytomegalovirus (cCMV) infection is the most common congenital viral infection and a leading non-genetic cause of sensorineural hearing loss (SNHL)1. Most (~90%) children with cCMV infection do not exhibit clinical abnormalities at birth (asymptomatic infection) and about 10% to 15% of these children and 40% to 60% of those with clinically apparent (symptomatic) infection develop permanent sequelae, predominantly SNHL2, 3. The pathogenesis of SNHL in cCMV infection remains largely unknown and it is thought that a combination of virologic and host immunologic factors contribute to SNHL and other sequelae.
CMV has a large genome and has been shown to be highly genetically diverse with polymorphisms scattered across the virus genome4–12. Infection with multiple genotypes (mixed infection) has been reported in immunocompromised patients and has been linked to poor clinical outcome5, 13, 14. We recently reported that mixed infection is common in infants with cCMV infection15. However, it is not known whether mixed infections play a role in newborn disease and SNHL in congenitally infected. The objective of this study is to determine the association between mixed infection and clinical findings at birth and hearing outcomes in children with cCMV infection.
Methods
Subjects and specimens
The study population consisted of 131 subjects from two cohorts of children with cCMV infection. A convenience sampling of saliva specimens collected at birth from 79 infants with cCMV infection who were identified by newborn screening as part of the ongoing National Institute on Deafness and Other Communication Disorders (NIDCD) CMV and Hearing Multicenter Screening (CHIMES) Study16. Specimens from infants with cCMV infection enrolled at all 7 sites were included and cCMV infection was confirmed in all 79 children by rapid culture of saliva or urine within the first 3 week of life. The results of genotyping of UL55, UL73 and UL75 from 28 of the 79 study children from the CHIMES cohort were included in a previous study15. Clinical and hearing data at birth are available for these subjects however, as the study is ongoing, the long-term hearing outcome is not yet available. Urine and blood samples collected within the first month of life from an additional cohort of 52 subjects with cCMV infection who were identified on newborn CMV screening at the University of Alabama at Birmingham (UAB) between 1994 and 200217 and had defined long-term hearing outcome data were analyzed. Samples were chosen based on the availability of adequate remnant saliva, urine and blood for analysis. Institutional review board approval was obtained at each study site. Written informed consent was obtained from a parent or legal guardian for their newborn’s enrollment in the study. Infants were classified as having symptomatic congenital CMV infection if they shed CMV and had any of the following clinical findings in the newborn period; petechial rash, jaundice with conjugated hyperbilirubinemia (direct bilirubin >2mg/dL), thrombocytopenia (<100,000/mm3), hepatosplenomegaly, microcephaly, seizures and chorioretinitis18. The mean duration of follow-up for children with defined long-term hearing outcome was 29.6 ± 22.9 months. Laboratory personnel were blinded to demographic characteristics, clinical findings, and previous genotyping results of study subjects.
Assessment of CMV genetic diversity
CMV virus diversity was evaluated by genotyping envelope glycoproteins, UL55 (gB), UL73 (gN), and UL75 (gH), and CMV-encoded cytokine/chemokine receptor homologues, US28 and UL144 in specimens from CHIMES Study infants. However, only UL55, UL73 and UL75 genes were analyzed in specimens from the 2nd cohort of 52 children because only limited amounts of archival samples were available. DNA was extracted from patient samples using commercial spin columns (Qiagen, Inc). For gN (UL73) genotyping, samples underwent polymerase chain reaction (PCR) to amplify the gN region using primers and conditions previously reported8. CMV US28 and UL144 genes were amplified using published primers19, 20. To reduce the possibility of PCR artifact, appropriate positive and negative controls were included with each PCR run. The amplified regions of UL73, UL144 and US28 were cloned into the TOPO TA cloning vector pCR 2.1 (Invitrogen Inc) and up to ten individual colonies screened for the presence of the insert8. The nucleotide sequences were aligned with published sequences of the UL73, UL144 and US28 genotypes from the GenBank to assign a genotype. Genotyping of UL55 (gB) and UL75 (gH) was performed by a genotype-specific real-time PCR using the TaqMan platform as described15, 21, 22.
