Abstract
Cells sense and respond to mechanical forces, regardless of whether the source is from a normal tissue matrix, an adjacent cell or a synthetic substrate. In recent years, cell response to surface rigidity has been extensively studied by modulating the elastic modulus of poly(ethylene glycol) (PEG)-based hydrogels. In the context of biomaterials, Poisson's ratio, another fundamental material property parameter has not been explored, primarily because of challenges involved in tuning the Poisson's ratio in biological scaffolds. Two-photon polymerization is used to fabricate suspended web structures that exhibit positive and negative Poisson's ratio (NPR), based on analytical models. NPR webs demonstrate biaxial expansion/compression behavior, as one or multiple cells apply local forces and move the structures. Unusual cell division on NPR structures is also demonstrated. This methodology can be used to tune the Poisson's ratio of several photocurable biomaterials and could have potential implications in the field of mechanobiology.
1. Introduction
Natural tissues and their cellular microenvironment have complex structural and biological heterogeneity and are constantly exposed to a myriad of forces. Materials scientists have investigated structure–function relationships of cell–materials interactions using various approaches. Cells generate contractile forces on their underlying substrate.[1,2] Over the years, researchers have altered the elastic modulus or stiffness of the substrate by modulating properties such as cross-link density.[3] Alterations in elastic modulus impact a variety of cell types in fundamentally different ways, including motility, gene expression, proliferation, and fate after differentiation.[4–10] These studies have increased our understanding of how cells experience forces, and provided insight into their biophysical mechanisms. However, another fundamental aspect of material properties, the Poisson's ratio, has been largely ignored.
The ability of a biomaterial scaffold to support and transmit cell and tissue forces can be quantitatively described by its elastic modulus and Poisson's ratio. Elastic modulus quantifies a scaffold's elastic behavior in the loading direction, while Poisson's ratio describes the degree to which the scaffold contracts or expands in the transverse direction, perpendicular to the loading direction (Figure 1A). Typically Poisson's ratio is assumed to be positive for all materials (≈0.3 to 0.5), even though materials with negative as well as zero values exist in nature. Positive Poisson's ratio (PPR) materials contract transversally when stretched, while a negative Poisson's ratio (NPR) material expands in both the axial and transverse directions. In nature, we come across several materials with a NPR or “auxetics”, Examples include crystalline materials,[11–16] carbon allotropes,[17] foams,[18–20] polymers and laminates,[21–24] and other extreme states of matter.[25–28] For naturally occurring materials, the NPR property seems to be an intrinsic property of the material and cannot be tuned according to specific applications. Tuning the Poisson's ratio requires control over the pore interconnectivity and internal architecture of a material, which can be quite difficult to manipulate. Man-made auxetics have been developed by incorporating rib-containing pores, which modify the shape and deformation mechanisms of polymers, transforming their mechanical behavior to exhibit NPR property.[18,20,22,29–33] These polymers demonstrate well-defined NPR behavior,[18,27,34,35] however, their strain-dependent response is process dependent. For example, polyurethane foams annealed in a compressed state naturally reorganize their cellular microstructure and exhibit NPR behavior.[18,29] However, processes such as annealing offer poor control over the reorganization of cellular microstructure comprising the foams, making it difficult to tune the Poisson's ratio according to specific applications. There is also evidence of negative and zero Poisson's ratio materials in biology.[11,36–42] To investigate cell-materials interactions, Poisson's ratio has to be precisely tuned, both in magnitude and polarity. Some tissue engineering applications require unique strain responses, wherein biomaterials having a negative or zero Poisson's ratio may be most suitable for emulating the behavior of native tissues. Recently, we developed micrometer-scale scaffolds which exhibit negative, positive and zero Poisson's ratio behavior in polyethylene diacrylate (PEGDA) biomaterials.[43–45] Although these scaffolds themselves exhibited NPR behavior, a single cell is not able to sense the effects of the altered Poisson's ratio at micrometer resolution (≈50–100 μm). In this study, we develop a methodology to develop suspended structures (≤1 μm) with tunable Poisson's ratios, which can be utilized to investigate cellular behavior.
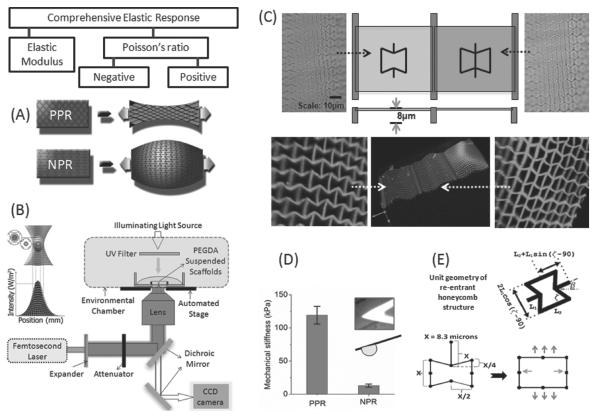
Figure 1.
