Abstract
The defining pathogenic feature of Parkinson’s disease is the age dependent loss of dopaminergic neurons. Mutations and inactivation of parkin, an ubiquitin E3 ligase, cause Parkinson’s disease through accumulation of pathogenic substrates. Here we show that transgenic overexpression of the parkin substrate, aminoacyl-tRNA synthetase complex interacting multifunctional protein-2 (AIMP2) leads to a selective, age-dependent progressive loss of dopaminergic neurons via activation of poly(ADP-ribose) polymerase-1 (PARP1). AIMP2 accumulation in vitro and in vivo results in PARP1 overactivation and dopaminergic cell toxicity via direct association of these proteins in the nucleus providing a new path to PARP1 activation other than DNA damage. Inhibition of PARP1 through gene deletion or drug inhibition reverses behavioral deficits and protects in vivo against dopamine neuron death in AIMP2 transgenic mice. These data indicate that brain permeable PARP inhibitors could be effective in delaying or preventing disease progression in Parkinson’s disease.
Mutations in the ubiquitin E3 ligase, Parkin, are an important cause of familial Parkinson’s disease 1,2. In general the majority of mutations in Parkin result in a loss of E3 ligase activity3–5. In the more common sporadic form of Parkinson’s disease, there may be a loss of parkin function due to S-nitrosylation, oxidative and dopaminergic stress, and phosphorylation by the stress activated kinase, c-Abl6–13. Parkin is a multifunctional E3 ligase that ubiquitinates proteins using different ubiquitin lysine linkages. Monoubiquitination of parkin substrates is thought to lead to alterations in receptor trafficking and cell signaling5. Polyubiquitination via lysine 63 or 29 linkages may play a role in inclusion body formation and autophagy14,15. Parkin substrates that are polyubiquitinated via lysine 48 linkages are degraded by the ubiquitin proteosome system. The loss of parkin function in Parkinson’s disease would be expected to impair the ubiquitin proteasome system clearance of lysine 48 substrates and interfere with other parkin E3 ligase functions16. Since parkin is inactivated in familial Parkinson’s disease with Parkin mutations, sporadic Parkinson’s disease, Parkin−/− mice and following 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) intoxication in mice9, substrates elevated in all four conditions are candidates for parkin mediated polyubiquitination via lysine 48 linkages and subsequent ubiquitin proteosome degradation.
Aminoacyl-tRNA synthetase complex interacting multifunctional protein-2 (AIMP2), also known as JTV-1 or P38, is a parkin substrate that is present in Lewy body inclusions of Parkinson’s disease substantia nigra17,18. AIMP2 is a strong candidate as a pathogenic parkin substrate that accumulates in Parkinson’s disease due to parkin inactivation since AIMP2 levels are elevated in the ventral midbrain in Parkin−/− mice, and post-mortem brain from patients with Parkin mutations or sporadic Parkinson’s disease 7,9,18. AIMP2 also accumulates in the MPTP model of Parkinson’s disease consistent with the notion that parkin is inactivated following MPTP intoxication9. If accumulation of a parkin substrate is important in the pathogenesis of Parkinson’s disease, transgenic overexpression in an animal model should lead to an age-dependent progressive degeneration of dopamine neurons.
To explore a potential biologic mechanism for AIMP2 and to validate its role as a pathogenic substrate in Parkinson’s disease, we generated a tetracycline regulated inducible transgenic mouse model19,20 of AIMP2 overexpression. AIMP2 overexpression at levels seen in Parkinson’s disease leads to an age-dependent degeneration of dopaminergic neurons that causes striatal dopaminergic deficits and impairment of motor coordination. AIMP2 toxicity is not mediated by its canonical function because gross protein translation was normal. Unexpectedly, AIMP2 directly activates Poly(ADP-ribose) polymerase-1 (PARP1), an important protein that plays a major role in the DNA damage response through poly(ADP-ribosylation) of PARP1 itself and chromatin associated proteins8,21. Excessive activation of PARP1 kills cells via the formation of poly(ADP-ribose) (PAR) polymer in a cell death mechanism designated parthanatos22. During parthanatos PAR polymer translocates from the nucleus to the mitochondria and binds apoptosis-inducing factor (AIF). PAR polymer binding to AIF facilitates the release of AIF from the mitochondria and translocation to the nucleus followed by large scale DNA fragmentation and nuclear condensation leading to the execution phase of parthanatos23,24. Knockout or inhibition of PARP1 completely prevents the degeneration of dopaminergic neurons due to AIMP2 overexpression. Thus, AIMP2 mediated dopaminergic cell death is mediated by parthanatos suggesting that PARP1 inhibition may be effective in delaying the progression of Parkinson’s disease.