Identification of new viral genotypes
The sequences of UL73, US28 and UL144 were aligned with respective published prototypes to determine novel mutations both at the nucleotide and the amino acid levels. A new genotype was assigned when a specific polymorphism resulting in an amino acid change was observed in more than one individual colony from a specimen.
Viral load measurements
CMV viral load was measured in saliva specimens from the CHIMES cohort (n=79 and compared between infants infected with a single virus strain and those with mixed infection. Viral load was derived from real-time PCR assay of saliva specimens as described previously23.
Statistical Analyses
The genotyping data from the entire study population of 131 infants was analyzed to determine the association between mixed infections and symptomatic disease and SNHL at birth. Only the data from the 52 children with long-term hearing outcome data was analyzed to examine whether mixed infections were associated with SNHL on follow-up. Statistical significance was determined using X2, Fisher’s exact test, and Wilcoxon rank sum test where appropriate.
Results
Study population characteristics
Of the 131 study children, 71 were male and 60 were female. Just over half were black (76/131, 58%), 51 (39%) were white, 2 (2%) were multiracial, and 2 (2%) were Asian. The demographic characteristics of the study infants in the CHIMES cohort were similar to the overall CHIMES study population described in previous reports16, 23.
Distribution of individual genotypes and the frequency of mixed infections
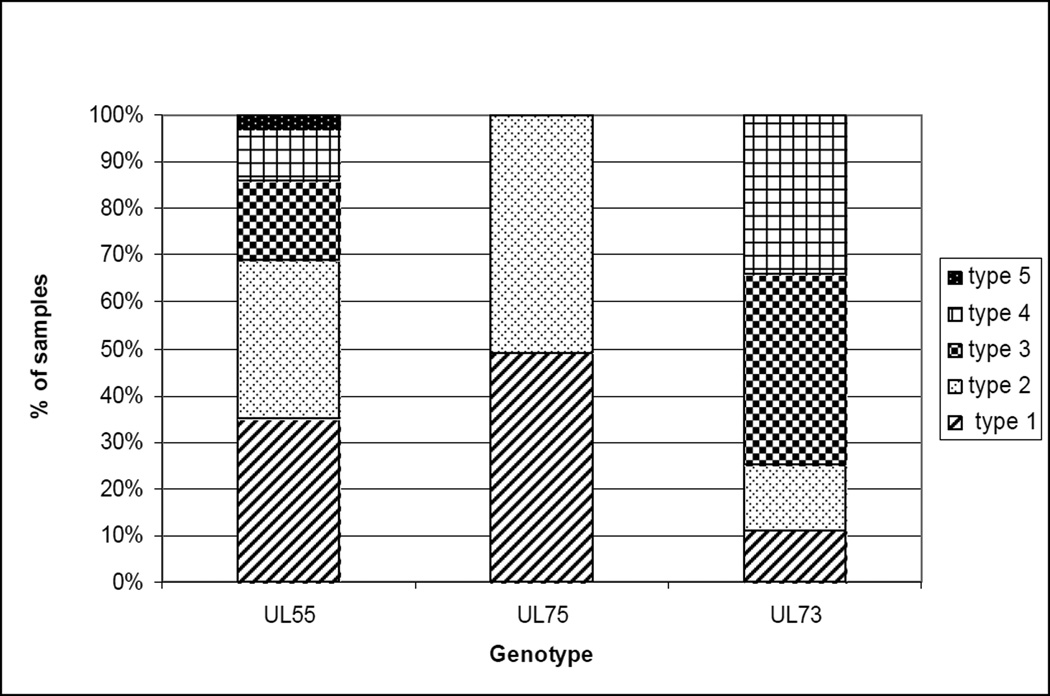
Specimens from 131 study infants with cCMV infection were analyzed to determine UL55, UL75 and UL73 genotypes. The UL55 gene could be amplified and the genotype assigned for 118/131 (90%) infants in the study. All five UL55 genotypes were represented in the cohort and distribution of individual types was as follows: type 1 in 42%, type 2 in 41%, type 3 in 21%, type 4 in 13% and type 5 in 3% of infants (Figure 1). Specimens from 24/118 (20%) infants contained multiple UL55 genotypes. UL75 genotyping could be accomplished in 118/131 (90%) study infants. Genotype 1 was detected in 56% of the infants and type 2 in 59% of infants. The specimens from 18 infants (15%) contained both UL75 genotypes. UL73 genotyping could be determined in 109/131 (83%) of infants enrolled in the study. All seven genotypes were represented and the distribution was as follows: type 1 in 14%, type 2 in 17%, type 3a in 27%, type 3b in 23%, type 4a in 12%, type 4b in 8% and type 4c in 19% of infants (Figure 1). Specimens from 15/109 (14%) children contained multiple UL73 genomic variants.