A) The elastic property of a biomaterial can be comprehensively expressed by the elastic modulus and the Poisson's ratio. Schematic shows a PPR material contracting transversally when axially stretched, while a NPR material expanding in both the axial and transverse directions. B) Schematic of the two-photon absorption process and the femtosecond laser fabrication set-up. C) Optical and confocal images of NPR and PPR suspended web structures with side supports using PEGDA biomaterial. D) AFM measurements using a 20 μ m bead measures the effective stiffness of NPR and PPR web structures in the z-direction. E) Re-entrant honeycomb configuration was adopted as the unit cell geometry for the NPR web, while an additional strut modification to the NPR structure served as the positive control. Schematic shows biaxial expansion of the NPR structure upon axial strains (arrows)
2. Results and Discussion
2.1. Fabrication of Tunable Poisson's Ratio Scaffolds
The re-entrant honeycomb unit cells design[34,46] was used to fabricate NPR suspended structures (Figure 1E). The re-entrant structure is formed by changing the four side angles (angle ζ) between the vertices (ribs) in a six-sided honeycomb (hexagon), Two rib lengths, L1 and L2, constrain the dimensions of the unit-cell, including angle ζ (the value of angle ζ is set by the rib-length ratio and is not arbitrarily set), and the ratio of the two rib lengths has a sizable influence on Poisson's ratio. Varying angle ζ alters the magnitude of Poisson's ratio, which gives Poisson's ratio its strain-dependent response. We have simplified the re-entrant model, by setting L1 = L2 = X = 8.3 μm, which exhibits a NPR response. Addition of an extra rib to the NPR design, inhibits free movements, and transforms the structure to exhibit a PPR response. Femtosecond-laser-induced two-photon polymerization was used to fabricate the NPR and PPR suspended web structures using photosensitive PEGDA biomaterial (Figure 1B). PEGDA biomaterial was chosen, because of its high water content and biocompatibility and tunable mechanical properties.[3,47] Photosensitive prepolymer solution was prepared by adding biocompatible initiator, lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP),[48] and acrylate-PEG RGDS peptide to 20% PEGDA solution. Methacrylated glass surface was coated with PEGDA solution without the peptide, to prevent cell adhesion onto the glass surface. A Ti:sapphire femtosecond laser was tightly focused onto the appropriate z-plane in the volume of the PEGDA solution, and 3D structures were fabricated by moving the stage in x-y-z directions. Structural support beams were incorporated at the ends of each web structure to ensure that the structure is suspended, and cell will be able to apply strains and freely move the structures. Similar NPR structures made on a flat surface can be actuated by addition/removal of water (Supporting Information Figure S1) and could be moved by the cells, however, the NPR structure movement could not be quantified in the absence of the supporting beams. Fixing the boundaries of the NPR structures also enables time-lapse imaging of cell movement and division. Visualization of the web-structures using scanning electron microscopy (SEM) resulted in collapsed web structures, even with critical point drying technique (Supporting Information Figure S2). As a result, the suspended structures were visualized by introducing PEG-RGD-FAM in the prepolymer solution[49] (Figure 1C). Fabrication methodology used in this work will be able to tune a wide range of elastic properties, by controlling the resolution of features via laser dosages, or by varying the prepolymer dilution (for example, mechanical properties of 80% NPR PEGDA structures will differ from both 20% NPR structures or 80% PEGDA slab structures). A 20 μm diameter glass colloidal particle attached to the atomic force microscopy (AFM) probe was used to compare the effective mechanical properties of NPR and PPR structures in the z-direction (Figure 1D). When a vertical forces is applied to the NPR web, the NPR features expand locally, yield, and conform to the bead surface, as opposed to the PPR web, which resists the vertical forces. This response possibly explains the significant differences in the mechanical properties between the NPR and PPR structures, even though both the structures were fabricated using 20% PEGDA.