RESULTS
Generation of tetracycline regulated inducible transgenic AIMP2 mice
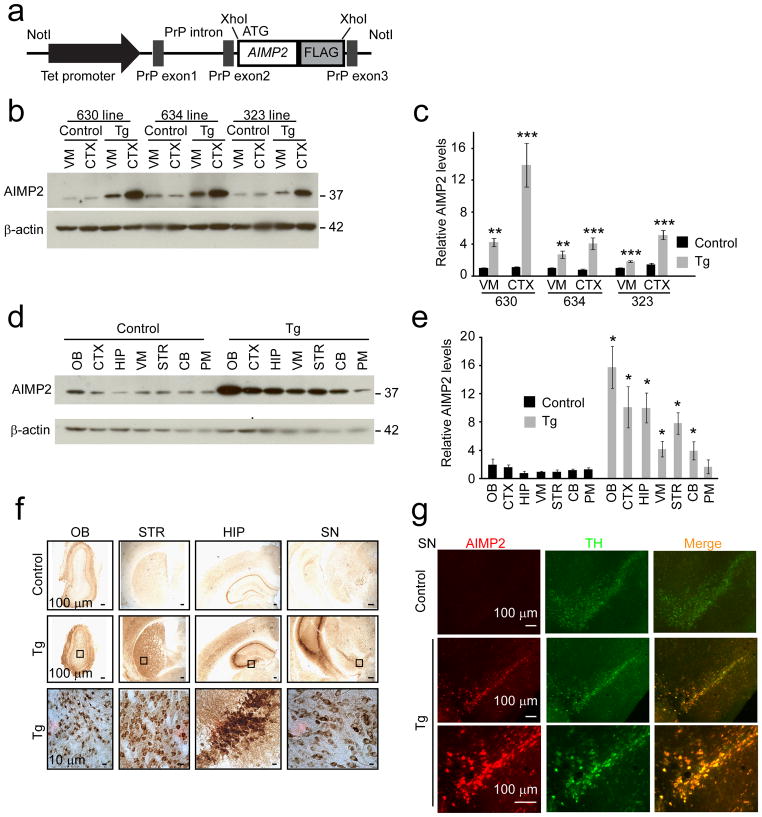
To investigate whether AIMP2 causes neuronal degeneration in vivo, we generated a conditional transgenic mouse model in which the expression of a C-terminal FLAG tagged human AIMP2 is under the control of a tetracycline responsive regulator (Fig. 1a). 29 founders expressing TetP-AIMP2 were identified via PCR screening for the tetracycline promoter (Supplementary Fig. 1a, b). Five of the highest copy number male founder mice were crossed with the CamKIIα-tTA transgenic mice25 and mice expressing both CamKIIα-tTA and AIMP2 were identified by PCR (Supplementary Fig. 1c).
Figure 1.
Generation and characterization of conditional AIMP2 transgenic mice.
(a) Schematic of the TetP-AIMP2-FLAG transgenic construct.
(b) Representative western blot of AIMP2 in cortex (CTX) and ventral midbrain (VM) of three lines (630, 634, and 323) of transgenic mice (Tg) and age-matched littermate controls (Control).
(c) Quantification of AIMP2 protein levels in ventral midbrain and CTX of AIMP2 transgenic mice and littermate controls from three lines normalized to β-actin, n = 3.
(d) Representative western blot of AIMP2 distribution in brain subregions from control and AIMP2 transgenic (630 line) mice (OB, olfactory bulb; CTX, cortex; HIP, hippocampus; VM, ventral midbrain; STR, striatum; CB, cerebellum; PM, pons and medulla).
(e) Quantification of AIMP2 distribution in mouse brains normalized to β-actin, n = 3.
(f) AIMP2 immunostaining of brain sections from AIMP2 transgenic mice and littermate controls. Magnified images are shown in the bottom panel to visualize AIMP2 staining in cell populations, n = 3. OB, olfactory bulb; HIP, hippocampus; STR, striatum; SN, substantia nigra.
(g) Immunofluorescent images of AIMP2 (red) and tyrosine hydroxylase (TH, green) from AIMP2 transgenic and control mice. High power view is shown in the bottom panel. Quantified data (c, e) are expressed as mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, unpaired two-tailed Student’s t test. β-actin was used as an internal loading control (b, d). Full length blots are presented in Supplementary Figure 14 (b, d).
Expression of AIMP2 was detected in ventral midbrain and cortex and compared to littermate controls expressing either CamKIIα-tTA or TetP-AIMP2 alone. The overexpression of AIMP2 is tetracycline responsive as doxycycline administration attenuates the upregulation of AIMP2 (Supplementary Fig. 1d). Line 630 overexpresses AIMP2 ~14 fold in the cortex and ~4 fold in the ventral midbrain (Fig. 1b, c). Line 634 overexpresses AIMP2 ~4 fold in the cortex and ~3 fold in the ventral midbrain (Fig. 1b, c). Line 323 overexpresses AIMP2 ~5 fold in the cortex and ~2 fold in the ventral midbrain (Fig. 1b, c). Since line 630 expressed the highest levels of AIMP2 it was selected for detailed characterization. Western blot analysis indicates that AIMP2 is overexpressed 4 to 15 fold throughout the forebrain, olfactory bulb, ventral midbrain and cerebellum with no overexpression in the pons and medulla (Fig. 1d, e). Immunohistochemistry with an anti-AIMP2 antibody suggests the majority of AIMP2 overexpression is in neurons (Fig. 1f). To determine if AIMP2 overexpression in the substantia nigra is localized to dopaminergic neurons, co-localization studies with AIMP2 and tyrosine hydroxylase were performed (Fig 1g). AIMP2 is overexpressed in the majority of tyrosine hydroxylase containing neurons within the substantia nigra (Fig 1g).