Figure 1.
The distribution of CMV UL55, UL73 and UL75 genotypes in saliva specimens from 131 infants with congenital CMV infection. All seven UL73 genotypes (gN-1, gN-2, gN-3a, gN-3b, gN-4a, gN-4b, gN-4c), all five UL55 genotypes (gB-1, gB-2, gB-3, gB-4, gB-5), and both UL75 genotypes (gH-1 and gH-2) were seen.
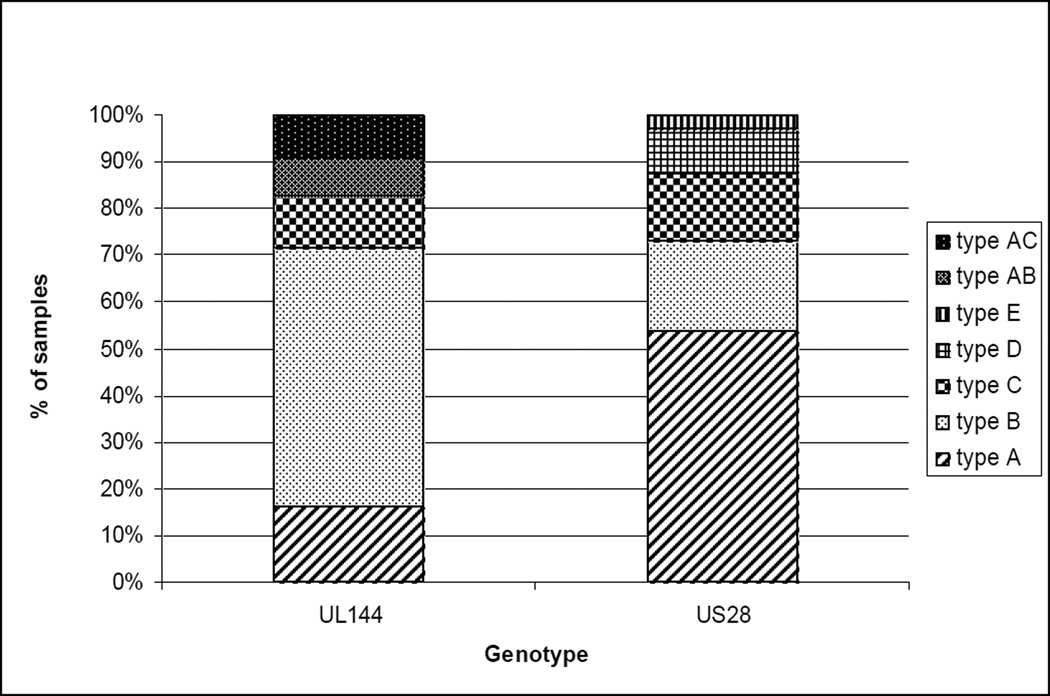
Saliva specimens from the 79 CHIMES study infants were also analyzed for UL144 and US28 genotypes. The UL144 gene could be amplified in 67/79 (85%) samples and genotype A1 was present in 9%, A2 in 9%, B1 in 15%, B2 in 9%, B3 in 12%, B4 in 22%, B5 in 1%, type C in 11%, type AB in 9% and type AC in 10% of infants (Figure 2). The US 28 gene was amplified in 61/79 (77%) specimens and genotype A1 was detected in 34%, A2 in 25%, A4 in 3%, B1 in 13%, B3 in 7%, C in 16%, D in 10% and a new type E in 3% of infants (Figure 2). Multiple UL144 and US28 genotypes were present in 7/67 (10%) and 7/61 (11%) of the specimens, respectively.