2.2. Characterization of Cell Response
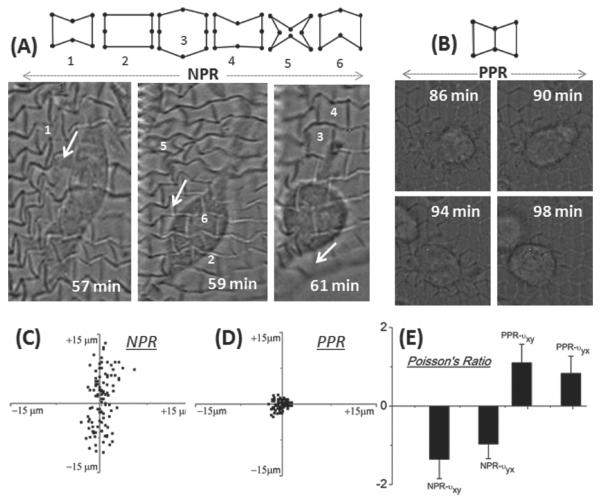
To evaluate cell response, 10T1/2 cells were seeded on the web structures and cell movement was captured using time-lapse microscopy. 10T1/2 are an anchorage dependent embryonic fibroblast cell line that differentiates into perivascular cells in vivo, typically used to stabilize a functional microvascular network. As cells move on NPR web structures, cells apply forces to their underlying substrate using focal adhesions, which deform the web in various configurations (Figure 2A). The web-points deformed by the cells return to their original positions when cells disengage and stop applying forces (Supporting Information Video S1). Cells are capable of deforming both NPR and PPR structures due to the intrinsic properties of a suspended web in a liquid medium. Due to its auxetic property, the NPR web responds and accommodates strains due to different cellular forces to a greater degree. When a NPR unit cell expands in the x- and y directions: for every 0.9 μm x displacement, there is a corresponding increase in y by ≈8 μm (Figure 1E), which explains the larger y displacement of web-point on NPR webs (Figure 2C). Cells can manipulate the PPR structures as well, however cells are not able to move the web-points to a greater degree (Figure 2B). For both the structures, there is some intrinsic movement, due to the suspended nature of the web in liquid medium (Figure 2D). The NPR web-points have the ability to move significantly more as compared to PPR web-points. The local yielding of NPR web due to cellular strains is a combination of rib bending (flexure), stretching, and hinging (angular deformations) of the position and arrangement of the ribs relative to each other.[31,34,46,50] The degree to which each structure deforms depends on unit cell geometry, the bulk material properties of the ribs and the direction of loading. According to the simple hinging model reported by Gibson and Ashby,[50] axial strain causes solely a change in angle ζ, while the magnitude of the negative Poisson's ratio depends upon both ζ and the ratio L2/L1, where the rib length ratio is assumed to stay. However, since multiple cells apply forces in many different directions on the NPR web, the local Poisson ratio changes depending on proximity to cells and force transmission from other cells on the web. Some web points experience cellular pulling forces from adjoining cells on the web, while others experience inward pulling forces by the cell, and web-points with no cellular forces remain neutral. Since the Poisson ratio is calculated by measuring the transverse displacement of web points when axial forces are applied, for this particular case, it is difficult to measure the Poisson's ratio of the overall structure, since multiple cells apply forces on unit structures in a variety of xy directions. Therefore, effective Poisson's ratios of unit NPR and PPR structures were calculated by selecting unit cells which deform by applying of cell-forces in either x- or y-axes, denoted by νxy and νyx (Figure 2E). The local expansion–contraction movement of the NPR web-points allows cellular force transmission. On the other hand, PPR web-points resist cellular forces to a greater degree and exhibit typical positive Poisson's ratio behavior. Careful control over cell concentration can be used to have single cell on each NPR web to precisely calculate local displacements and strains (Supporting Information Video S2). Structures from the literature, such as star honeycomb, chirals, rotating triangle or square units, structures formed from lozenge grids, etc., can be incorporated to design and tune biomaterials with precise strain-dependent NPR response.[51]
Figure 2.
A) The suspended structure exhibits a combination of rib bending or flexure, stretching, and hinging (angular deformations) as a 10T1/2 cell moves across the web structure (1–6). White arrows indicate the direction of movement with associated time stamps. B) The PPR web structure resists cells movements. C,D) Frame-to-frame trajectory of multiple webpoints after 10T1/2 cell seeding on PPR and NPR structures. Data pooled from different locations on the web structures for 2 h with 8 min intervals. Web-points of the NPR structures move significantly more as compared to PPR web-points. E) Graph shows effective Poisson's ratio upon cellular strains in the x- and y-directions.