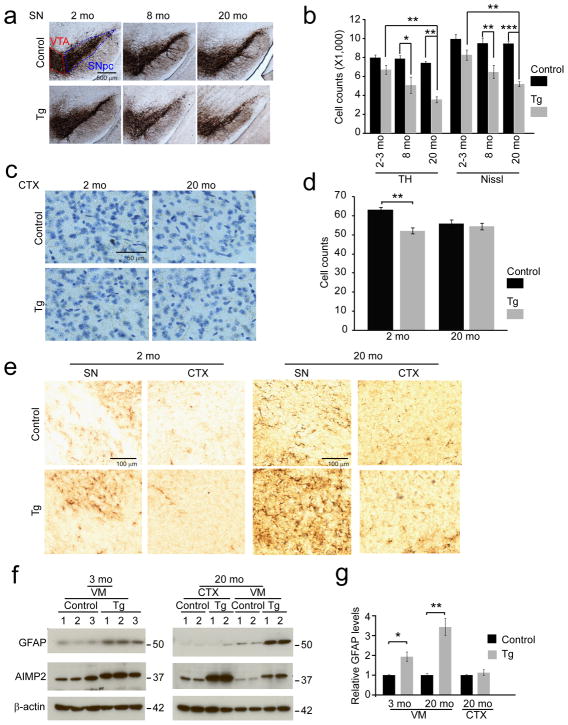
Selective age-dependent dopaminergic degeneration in AIMP2 transgenic mice
All lines of AIMP2 transgenic mice are viable and develop normally. AIMP2 transgenic mice appear to have comparable body weight compared to litter mate control mice for up to three months of age. Starting at five months of age, the AIMP2 mice gain less body weight compared to litter mate controls and have a significant reduction in body weight at 14 and 20 months of age (Supplementary Fig. 2). To determine the effect of AIMP2 expression on dopamine neuronal viability, tyrosine hydroxylase immunoreactivity and Nissl staining in the substantia nigra zona compacta (SNpc) was monitored by unbiased stereologic methods. At 2–3 months of age there is a trend towards a loss of tyrosine hydroxylase positive and Nissl stained neurons. At 8 months of age there is a 37% loss that progresses to a loss of 53% of tyrosine hydroxylase-positive and Nissl stained neurons in the SNpc at 20 months (Fig. 2a, b). In addition, approximately 28% of dopamine neurons in the ventral tegmental area (VTA) degenerate in AIMP2 transgenic mice at 20 months of age (Supplementary Fig. 3a) consistent with the loss of VTA dopamine neurons in Parkinson’s disease26. Thus, VTA dopamine neurons are less sensitive to AIMP2 toxicity compared to those in the SNpc similar to the differential vulnerability of the SNpc and VTA dopamine neurons in Parkinson’s disease26. To further confirm that dopamine neuronal loss in the SNpc depends on AIMP2 accumulation, another transgenic line expressing AIMP2 at lower levels (line 634) was also characterized. At 10 months of age there is a significant 21% loss of dopamine neurons in this second line of AIMP2 transgenic mice (Supplementary Fig. 3b, c).
Figure 2.
AIMP2 accumulation leads to a progressive and selective degeneration of dopaminergic neurons in the substantia nigra of transgenic mice.
(a) Representative tyrosine hydroxylase (TH) immunohistochemistry of the substantia nigra (SN) of AIMP2 transgenic and age-matched littermate controls. Substantia nigra pars compacta (SNpc), ventral tegmental area (VTA).
(b) Stereological assessment of tyrosine hydroxylase and Nissl positive neurons in the substantia nigra (n = 9 per group at 2–3 months; n = 5 Control, n = 7 transgenic at 8 months; n = 6 per group at 20 months).
(c) Representative images of Nissl-stained cortex.
(d) Stereological assessment of Nissl positive cortical neurons (n = 3 per group)
(e) Representative GFAP immunohistochemistry of the substantia nigra (SN) and CTX of the AIMP2 transgenic and age-matched littermate control mice.
(f) Representative western blots of GFAP, AIMP2 and β-actin in the CTX and the ventral midbrain of 2 month and 20 month old AIMP2 transgenic and littermate control mice.
(g) Quantification of GFAP protein levels normalized to β-actin (n = 3 per group of 2 month old; n = 4 per group of 20 month old).
Quantified data (b, d, and g) are expressed as mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, Kruskal-Wallis ANOVA test (TH counts of Fig. 2b), analysis of variance (ANOVA) test followed by Student-Newman-Keuls post-hoc analysis (Nissl counts of Fig. 2b, d), unpaired two-tailed Student t test (g). Full length blots are presented in Supplementary Figure 14 (f).
In the highest AIMP2 expressing line no substantial loss of neurons was identified in other brain regions. In the cortex, which overexpresses AIMP2 ~14 fold there is a small 10% reduction of neurons at 2 months of age while there was no substantial difference in neuron number at 20 months of age between AIMP2 transgenics and age matched littermate controls, suggesting that the degeneration of dopamine neurons induced by AIMP2 is selective (Fig. 2c, d). To evaluate if there are other neuropathologic abnormalities, the brains of AIMP2 mice and littermate controls were examined for astrocytosis. In 20 month old AIMP2 transgenic mice there is a substantial increase in glial fibrillary acidic protein (GFAP) immunoreactivity in the substantia nigra consistent with the loss of dopaminergic neurons (Fig. 2e). There is no substantial difference in GFAP immunoreactivity in the cortex between the AIMP2 transgenic mice and age matched littermate controls (Fig. 2e). Western blot analyses of the levels of GFAP confirm that there is a significant, greater than 3 fold and 2 fold, upregulation of GFAP in the substantia nigra of 20 month and 3 month old AIMP2 transgenic mice, respectively, but no significant change in the cortex (Fig. 2f, g).