Figure 2.
The distribution of CMV UL144 and US28 genotypes in saliva specimens from 79 CHIMES infants with congenital CMV infection. All five UL144 genotypes (A, B, C, AB and AC) all four known US28 genotypes (A, B, C, and D) and a new US28 type ‘E’ (in 2 infants) were seen.
Mixed infections and clinical outcomes
Of the 131 infants in the study, 37 (28%) were symptomatic at birth and 24 (18%) had hearing loss at birth and two additional children developed late-onset hearing loss. Mixed infection, defined as the presence of multiple genotypes of any of the five genes (UL55, UL73, UL75, US28 and UL144) examined, was observed in 59 (45%) study infants. Mixed infection was seen in 19 symptomatic infants and forty asymptomatic infants (p=0.43). Eleven infants (46 %) with SNHL at birth shed multiple CMV genotypes at birth compared with forty-eight infants (46%) with normal hearing at birth (p = 0.99) (Table 1). No particular genotype of UL55, UL73, UL75 or US28 was associated with the presence of symptoms or SNHL at birth (data not shown). Among CHIMES study infants, UL144 subtype C was more prevalent in symptomatic (6/20, 30%) babies compared to asymptomatic infants (2/47, 4%) (p=0.007). However, there was no association between a specific UL144 subtype and hearing loss at birth.
Table 1.
| Children with congenital CMV infection (n=131) |
Genotypes | |
|---|---|---|
| Single | Multiple | |
| Asymptomatic (n=94) | 54 (57%) | 40 (43%) |
| Symptomatic (n=37) | 18 (49%) | 19 (51%) |
| Hearing loss (n=26) | 15 (58%) | 11 (42%) |
| Normal hearing (n=105) | 57 (54%) | 48 (46%) |
Viral load between mixed vs single type
Of the 79 infants in the CHIMES cohort (n=79), 55 infants were infected with single virus strain and 24 infants had mixed infection. Viral load in the group of infants infected with a single virus strain displayed a median of 7.80e6ge/ml (interquartile range, 8.40e5 to 5.0e7ge/ml) was not statistically different (p=0.93) from that in the group with mixed infection having median of 8.10e6ge/ml (interquartile range, 1.68e6 to 4.05e7ge/ml).
Long term hearing outcomes
In the cohort of 52 infants who were followed longitudinally and for whom hearing outcome was known, five children had hearing loss (three at birth and two additional children developed late onset hearing loss at 12.4 months and at 16.4 months of age, respectively. Three of the five children with hearing loss in this cohort had infection with multiple strains compared with 16/48 children with normal hearing (p = 0.23).
New genotypes of UL73 and US28
Comparison of UL73 sequences with existing prototypes showed several new subtypes of the previously described UL73 genotypes based on polymorphisms within the first 100 amino acids of the N-terminal region (see Fig. A, Supplemental Digital Content 1, http://links.lww.com/INF/B587). Similarly, an analysis of US28 sequences showed one new type ‘E’ (see Fig. B, Supplemental Digital Content 1, http://links.lww.com/INF/B587).
Discussion
The current study examined the association between infection with multiple CMV strains, or mixed infection, and disease in cCMV infection utilizing two cohorts of congenitally infected children. Viral strain diversity was examined by determining the genetic diversity of five CMV genes; UL55, UL73, UL75, UL144, and US28. The envelope glycoproteins, UL55, UL73 and UL75 are major targets of virus neutralizing antibodies and the variability within these genes has been examined in many previous studies4, 5, 8, 9, 15. The US28 and UL144 are cytokine/chemokine receptor homologues and thought to play an important role in evasion of immune surveillance against CMV24, 25. Overall, mixed infection based on the presence of more than one genotype of at least one of the five CMV genes was observed in 45% of study infants confirming previous observations that mixed infection is a common occurrence in cCMV infection15. The findings of the current study also demonstrate that mixed infection was neither associated with symptomatic infection nor SNHL at birth. In addition, the findings from the smaller cohort of congenitally infected children with defined long-term hearing outcomes suggests that mixed infections were not associated with SNHL on long-term follow-up. However, this group included only five infants with SNHL and therefore, such an association cannot be ruled out with certainty. The results from a larger prospective follow-up study of children identified on newborn CMV screening, such as the ongoing CHIMES study, would provide more definitive information on the role of mixed infection in outcome in cCMV infection, particularly with the use of newer deep sequencing technology. However, the follow-up of infants enrolled in the CHIMES study is not expected to be complete for at least three more years.