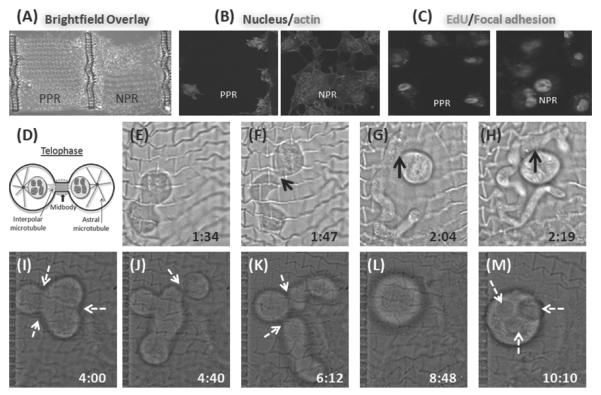
Immunofluoscence staining was carried out to evaluate cell adhesion and proliferation on both NPR and PPR web structures. On the NPR web, the cells seem to be randomly oriented, evident from actin staining (white arrow) (Figure 3A–C). The pseudopods of the spread cells do not align along the suspended NPR struts. Cells on the PPR web are not able to spread in the first two hours post-seeding, and most of the cells adhere to the support structures (Supporting Information Video S3,4). Cells adhere to both structures through integrin-mediated adhesions. Vinculin labeling is seen throughout the cell structure and indicated small nascent adhesions while strong labeling on some of the locations indicate larger (> 1 μm) focal adhesions. Cells proliferate well (≈ 100%) on both PPR and NPR structures. Differences in focal adhesions between the NPR and PPR structures were not significant. Tuning the size of the structures using varying laser dosage and/or prepolymer concentration has the potential to be used as a way to have direct control over adhesion sites. Cells seeded on NPR and PPR topologies attached to the glass substrate also demonstrate high adhesion and proliferation (Supporting Information Figure S3). Mechanical forces play an important role in cell division.[52] Cell division, a critical aspect of all multicellular organisms, requires segregation of chromosome facilated by physical division of the cytoplasm, resulting in separate daughter cells. In normal cell division, during telophase, the cleavage furrow ingresses resulting in compression of the central spindle to form an intercellular microtubule bridge called the midbody. The midbody is severed, resulting in the final separation of daughter cells (Figure 3D). Time-lapse experiments revealed unusual cell division of 10T1/2 cells on NPR structures (Figure 3). On NPR webs, cleavage furrow ingression began normally, however, the resulting daughter cells remain attached to each other for the duration of the experiment (≈ 12 h). In one case, (Figure 3E–H) one cell resulted in formation of a long structure, and probably underwent apoptosis after being unable to complete abscission. In another case (Figure 3I–M), a cell underwent midbody regressed after failing to separate, giving rise to a multinucleated cell. The cell then re-entered mitosis, however, the midbody persisted and did not break off throughout the duration of experiment. In one case, one of the nascent daughter cell was on top of the NPR web while the other was under the web, maintaining the midbody connection throughout the duration of the experiment. (Supporting Information Figure S4–6 and Video S5). Since cell division failure can cause genetic instability, ultimately leading to cancer[53] researchers have elucidated molecular mechanism of abnormal cell division using several midbody components. Cytokinesis failure and associated behavior has been reported following depletion of components of the cortical cytoskeleton, such as anillin, septins, and formins.[54–56] Our observations suggest that unusual cell division can be induced solely by the NPR structure, in absence of any external biochemical manipulations to the cell components. Fluorescent time-lapse images were also taken to monitor the nucleus activity of the cells using Hoechst 33342 (Supporting Information Figure S7). Arrows indicate symmetric as well as asymmetric furrow formation during cell division on NPR webs. During cell movement, bleb formation and constantly extending and retracting protrusions are also seen on the NPR web (Supporting Information Video S6,7). The NPR property affects the spatial distribution of adhesive contacts a cell experiences during cell division and movement, causing abnormal cell division.
Figure 3.
A–C) Immunostaining of 10T1/2 cells for adhesion (nucleus, actin, and focal adhesions) and proliferation (EdU). D) Schematic illustration showing cell in telophase: the cleavage furrow ingresses, compresses the midzone, and creates an intercellular bridge containing a microtubule midbody. In normal cell division, the bridge is resolved creating two daughter cells. The NPR structure induces aberrant cell-division response: E–H) Abnormal cell-division initiates and results in the formation of a long structure. Black arrow indicates persistent mid-body connection. I–M) Multiple sites of symmetric as well as asymmetric furrow formation during cell division on the NPR webs (dotted white arrows).
3. Conclusion
Most tissue cells not only adhere to but also pull on their microenvironment and thereby respond to the mechanical properties of the substrates through cytoskeletal reorganization and associated processes. So far only one component of the mechanical property, the elastic modulus, has been investigated. This work potentially opens doors to fabricating biomaterials with tunable Poisson's ratio while keeping the bulk elastic modulus constant. We envision that this technique will be used to investigate effects of altering the Poisson's ratio on a variety of cellular aspects including morphology, gene expression, and migration using different cell types. Due to its importance in fundamental biological processes, such as tumor metastasis, wound healing and morphogenesis, cell migration has been extensive studied on a substrate with varying mechanical properties. Present understanding is that cellular forces are concentrated at focal adhesions (FA), whose size can be correlated to the amount of forces exerted by the cells on their underlying substrate. Recent reports acknowledge the fundamentally different cellular response on larger than micron and submicron scale features.[57] Methodology developed in this work, can control the resolution as well as have local yielding of the rib-structures thereby potentially controlling the size and spacing of FA sites. Cell response to a substrate with a gradient of elastic modulus[7] has also been investigated, and it is found that cells typically migrate preferentially toward stiffer regions. Similarly, a gradient Poisson's ratio structure can be fabricated (Supporting Information Figure S8), which will transmit cellular forces across a range of Poisson's ratio on a single substrate. A wealth of information about how cells apply forces has been gained by modulating the elastic modulus of the underlying substrate. It is expected that tuning the Poisson's ratio allows one to identify several components involved in mechanosensing. The platform developed in this work will foster experiment and contribute to the broad field of mechanobiology.