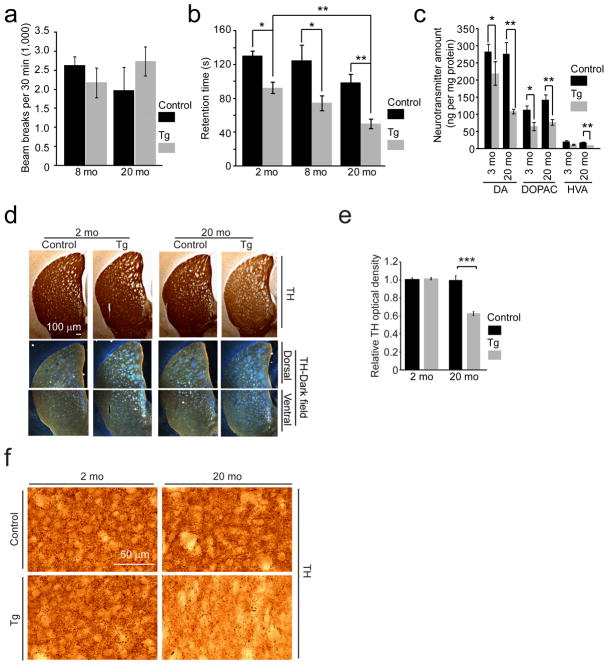
Behavioral and dopaminergic deficits in AIMP2 transgenic mice
The motor function of AIMP2 transgenic mice was examined by rotarod and open field testing. AIMP2 transgenic mice perform normally in the open field test compared to age matched littermate controls at 8 and 20 months of age (Fig. 3a). In an accelerating rotarod paradigm, AIMP2 transgenic mice have a progressive loss of the ability to remain on the rod compared to age matched littermate controls (Fig. 3b). In a dopamine-sensitive pole test, AIMP2 transgenic mice exhibit deficits in climbing down the pole as assessed at 20 months of age (time to ground in pole test, mean ± s.e.m.: 43.8±11.2 s for Control and 97.9 ± 10.2 s for transgenic mice, P < 0.05 determined by Mann-Whitney test. n=13 Control and 8 transgenic mice). HPLC analysis of dopamine and its metabolites, 3,4-dihydroxyphenylacetic acid (DOPAC) and homovanillic acid (HVA), was performed to monitor dopaminergic integrity. At 3 months of age AIMP2 transgenic mice have a 20% reduction in dopamine, DOPAC and HVA compared to age matched littermate controls. The loss of dopamine is progressive with a 60% reduction of dopamine content at 20 months of age in the AIMP2 transgenic mice. DOPAC and HVA remain significantly reduced at 20 months of age (Fig. 3c).
Figure 3.
Progressive motor deficit, reduction of dopamine and dopaminergic fiber densities in the striatum of AIMP2 transgenic mice.
(a) Assessment of spontaneous exploration in an open field chamber for AIMP2 transgenic and littermate control mice (n = 7 Control and n = 8 Tg at 8 months; n = 6 Control and n =8 Tg at 20 months).
(b) Assessment of retention time in an accelerated rotarod test (n = 11 Control, n = 5 Tg at 2 months; n = 8 per group at 8 months; n = 13 Control, n = 8 Tg at 20 months)
(c) High performance liquid chromatography (HPLC) assessment of the striatal content of dopamine and its metabolites, 3,4-dihydroxyphenylacetic acid (DOPAC) and homovanillic acid (HVA) (n = 5 for Control and n = 6 for Tg at 3 months, n = 10 for Control and n = 6 for Tg at 20 months).
(d) Tyrosine hydroxylase immunohistochemistry of the striatum. Darkfield photomicrographs (bottom panels) of dorsal and ventral striatum.
(e) Quantification of optical densities of (d) (n = 3 Control, n = 4 Tg). Optical density of the striatum was subtracted by background before normalization. Densitometric analysis was processed using ImageJ software.
(f) High power view of tyrosine hydroxylase-positive dopaminergic nerve fibers in the striatum.
Quantified data (a, b, c, e) were expressed as mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, unpaired two-tailed Student’s t test (a, e), or Kruskal-Wallis ANOVA test (b, c).
Assessment of the (DOPAC + HVA)/dopamine ratio suggests that there is a trend toward increased dopamine metabolism in the AIMP2 transgenic mice (data not shown). Dopamine terminal density in the striatum was assessed in the AIMP2 transgenics compared to age matched littermate controls. There is a 40% reduction of dopamine terminal density in 20 month old AIMP2 transgenic mice (Fig. 3d, e, f). Taken together these data indicate that expression of AIMP2 leads to an age dependent loss of dopaminergic neurons accompanied by motor deficits.