Several previous studies have reported the occurrence of mixed infection by examining genetic diversity of different CMV loci in children with congenital infection15, 19, 26–28 and in immunocompromised patients5, 29, 30. However, most previous studies of children with cCMV infection were limited by small sample size19, 27, 31, 32. In addition, many of the published reports have examined culture propagated virus isolates which could select for a single virus strain and thus decrease the chances of identifying multiple virus strains or genotypes19, 33. To minimize the underestimation of the extent of virus diversity, we analyzed saliva samples from infants with cCMV infection identified on newborn screening16 and specimens collected from an additional cohort of 52 children to determine the role of mixed infection in newborn disease and long-term outcome17. The differences in methodologies between previous studies and our study likely explains the fact that nearly half (45%) of infants in this study were shedding more than one genotype of CMV.
Studies examining the association between CMV genotypes and clinical outcomes have reported conflicting results, likely due to selected patient populations and smaller number of subjects studied19, 32, 34. Arav-Boger et al. reported that UL144 genotype B was more frequent in infants with asymptomatic cCMV infection whereas UL144 genotypes A and C were associated with symptomatic infection19. We also observed that infection with UL144 genotype C was significantly more frequent in symptomatic (6/20) infants than those with asymptomatic infection (2/47; p=0.007). However, this association needs to be confirmed in future studies with larger sample size. In a more recent study, Pignatelli et al. examined the association between UL73 genotypes and clinical outcomes in a cohort of 74 infants with cCMV infection and reported that UL73 genotypes 1 and 3a were associated with favorable outcome and UL73 type 4 with symptomatic infection, abnormal imaging results and long-term sequelae34. In our study in which UL73 genotyping was accomplished in 109 subjects, the relationship between UL73 genotypes and disease could not be confirmed. Further, we did not observe an association between specific UL55, UL75, and US28 genotypes and clinical outcome at birth or at long-term follow-up.
Given the large size of the CMV genome, our finding of lack of an association between mixed infection and clinical outcome should be interpreted with caution because polymorphisms were only examined in five CMV genes. Furthermore, emerging data from studies utilizing deep sequencing technology suggest that the true extent of genetic diversity among CMV strains in infected individuals including infants with cCMV infection is much greater than previously recognized12. Therefore, it is possible that mixed infections occur even more commonly and the exact of role of infection with multiple virus strains in the pathogenesis of cCMV infection is yet to be defined. However, deep sequencing technology continues to evolve and the high cost of this methodology at the current time makes it difficult to conduct studies with large numbers of clinical samples.
We were unable to determine the subtypes of CMV genes in some of the specimens in this study. UL55 and UL75 genotypes could not be ascertained using the genotype-specific real-time PCR in specimens from 13 study infants. The UL73 gene could only be amplified by qualitative PCR in specimens from 109/131 children. It is possible that the inability to amplify UL55 and UL75 by the genotype-specific PCR in some samples could be due to low concentration or degradation of viral DNA. Another limitation of our study is that only samples from some of the infants participating in the CHIMES study were analyzed for US28 and UL144 genotypes because of the limited amount of the archival sample available in these study children.
In conclusion, the current study confirms the observation that infection with multiple strains is a common occurrence in congenital CMV infection. In addition, we demonstrate that mixed infection is not predictive of symptoms or hearing loss at birth. Results from large prospective studies with long-term follow-up of infants identified on newborn screening and studies utilizing deep sequencing technology will provide more definitive information regarding the significance of infection with multiple strains and sequelae including SNHL.