4. Experimental Section
Preparation of Sample Substrate
Glass coverslips (Electron Microscopy Sciences, Hatfield, PA; 0.08–0.13 μm) were cleaned for 5 min in piranha solution (35 mL sulfuric acid and 15 mL hydrogen peroxide), washed 5 times with deionized (DI) water, and immersed in ethanol (5 min). Methacrylation solution was prepared by mixing together 1 mL 3-(trimethoxysilyl)-propyl methacrylate, 50 mL ethanol, and 6 mL of 1:10 glacial acetic acid (acetic acid in ethanol). Dried coverslips were immersed in the methacrylation solution on a shaker overnight, cleaned with ethanol and dried with compressed air. Methyacrylated glass coverslips were coated with a thin layer of 20% PEGDA (80% water) as follows. PEGDA solution was sandwiched between one piece of methacrylated coverslip and one piece of untreated coverslip. The untreated coverslip was cleaned with ethanol before use. A pipettor was used to transfer 7 μL PEGDA onto the surface of methacrylated coverslip and the untreated coverslip was covered on top of the methacrylated coverslip. The sandwich structure was placed on an experiment table for 3 min to stabilize, and irradiated using a UV lamp (1 min) to polymerize the PEGDA layer and the two pieces were detached. A 10 mm × 10 mm hole was cut on the bottom of a 35 mm plastic petri dish with a single edge cutter blade and the PEGDA coated coverslip was glued (dipentaerythritol pentaacrylate, Sartomer) with 1% photoinitiator (Irgacure 819) onto the bottom of the petri dish using UV polymerization.
Femtosecond Laser Fabrication of the Webs
Figure 1B shows the schematic drawing of the femtosecond laser fabrication system setup. The laser source was a Ti:sapphire femtosecond laser (Vitesse, Coherent Inc., Santa Clara, CA) producing 100-femtosecond wide pulses at a repetition rate of 80 MHz with a maximum power of 350 mW. The central wavelength of the laser was 800 nm and the diameter of the beam out of the laser head was 1 mm. The laser beam was expanded by a 4× beam expander to make full use of the aperture of the objective lens. Then the laser beam was guided by a group of mirrors into an inverted microscope (Nikon Eclipse Ti, Nikon Instruments Inc., Melville, NY) and focused by an oil-immersion objective lens (Plan Apo VC 100x, Nikon Instruments Inc., Melville, NY) onto the sample which was mounted on a motorized stage (Applied Scientific Instrumentation, Eugene, OR). The laser power could be continuously adjusted by an attenuator (NIR Polarizer, Edmund Optics Inc. Barrington, NJ). The scanning of the sample was implemented by simultaneously controlling the stage and an electrical-motorized shutter (Sutter Instrument Company, Novato, CA) using the microscope software. The laser power employed for the fabrication was 50 mW, measured by a power meter (PowerMax 500D, Molectron Detector Inc., Portland, OR) before the laser beam enters the objective lens. The scanning process was monitored in situ by a charge-coupled device (CCD) camera (Qimaging, Surrey, BC, Canada).
Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) photoinitiator was prepared as previously described.[48] Pre-polymer solution was prepared as follows. 20% PEGDA (MW 700, Sigma-Aldrich) was mixed with DI water (1:4, 4% (w/v)), LAP (2%), acrylate-PEG RGDS peptide (5 mM), and vortexed (5 min) and filtered using a 0.22 μm pore size syringe filter. Before making the structures, a drop of immersion oil was placed on the objective lens. A pipettor was used to place 20 μL of the 20% PEGDA on the bottom of the glass petri dish. The petri dish was then secured on the stage by two strips of double-side tape. A series of dots were made by the laser beam to locate the z-position of the coverslip surface. An array of seven supporting walls was fabricated. The supporting wall was an 8-layer wood-pile structure with the size of 120 μm (length) × 6 μm (width) × 12 μm (height) using stage speed of 0.05 mm/s and a laser power of 70 mW. Suspended NPR and PPR structures were fabricated with the sides of the structures attached to the supporting walls using a stage speed of 0.65 mm/s and laser power of 45 mW. Unpolymerized PEGDA was washed away using DI water, and the petri dish was filled with phosphate buffered saline (PBS) that contained 2% penicillin-streptomycin (PS), and stored at 37 °C. Fluorescence-labeled webs were fabricated using similar conditions using FAM-labelled PEG-RGD[49] (FAM 0.5 mg/mL)
Time-Lapse Imaging
10T1/2 cells (American Type Culture Collection, Manassas, VA) were maintained in Dulbecco's modified Eagle's medium with high glucose (DMEM) (Gibco, North Andover, MA) supplemented with 10% fetal bovine serum (FBS). The cells were cultured in an incubator with 5% CO2 concentration and 37 °C. The culture medium was replenished every other day and the cells were split as necessary. Prior to cell seeding, the petri dish with the webs was washed twice with PBS. Then it was filled with 3 mL pre-warmed DMEM supplemented with 10% FBS and 1% PS. Throughout this process, caution was taken to keep the webs in liquid to prevent them from collapsing. An ethanol-washable marker was used to mark the position of the webs on the bottom of the petri dish to facilitate cell seeding. A drop of 10 μL medium (seeding density: 5000 cells per petri dish) was carefully added on top of the webs.