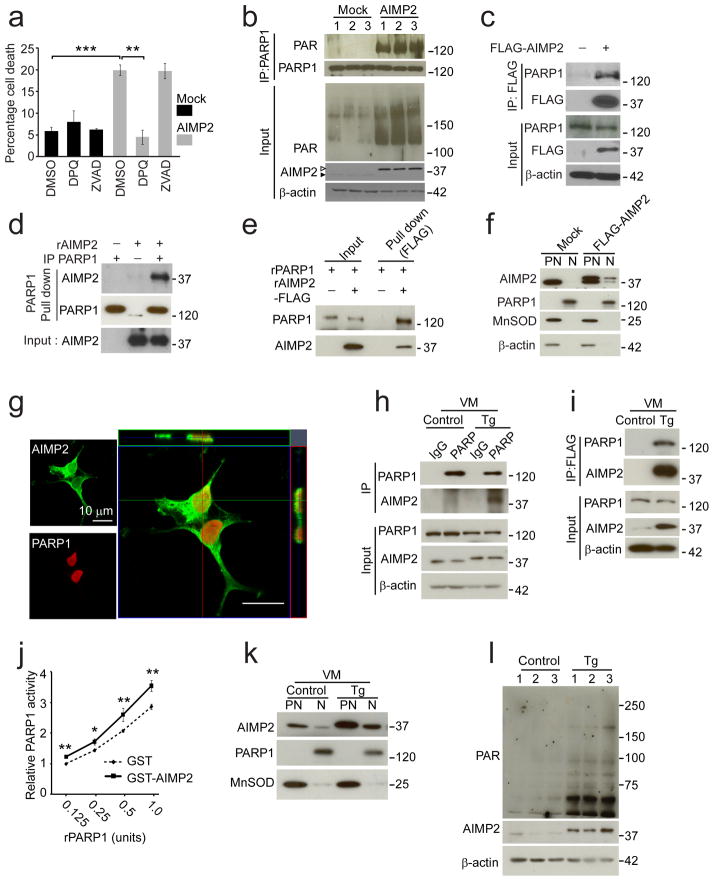
AIMP2 accumulation leads to PARP1 activation
To explore the potential mechanism by which AIMP2 overexpression leads to neurodegeneration a conditional AIMP2 PC12 cell model9 was developed by expressing AIMP2 under the tetracycline responsive promoter (Supplementary Fig. 4a). The canonical function of AIMP2 is in aminoacyl-tRNA synthesis which is essential for peptide extension during protein synthesis27. To determine whether AIMP2 overexpression affects protein synthesis, 35S-methionine incorporation into proteins was monitored in the stable PC12-conditional AIMP2 cell model in the presence or absence of doxycyline. No substantial difference in protein synthesis is observed between doxycyline stimulated or unstimulated conditions (Supplementary Fig. 4b). To determine whether AIMP2 overexpression might regulate stress induced protein synthesis the effects of H2O2, thapsigargin and MPP+ were evaluated in the presence or absence of doxycyline. No substantial difference in protein synthesis was observed under these conditions. 200 μM H2O2 suppresses protein synthesis as previously reported28 and serves as a positive control (Supplementary Fig. 4b). Since AIMP2 overexpression did not have any observable effects on overall protein synthesis, cell death was then monitored in a transient conditional AIMP2 overexpression model in SH-SY5Y cells. In these cellular models, AIMP2 overexpression causes approximately 20% cell death that is caspase-independent as the broad spectrum caspase inhibitor, ZVAD, does not prevent cell death. However, AIMP2 toxicity is PARP dependent as the PARP inhibitor DPQ completely prevents cell death (Fig. 4a).
Figure 4.
AIMP2 accumulation leads to PARP1 activation and cell death.
(a) Viability of SH-SY5Y cells transfected with AIMP2 and mock DNA. DPQ, 10 μM; ZVAD, 50 μM. n = 4 per group.
(b) Western blot analysis of PARP1 self-PARsylation and PAR conjugation to proteins in SH-SY5Y cells (Empty triangle, FLAG-AIMP2; filled triangle, endogenous AIMP2).
(c) Immunoprecipitation of FLAG-AIMP2 and endogenous PARP1 in SH-SY5Y cells.
(d) Anti-PARP1 pull-down of recombinant AIMP2 (rAIMP2) and immunopurified PARP1 (IP PARP1).
(e) Anti-FLAG pull-down of recombinant PARP1 (rPARP1) and recombinant AIMP2-FLAG (rAIMP2-FLAG).
(f) Subcellular localization of AIMP2 and PARP1 in SH-SY5Y cells.
(g) Confocal microscopic images in SH-SY5Y cells transfected with FLAG-AIMP2.
(h) Co-immunoprecipitation of AIMP2 using PARP1 antibody in the ventral midbrain (VM) of mice.
(i) Co-immunoprecipitation of PARP1 using FLAG antibody in the ventral midbrain of mice.
(j) In vitro PARP1 activation assay (n = 5 per group).
(k) Subcellular localization of AIMP2 and PARP1 in the ventral midbrain of mice.
(l) Western blots of AIMP2 and PAR in the ventral midbrain of 12 month old mice.