Supplementary Material
Acknowledgements
We are indebted to our medical and nursing colleagues and the infants and their parents who agreed to take part in this study. We would like to thank the following members of the CHIMES study team for their contributions:
University of Alabama at Birmingham Health System: Nitin Arora, MD, MPH; Belinda Blackstone, MS, CCC-A; Jennifer Blumenthal, MD; William J. Britt MD; Alice Brumbach, MSN; Steven Febres-Cordero; Mirjam Kempf, PhD; Faye McCollister, EdD; Emily Mixon, MPH; Noelle Nicholls; Misty Purser, BS; Julie Woodruff, AuD.
Carolinas Medical Center: Amina Ahmed, MD; Edie Cox, AuD; Nubia Flores; Anita Hoyt, Milly Ricart; Lisa Schneider, AuD; Jennifer West, RN, BSN.
Children’s Hospital of Pittsburgh of UPMC: Jena Colaberardino, BA; Kate Faunce; Noreen Jeffrey, RN; Sabrina Malik; Marian Michaels, MD; Gretchen E. Probst MAT, CCC-A; Chad Stephens; Diane Sabo, PhD.
Saint Peter’s University Hospital Maria Class, RN; Kristina Feja, MD; Christy Glick, MA, CCC-A; Robert W. Tolan Jr, MD.
University of Cincinnati and Cincinnati Children's Hospital Medical Center: David I. Bernstein, MD, MA; Daniel Choo, MD; Kate Catalanotto, RN, BSN, CCRC; Patty Kern, Kurt Schibler, MD; Maureen Sullivan-Mahoney, Au.D; Stacie Wethington. RN, CCRC.
University of Mississippi Medical Center: Kathy Irving, AuD; Delia Owens, RN; April L. Palmer, MD; Suzanne Roark, AuD; Mindy Ware, AuD.
University of Texas Southwestern Medical Center at Dallas, Parkland Health & Hospital System and Children's Medical Center Dallas: Cathy Boatman, MS, CIMI; Jessica Esquivel; Gregory L. Jackson, MD, MBA; Kathy Katz-Gaynor, April Liehr Townsley, MA, CCC-A; Asuncion Mejías, MD; Kristine E. Owen, AuD, CCC-A; Peter S. Roland, MD; Oscar Rosado, MD; Pablo J. Sanchez, MD; Angela G. Shoup, PhD; David Sosa; Erica Santoyo, Jessica Santoyo; Elizabeth K. Stehel, MD; Lizette Torres, RN; Fiker Zeray, RN, MS, CPNP.
Sources of Financial Support: This study was supported by the National Institute on Deafness and Other Communication Disorders (N01 DC50008 and K23 DC008539).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: No authors have any conflict of interest to disclose.