Time-lapse images were taken in a microscope (Nikon) with a cell culture chamber (In Vivo Scientific Inc.) after the cells were seeded on the webs. The CO2 concentration and the temperature in the cell culture chamber were maintained at 5% and 37 °C, respectively. The bottom of the chamber was filled with DI water to maintain the humidity. The heater of the chamber was turned on 4 h before the time-lapse imaging to achieve a stable 37 °C environment. A 40× objective lens was used to take the time-lapse images every 2 min at a range of z-positions with a step of 2 μm. With the same setup, fluorescent time-lapse images were taken to monitor the nucleus activity of the cells using Hoechst 33342 (1 μg/mL).
Immunofluorescence Labeling
EdU staining was conducted with the Click-iT EdU imaging kit (Invitrogen, Carlsbad, CA) following the protocol provided by the manufacturer. After the cells were seeded on the web for 12 h, EdU was added into the medium at 10 μM and the cells were cultured in the incubator for another 12 h. After the incubation, the medium was removed and the sample was fixed with 0.5 mL of 4% formaldehyde in PBS for 15 min at room temperature. Formaldehyde was removed and the petri dish was rinsed twice with 1 mL of 2% BSA in PBS for 5 min each. After being permeablized with 1 mL of 0.5% Triton X-100 in PBS for 20 min, the sample was again rinsed twice with 1 mL of 2% BSA in PBS for 5 min each. Then the sample was incubated in the Click-iT reaction cocktail which contained Click-iT reaction buffer, CuSO4 , Alexa Fluor azide, and reaction buffer additive for 30 min at room temperature, protected from light. After the reaction cocktail was removed, the sample was rinsed once again with 1 mL of 2% BSA in PBS for 5 min.
The sample was incubated with 2 mL of 5 μg/mL Hoechst 33342 in PBS for 30 min at room temperature for DNA staining. Hoechst solution was removed and the sample was washed twice with 3 mL of PBS. For focal adhesion staining, the sample was first blocked for 60 min in blocking buffer containing 2% BSA, 0.3% Triton X-100, and 97.7% PBS. Primary antibody solution, (0.5% monoclonal anti-vinculin, Sigma, in blocking buffer), was applied onto the sample and the sample was incubated overnight at 4 °C. Sample was rinsed and incubated in the secondary antibody solution (1% Alexa Fluor 647 goat anti-mouse IgG in blocking buffer) for 60 min at room temperature. Cell nucleus, proliferation (EdU), actin filaments, and focal adhesion architecture were examined by immunofluorescence confocal microscopy. Cell were stained for nuclear DNA, proliferation marker EdU, focal adhesion protein vinculin, and cytoskeletal filamentous actin. 3D image stacks were acquired using a confocal system (Olympus FV1000) mounted on a Olympus IX81 inverted optical microscope. 3D images were processed using a combination of Volocity imaging software and Image J (National Institute of Health).
Strain Analysis
Time-lapse images were digitized and x- and y-displacements of various webpoints were determined based on the undeformed in-plane dimensions of the structures. Since webpoints on NPR and PPR structure experinced forces in different directions from multiple cells, effective Poisson's ratios of individual unit-cells were calculated. Unit cells were chosen where cells apply forces in either x- or y-directions. The transverse elastic deformation of the unit NPR and PPR webpoints was calculated by resulting strains (in x- or y-directions). Axial and transverse strains and Poisson's ratios of selected unit cells were estimated by measuring the displacement of webpoints, using Equation 1.[58]
| (1) |
where υy and υx are strains in y-and x-directions. In-plane values of Poisson's ratio resulting from in-plane cellular strains were calculated using a 2D Cartesian coordinate system with orthogonal x- and y-axes
Effective Stiffness
The AFM indentation studies were performed with a commercial instrument (multimode microscope, Veeco Inc., Digital Instruments, Santa Barbara, CA) mounted on an inverted optical microscope. This set-up enabled the positioning of the AFM tip on the NPR and PPR regions of the suspended web structures. A standard sharp silicon nitride tip could not be used, because of the nature of the open suspended web structures. Soft contact tips (PNP-TR silicon nitride probe) with a 20 μm diameter glass colloidal particle (force constant ≈0.08 N/m), was used for all force measurements. Force curves were taken on each web structure using a 4 × 4 grid array (16 points per web, three PPR and NPR webs with varying indentation depths) in aqueous mode using a fluid cell with PBS at room temperature. Force curve data were extracted from AFM files and analyzed off-line. The Young's modulus (E) was computed from approaching force curves using a Hertzian model with any arbitrary two points, (z1, d1) and (z2, d2), on the force curve, using the following equation:
where υ is the Poisson ratio (since the force was applied in z-direction and Poisson's ratio was altered in the x-y plan, the Hertz model was used and the Poisson's ratio was 0.3); k is the spring constant of cantilever, R is the tip radius (10 μm). E was determined using the entire region of the compression curve, averaging values by using a five-point moving filter (slope of the curve every 5 points).