Quantified data (a, j) are expressed as mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, (a) ANOVA test followed by Student-Newman-Keuls post-hoc analysis, or (j) Mann-Whitney U test. Similar results were reproduced in multiple experiments (c, d, e, f, h, i, k). β-actin serves as a loading control (b, c, h, i, l). Full length blots are presented in Supplementary Figure 14 (b–f, h–i, k–l).
AIMP2 overexpression activates PARP and leads to a 3.5 fold increase in PAR levels (Relative levels, mean ± s.e.m.: 1.4 ± 0.2 for Mock and 3.9 ± 0.3 for AIMP2, P < 0.01 determined by unpaired student’s t test) and a greater 5 fold increase in PARsylated PARP1 (Relative levels, mean ± s.e.m.: 1.2 ± 0.4 for Mock and 6.1 ± 1.0 for AIMP2, P < 0.05 determined by unpaired student’s t test) (Fig. 4b). To determine whether AIMP2 interacts with PARP1 directly, co-immunoprecipitation experiments were conducted and show that immunoprecipitation of FLAG-AIMP2 in SH-SY5Y cells pulls down endogenous PARP1 (Fig. 4c). In an immunopurified PARP1 pull down assay recombinant AIMP2 binds to immunopurified PARP1 (Fig. 4d). Recombinant AIMP2-FLAG also pulls down recombinant PARP1 in an immunoprecipitation with an anti-FLAG antibody (Fig. 4e). Taken together these data indicate that AIMP2 and PARP1 are interacting proteins.
Deletion analysis reveals that PARP1 interacts with amino acids 1-83 of AIMP2 (Supplementary Fig. 5a) and AIMP2 interacts with the automodification domain of PARP1 (Supplementary Fig. 5b). While AIMP2 is primarily enriched in the post-nuclear fraction, there is also a smaller pool present in the nuclear fraction available to interact with PARP1 as determined by subcellular fractionation and monitored by AIMP2 and the nuclear marker, PARP1, and the mitochondrial marker, manganese superoxide dismutase (MnSOD), immunoreactivity (Fig. 4f). Confocal image analysis shows that AIMP2 is primarily localized to the cytosol but a small percentage is found within the nucleus (Fig. 4g). In the ventral midbrain of AIMP2 transgenic mice immunoprecipitated FLAG-AIMP2 pulls down PARP1 and immunoprecipitation of PARP1 pulls down FLAG-AIMP2 (Fig. 4h, i). The interaction of AIMP2 and PARP1 in the ventral midbrain in vivo was also confirmed in transgenic line 322, which overexpresses AIMP2 at levels similar to the adult conditional Parkin−/− mouse brains (Supplementary Fig. 6a and see Supplementary Fig. 10a, b). To ascertain whether AIMP2 binding to PARP1 directly activates PARP1, a PARP ribosylation activity assay was performed in the presence of recombinant GST-AIMP2 versus GST as a control. GST-AIMP2 significantly increases the ribosylation activity of PARP1 (Fig. 4j).
Next the subcellular localization of AIMP2 was monitored in the ventral midbrain of AIMP2 transgenic mice versus age matched littermate controls. AIMP2 is found in both the nuclear and post-nuclear fraction as monitored by PARP1 and MnSOD immunoreactivity, respectively (Fig. 4k). Accompanying the nuclear localization of AIMP2 there is an approximate 2.5 fold increase in the level of PAR (Relative levels, mean ± s.e.m.: 1.1 ± 0.1 for Control and 2.4 ± 0.3 for transgenic, P < 0.05 determined by unpaired student’s t test. n=6 per group) in the ventral midbrain of AIMP2 transgenics as compared to age matched littermate controls (Fig. 4l). The association of overexpressed AIMP2 and nuclear PARP1 appears region specific since we failed to detect nuclear translocation of AIMP2 or interaction of AIMP2 and PARP1 in cortical tissues of AIMP2 transgenic mice (Supplementary Fig. 6b, c). Consistent with these results, no significant change in PAR levels is observed in the cortex of AIMP2 transgenic mice versus age matched control mice (Supplementary Fig. 7a, b).
To further explore the AIMP2-PARP1 pathway, SH-SY5Y cells were treated with hydrogen peroxide or N-methyl-N′-nitro-N′-nitrosoguanidine (MNNG), a DNA damaging reagent and strong PARP1 activator. Following treatment with hydrogen peroxide or MNNG, endogenous AIMP2 translocates into the nucleus (Supplementary Fig. 8a) leading to a strong association with PARP1 and subsequent PAR production (Supplementary Fig. 8b). Taken together these data indicates that AIMP2 interacts with PARP1 in the nucleus leading to PARP1 activation and PAR formation in dopaminergic neurons of the substantia nigra.