References
- 1.Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2.Dahle AJ, Fowler KB, Wright JD, Boppana SB, Britt WJ, Pass RF. Longitudinal investigations of hearing disorders in children with congenital cytomegalovirus. J Am Acad Audiol. 2000;11:283–290. [PubMed] [Google Scholar]
- 3.Britt WJ. Cytomegalovirus. In: Reminghton JS, Klein JO, Wilson CB, Nizet V, Maldonaldo Y, editors. Infectious Diseases of the Fetus and Newborn Infant. 7th ed. Philadelphia: Elsevier Saunders; 2011. pp. 706–755. [Google Scholar]
- 4.Chou SW. Differentiation of cytomegalovirus strains by restriction analysis of DNA sequences amplified from clinical specimens. J Infect Dis. 1990;162:738–742. doi: 10.1093/infdis/162.3.738. [DOI] [PubMed] [Google Scholar]
- 5.Coaquette A, Bourgeois A, Dirand C, Varin A, Chen W, Herbein G. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin Infect Dis. 2004;39:155–161. doi: 10.1086/421496. [DOI] [PubMed] [Google Scholar]
- 6.Fries BC, Chou S, Boeckh M, Torok-Storb B. Frequency distribution of cytomegalovirus envelope glycoprotein genotypes in bone marrow transplant recipients. J Infect Dis. 1994;169:769–774. doi: 10.1093/infdis/169.4.769. [DOI] [PubMed] [Google Scholar]
- 7.Murphy E, Yu D, Grimwood J, et al. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci U S A. 2003;100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novak Z, Ross SA, Patro RK, et al. Cytomegalovirus strain diversity in seropositive women. J Clin Microbiol. 2008;46:882–886. doi: 10.1128/JCM.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pignatelli S, Dal Monte P, Rossini G, et al. Human cytomegalovirus glycoprotein N (gpUL73-gN) genomic variants: identification of a novel subgroup, geographical distribution and evidence of positive selective pressure. J Gen Virol. 2003;84:647–655. doi: 10.1099/vir.0.18704-0. [DOI] [PubMed] [Google Scholar]
- 10.Puchhammer-Stockl E, Gorzer I, Zoufaly A, et al. Emergence of multiple cytomegalovirus strains in blood and lung of lung transplant recipients. Transplantation. 2006;81:187–194. doi: 10.1097/01.tp.0000194858.50812.cb. [DOI] [PubMed] [Google Scholar]
- 11.Rasmussen L, Geissler A, Cowan C, Chase A, Winters M. The genes encoding the gCIII complex of human cytomegalovirus exist in highly diverse combinations in clinical isolates. J Virol. 2002;76:10841–10848. doi: 10.1128/JVI.76.21.10841-10848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Renzette N, Bhattacharjee B, Jensen JD, Gibson L, Kowalik TF. Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog. 2011;7:e1001344. doi: 10.1371/journal.ppat.1001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lisboa LF, Tong Y, Kumar D, et al. Analysis and clinical correlation of genetic variation in cytomegalovirus. Transpl Infect Dis. 2011;14:132–140. doi: 10.1111/j.1399-3062.2011.00685.x. [DOI] [PubMed] [Google Scholar]
- 14.Manuel O, Asberg A, Pang X, et al. Impact of genetic polymorphisms in cytomegalovirus glycoprotein B on outcomes in solid-organ transplant recipients with cytomegalovirus disease. Clin Infect Dis. 2009;49:1160–1166. doi: 10.1086/605633. [DOI] [PubMed] [Google Scholar]
- 15.Ross SA, Novak Z, Pati S, et al. Mixed infection and strain diversity in congenital cytomegalovirus infection. J Infect Dis. 2011;204:1003–1007. doi: 10.1093/infdis/jir457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boppana SB, Ross SA, Novak Z, et al. Dried blood spot real-time polymerase chain reaction assays to screen newborns for congenital cytomegalovirus infection. JAMA. 2010;303:1375–1382. doi: 10.1001/jama.2010.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross SA, Novak Z, Kumbla RA, Zhang K, Fowler KB, Boppana S. GJB2 and GJB6 mutations in children with congenital cytomegalovirus infection. Pediatr Res. 2007;61:687–691. doi: 10.1203/pdr.0b013e3180536609. [DOI] [PubMed] [Google Scholar]
- 18.Boppana SB, Pass RF, Britt WJ, Stagno S, Alford CA. Symptomatic congenital cytomegalovirus infection: neonatal morbidity and mortality. Pediatr Infect Dis J. 1992;11:93–99. doi: 10.1097/00006454-199202000-00007. [DOI] [PubMed] [Google Scholar]