Supplementary Material
Acknowledgements
The project described was supported by Award Number R01EB012597 from the National Institute of Biomedical Imaging and Bioengineering and grants (CMMI-1130894 and CMMI-1120795) from the National Science Foundation. The UCSD Neuroscience Microscopy Shared Facility was supported by Grant P30 (NS047101).
Footnotes
Supporting Information Supporting Information is available from the Wiley Online Library or from the author.
References
- [1].Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Am. J. Physiol. Cell Physiol. 2002;282:C595. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- [2].Dembo M, Wang Y-L. Biophys. J. 1999;76:2307. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Khademhosseini A, Langer R. Biomaterials. 2007;28:5087. doi: 10.1016/j.biomaterials.2007.07.021. [DOI] [PubMed] [Google Scholar]
- [4].Discher DE, Janmey P, Wang Y.-l. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- [5].Janmey PA, Winer JP, Murray ME, Wen Q. Cell Motil. Cytoskeleton. 2009;66:597. doi: 10.1002/cm.20382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pelham RJ, Wang Y.-l. Proc. Natl. Acad. Sci. USA. 1997;94:13661. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lo C-M, Wang H-B, Dembo M, Wang Y.-l. Biophys. J. 2000;79:144. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- [9].Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Cancer Cell. 2005;8:241. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- [10].Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ. Nat. Mater. 2010;9:518. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Williams JL, Lewis JL. J. Biomech. Eng. 1982;104:50. doi: 10.1115/1.3138303. [DOI] [PubMed] [Google Scholar]
- [12].Keskar NR, Chelikowsky JR. Nature. 1992;358:222. [Google Scholar]
- [13].Baughman RH, Shacklette JM, Zakhidov AA, Stafstrom S. Nature. 1998;392:362. [Google Scholar]
- [14].Gunton DJ, Saunders GA. J. Mater. Sci. 1972;7:1061. [Google Scholar]
- [15].Baughman RH, Galvao DS. Nature. 1993;365:735. [Google Scholar]
- [16].Gardner GB, Venkataraman D, Moore JS, Lee S. Nature. 1995;374:792. [Google Scholar]
- [17].Hall LJ, Coluci VR, Galvao DS, Kozlov ME, Zhang M, Dantas SO, Baughman RH. Science. 2008;320:504. doi: 10.1126/science.1149815. [DOI] [PubMed] [Google Scholar]
- [18].Lakes R. Science. 1987;235:1038. doi: 10.1126/science.235.4792.1038. [DOI] [PubMed] [Google Scholar]
- [19].Choi JB, Lakes RS. J. Mater. Sci. 1992;27:5375. [Google Scholar]
- [20].Choi JB, Lakes RS. J. Mater. Sci. 1992;27:4678. [Google Scholar]
- [21].Milton GW. J. Mech. Phys. Solids. 1992;40:1105. [Google Scholar]
- [22].Alderson KL, Evans KE. Polymer. 1992;33:4435. [Google Scholar]
- [23].Caddock BD, Evans KE. J. Phys. D Appl. Phys. 1989;22:1877. [Google Scholar]
- [24].Evans KE, Caddock BD. J. Phys. D Appl. Phys. 1989;22:1883. [Google Scholar]
- [25].Baughman RH, Dantas SO, Stafstrom S, Zakhidov AA, Mitchell TB, Dubin DHE. Science. 2000;288:2018. doi: 10.1126/science.288.5473.2018. [DOI] [PubMed] [Google Scholar]
- [26].Lakes R. Science. 2000;288:1976. doi: 10.1126/science.288.5473.1976. [DOI] [PubMed] [Google Scholar]
- [27].Evans KE, Alderson A. Adv. Mater. 2000;12:617. [Google Scholar]
- [28].Lakes R. Nature. 1992;358:713. [Google Scholar]
- [29].Burns S. Science. 1987;238:551. doi: 10.1126/science.238.4826.551. [DOI] [PubMed] [Google Scholar]
- [30].Baughman RH, Stafstram S, Cui C, Dantas SO. Science. 1998;279:1522. doi: 10.1126/science.279.5356.1522. [DOI] [PubMed] [Google Scholar]
- [31].Evans KE, Nkansah MA, Hutchinson IJ, Rogers SC. Nature. 1991;353:124. [Google Scholar]