AIMP2 mediated degeneration of dopamine neurons occurs through parthanatos
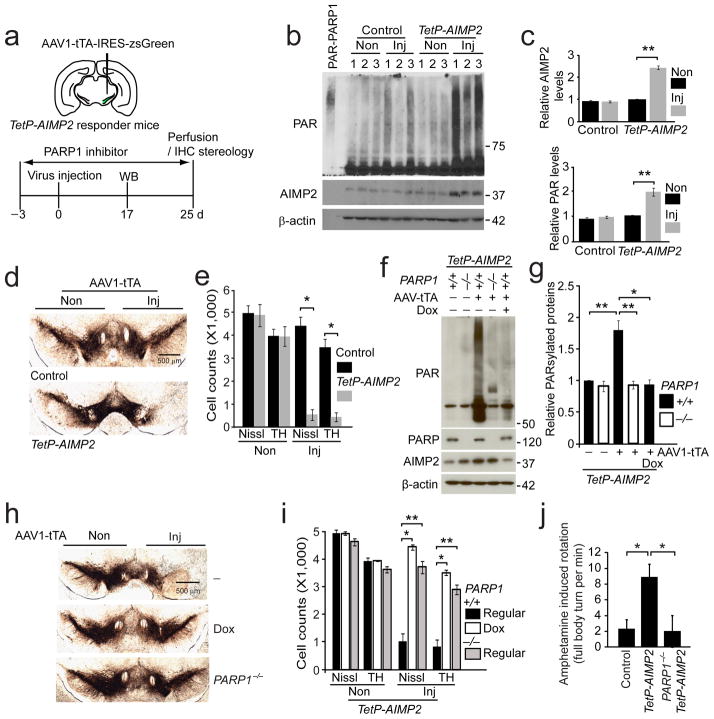
Since the CamKIIα-tTA driven expression of AIMP2 results in a broad expression of AIMP2 in the mouse forebrain and the degeneration of dopamine neurons in the midbrain takes 8 to 20 months, we sought to develop a conditional model where AIMP2 overexpression is largely confined to the ventral midbrain and loss of dopamine neurons occurs on a shorter time scale. Accordingly an adeno-associated virus type 1 (AAV1) vector was constructed with the chicken β-actin promoter driving tTA with an IRES2 zsGreen for monitoring expression (AAV1-tTA-IRES-zsGreen) (Supplementary Fig. 9a). AAV1-tTA-IRES-zsGreen was stereotaxically injected into the ventral midbrain of TetP-AIMP2 transgenic mice or non-transgenic control mice (Fig. 5a). The AAV1-tTA-IRES-zsGreen robustly transduces the ventral midbrain of non-transgenic mice with a majority of dopaminergic neurons transduced as monitored by colocalization of TH immunostaining with zsGreen (Supplementary Fig. 9b). 17 days after injection, AIMP2 levels were monitored by immunoblot and AIMP2 levels are increased by greater than 2 fold (Fig. 5b, c). To determine if these mice exhibit comparable levels of AIMP2 expression as in Parkin−/− mice, the level of AIMP2 was assessed in adult conditional Parkin−/− mice. AIMP2 is overexpressed approximately 2 fold in adult parkin conditional knockout mice (Supplementary Fig. 10a, b). At 25 days post injection the integrity of dopamine neurons as assessed by tyrosine hydroxylase immunoreactivity was monitored and notably there is robust loss of tyrosine hydroxylase immunoreactivty in the substantia nigra zona compacta (Fig. 5d). Unbiased stereologic analysis of tyrosine hydroxylase immunoreactive and Nissl stained neurons reveals a greater than 95% loss of dopamine neurons (Fig. 5e). Consistent with the lack of cortical neurotoxicity in the AIMP2 transgenic mice, virally induced AIMP2 overexpression in the cortex of TetP-AIMP2 mice (at similar levels of overexpression of AIMP2 in the ventral midbrain) failed to activate PARP1 (Supplementary Fig. 11a, b, c) and there was no obvious degeneration of cortical neurons (Supplementary Fig. 11d, e). To determine whether parthanatos plays a role in AIMP2 induced dopamine cell loss, PARP1 activation was monitored 17 days after the AAV1-tTA-IRES-zsGreen injection. PAR levels are increased by 2 fold (Fig. 5b, c). Next the TetP-AIMP2 transgenic mice were crossed with PARP1−/− mice to create TetP-AIMP2/PARP1−/− mice (Supplementary Fig. 1e). These TetP-AIMP2/PARP1−/− mice were stereotaxically injected with AAV1-tTA-IRES-zsGreen and dopamine cell loss was monitored and compared to TetP-AIMP2 with and without doxycycline treatment. AIMP2 induction, and PARP1 activation were examined via western blot analysis using AIMP2, and PAR antibodies (Fig. 5f). AIMP2 induction was efficiently suppressed by doxycycline in TetP-AIMP2 mice and the elevation of PARsylation proteins in the injected side of TetP-AIMP2 is also reduced to basal levels (Fig. 5f, g). Knockout of PARP1 completely rescues the degeneration of dopamine neurons similar to the absence of loss of dopamine neurons in the presence of doxycycline, which prevents the expression of AIMP2 (Fig. 5h, i). PARP1 activation was monitored by PAR immunoreactivity in AAV1-tTA-IRES-zsGreen injected TetP-AIMP2 transgenic mice versus TetP-AIMP2/PARP1−/− mice. There is an approximate 2 fold increase in PAR observed in the TetP-AIMP2 transgenics but no increase is found in the TetP-AIMP2/PARP1−/− mice (Fig. 5f, g). Since the injection is unilateral, amphetamine induced rotation was utilized as a functional behavioral readout of dopaminergic degeneration. AIMP2 overexpression leads to an 8 fold increase in rotational behavior consistent with the loss of dopamine neurons. Knockout of PARP1 prevents amphetamine induced rotation indicating that the dopamine neurons are spared and functional (Fig. 5j).