- 19.Arav-Boger R, Willoughby RE, Pass RF, et al. Polymorphisms of the cytomegalovirus (CMV)-encoded tumor necrosis factor-alpha and beta-chemokine receptors in congenital CMV disease. J Infect Dis. 2002;186:1057–1064. doi: 10.1086/344238. [DOI] [PubMed] [Google Scholar]
- 20.Yan H, Koyano S, Inami Y, et al. Genetic variations in the gB, UL144 and UL149 genes of human cytomegalovirus strains collected from congenitally and postnatally infected Japanese children. Arch Virol. 2008;153:667–674. doi: 10.1007/s00705-008-0044-7. [DOI] [PubMed] [Google Scholar]
- 21.Gorzer I, Kerschner H, Jaksch P, et al. Virus load dynamics of individual CMV-genotypes in lung transplant recipients with mixed-genotype infections. J Med Virol. 2008;80:1405–1414. doi: 10.1002/jmv.21225. [DOI] [PubMed] [Google Scholar]
- 22.Pang X, Humar A, Preiksaitis JK. Concurrent genotyping and quantitation of cytomegalovirus gB genotypes in solid-organ-transplant recipients by use of a real-time PCR assay. J Clin Microbiol. 2008;46:4004–4010. doi: 10.1128/JCM.01341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boppana SB, Ross SA, Shimamura M, et al. Saliva polymerase-chain-reaction assay for cytomegalovirus screening in newborns. N Engl J Med. 2011;364:2111–2118. doi: 10.1056/NEJMoa1006561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benedict CA, Butrovich KD, Lurain NS, et al. Cutting edge: a novel viral TNF receptor superfamily member in virulent strains of human cytomegalovirus. J Immunol. 1999;162:6967–6970. [PubMed] [Google Scholar]
- 25.Bodaghi B, Jones TR, Zipeto D, et al. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J Exp Med. 1998;188:855–866. doi: 10.1084/jem.188.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bale JF, Jr, Murph JR, Demmler GJ, Dawson J, Miller JE, Petheram SJ. Intrauterine cytomegalovirus infection and glycoprotein B genotypes. J Infect Dis. 2000;182:933–936. doi: 10.1086/315770. [DOI] [PubMed] [Google Scholar]
- 27.de Vries JJ, Wessels E, Korver AM, et al. Rapid genotyping of cytomegalovirus in dried blood spots by multiplex real-time PCR assays targeting the envelope glycoprotein gB and gH genes. J Clin Microbiol. 2011;50:232–237. doi: 10.1128/JCM.05253-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu ZS, Zou CC, Zheng JY, Zhao ZY. Cytomegalovirus gB genotype and clinical features in Chinese infants with congenital infections. Intervirology. 2006;49:281–285. doi: 10.1159/000093458. [DOI] [PubMed] [Google Scholar]
- 29.Humar A, Kumar D, Gilbert C, Boivin G. Cytomegalovirus (CMV) glycoprotein B genotypes and response to antiviral therapy, in solid-organ-transplant recipients with CMV disease. J Infect Dis. 2003;188:581–584. doi: 10.1086/377002. [DOI] [PubMed] [Google Scholar]
- 30.Sarcinella L, Mazzulli T, Willey B, Humar A. Cytomegalovirus glycoprotein B genotype does not correlate with outcomes in liver transplant patients. J Clin Virol. 2002;24:99–105. doi: 10.1016/s1386-6532(01)00238-4. [DOI] [PubMed] [Google Scholar]
- 31.Paradowska E, Studzinska M, Nowakowska D, et al. Distribution of UL144, US28 and UL55 genotypes in Polish newborns with congenital cytomegalovirus infections. Eur J Clin Microbiol Infect Dis. 2011;31:1335–1345. doi: 10.1007/s10096-011-1447-z. [DOI] [PubMed] [Google Scholar]
- 32.Waters A, Hassan J, De Gascun C, et al. Human cytomegalovirus UL144 is associated with viremia and infant development sequelae in congenital infection. J Clin Microbiol. 2010;48:3956–3962. doi: 10.1128/JCM.01133-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arav-Boger R, Battaglia CA, Lazzarotto T, et al. Cytomegalovirus (CMV)-encoded UL144 (truncated tumor necrosis factor receptor) and outcome of congenital CMV infection. J Infect Dis. 2006;194:464–473. doi: 10.1086/505427. [DOI] [PubMed] [Google Scholar]
- 34.Pignatelli S, Lazzarotto T, Gatto MR, et al. Cytomegalovirus gN genotypes distribution among congenitally infected newborns and their relationship with symptoms at birth and sequelae. Clin Infect Dis. 2010;51:33–41. doi: 10.1086/653423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.