- [32].Rothenburg L, Berlin AI, Bathurst RJ. Nature. 1991;354:470. [Google Scholar]
- [33].Choi JB, Lakes RS. J. Compos. Mater. 1995;29:113. [Google Scholar]
- [34].Gibson LJ, Ashby MF, Schajer GS, Robertson CI. Proc. R. Soc. London, Ser. A. 1982;382:25. [Google Scholar]
- [35].Almgren RF. J. Elast. 1985;15:427. [Google Scholar]
- [36].Chen XG, Brodland GW. J. Mech. Behav. Biomed. Mater. 2009;2:494. doi: 10.1016/j.jmbbm.2008.12.004. [DOI] [PubMed] [Google Scholar]
- [37].Timmins LH, Wu QF, Yeh AT, Moore JE, Greenwald SE. Am. J. Physiol.–Heart Circ. Physiol. 2010;298:H1537. doi: 10.1152/ajpheart.00891.2009. [DOI] [PubMed] [Google Scholar]
- [38].Burriesci G, Bergamasco G. US patent 162112. 2007
- [39].Lin CY, Kikuchi N, Hollister SJ. J. Biomech. 2004;37:623. doi: 10.1016/j.jbiomech.2003.09.029. [DOI] [PubMed] [Google Scholar]
- [40].Lakes R. Nature. 2001;414:503. doi: 10.1038/35107190. [DOI] [PubMed] [Google Scholar]
- [41].Jackman RJ, Brittain ST, Adams A, Prentiss MG, Whitesides GM. Science. 1998;280:2089. doi: 10.1126/science.280.5372.2089. [DOI] [PubMed] [Google Scholar]
- [42].Veronda DR, Westmann RA. J. Biomech. 1970;3:111. doi: 10.1016/0021-9290(70)90055-2. [DOI] [PubMed] [Google Scholar]
- [43].Soman P, Fozdar DY, Lee JW, Phadke A, Varghese S, Chen S. Soft Matter. 2012;8:4946. doi: 10.1039/C2SM07354D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Soman P, Lee JW, Phadke A, Varghese S, Chen S. Acta Biomater. 2012;8:2587. doi: 10.1016/j.actbio.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Fozdar DY, Soman P, Lee JW, Han L-H, Chen S. Adv. Funct. Mater. 2011;21:2712. doi: 10.1002/adfm.201002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Masters IG, Evans KE. Compos. Struct. 1996;35:403. [Google Scholar]
- [47].Lee KY, Mooney DJ. Chem. Rev. 2001;101:1869. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- [48].Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Biomaterials. 2009;30:6702. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hahn MS, Taite LJ, Moon JJ, Rowland MC, Ruffino KA, West JL. Biomaterials. 2006;27:2519. doi: 10.1016/j.biomaterials.2005.11.045. [DOI] [PubMed] [Google Scholar]
- [50].Gibson LJ, Ashby MF. Cellular solids: structure and properties. Cambridge University Press; Cambridge, UK: 1997. [Google Scholar]
- [51].Greaves GN, Greer AL, Lakes RS, Rouxel T. Nat. Mater. 2011;10:823. doi: 10.1038/nmat3134. [DOI] [PubMed] [Google Scholar]
- [52].Itabashi T, Terada Y, Kuwana K, Kan T, Shimoyama I, Ishiwata S. i. Proc. Natl. Acad. Sci. USA. 2012;109:7320. doi: 10.1073/pnas.1116749109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Nature. 2005;437:1043. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- [54].Estey MP, Di Ciano-Oliveira C, Froese CD, Bejide MT, Trimble WS. J. Cell. Biol. 2010;191:741. doi: 10.1083/jcb.201006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dean SO, Rogers SL, Stuurman N, Vale RD, Spudich JA. Proc. Natl. Acad. Sci. USA. 2005;102:13473. doi: 10.1073/pnas.0506810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Straight AF, Field CM, Mitchison TJ. Molec. Biol. Cell. 2005;16:193. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ghassemi S, Meacci G, Liu S, Gondarenko AA, Mathur A, Roca-Cusachs P, Sheetz MP, Hone J. Proc. Natl. Acad. Sci. USA. 2012 doi: 10.1073/pnas.1119886109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Eisenstadt MM. Introduction to mechanical properties of materials. Macmillan; New York: 1971. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.