Figure 5.
AIMP2-induced degeneration of dopamine neurons is rescued by genetic deletion of PARP1.
(a) Schematic of time course of experiments.
(b) Western blots of AIMP2 and PAR in the ventral midbrains of AAV-tTA injected (Inj) or non-injected (Non) side of control and TetP-AIMP2 mice.
(c) Quantification of protein levels in panel (b) normalized to β-actin (n = 3 per group)
(d) Tyrosine hydroxylase immunohistochemistry of the substantia nigra 25 days after AAV-tTA injection.
(e) Stereological assessment of tyrosine hydroxylase or Nissl-positive dopaminergic neurons (n = 4 per group).
(f) Western blot analysis of indicated proteins in the ventral midbrain of mouse groups in panel (g).
(g) Quantification of PARsylated proteins from panel (h) normalized to β-actin (n = 3 Dox group, and n = 6 other groups).
(h) Tyrosine hydroxylase immunohistochemistry of the indicated mouse groups. Dox, doxycycline diet.
(i) Stereological assessment of tyrosine hydroxylase or Nissl-positive dopaminergic neurons in mice shown in panel (h) (n = 4 for Dox and n = 5 other groups).
(j) Amphetamine induced ipsilateral rotations (n = 7 for PARP1−/− and n = 5 for other groups)
Quantification of data (c, e, g, i, j) expressed as mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, unpaired two-tailed Student’s t test (c), Kruskal-Wallis ANOVA test followed by Mann-Whitney U post-hoc analysis (e, g, i, j). β-actin serves as a loading control (b, f). Full length blots are presented in Supplementary Figure 14 (b, f).
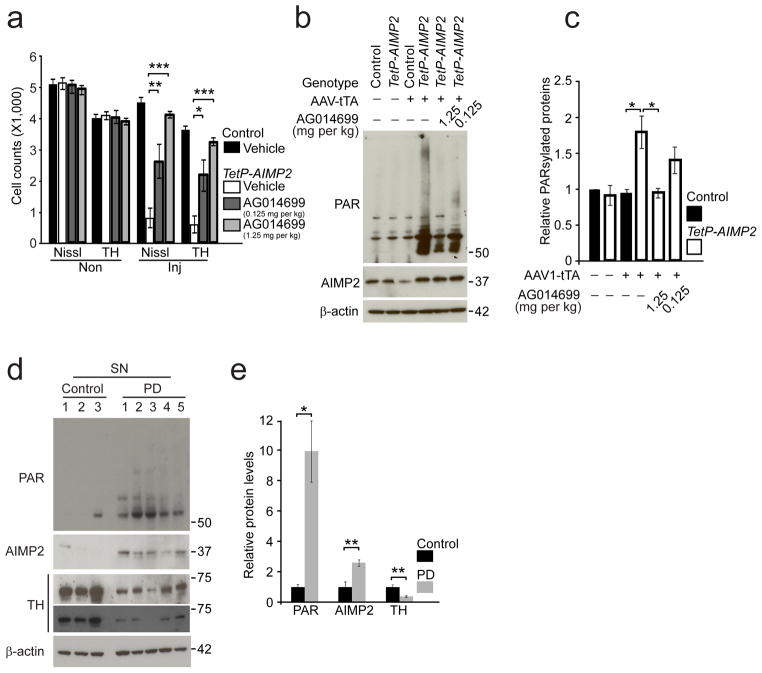
The PARP inhibitor, AG01469929 was administered to Tet-AIMP2 mice three days prior to injection of AAV1-tTA-IRES-zsGreen and then was continuously administered until the mice were analyzed (Fig. 5a). AG014699 in a dose dependent manner prevents the degeneration of dopamine neurons as monitored by unbiased stereologic counting of tyrosine hydroxylase- and Nissl- positive neurons (Fig. 6a). AG014699 reduces PARP activation as monitored by PAR immunoreactivity (Fig. 6b, c). To evaluate the potential pathophysiologic relevance of PARP1 activation and PAR accumulation in human disease, PAR was monitored in human postmortem substantia nigra brain tissue from Parkinson’s disease patients and age-matched controls (Supplementary Table 1). It is well known that substantial numbers of dopamine neurons are present in patients with Parkinson’s disease 26,30, which we confirm by the demonstration of TH immunoreactivity via immunoblot analysis (Fig. 6d, e). As previously reported there is a 2–3 fold increase in AIMP2 levels in the substantia nigra of Parkinson’s disease patients as compared to control, similar to the level of overexpression of AIMP2 in the models reported here. Accompanying the increase in AIMP2 there is a ~10 fold increase in PAR levels (Fig. 6d, e). It has been previously reported that there is no increase in the level of AIMP2 in areas of the Parkinson’s disease brain that are relatively non-affected such as the cortex7,9. There is no increase in PAR in the cortex of Parkinson’s disease patients (Supplementary Fig. 12) suggesting that the increase in the substantia nigra is due to the elevation of AIMP2 levels. Taken together these data suggest that elevation of AIMP2 leads to pathologic activation of PARP1 and accumulation of PAR that results in loss of dopaminergic neurons (Supplementary Fig. 13). This signaling may participate in the disease process of sporadic Parkinson’s disease.
Figure 6.
PARP inhibition protects against AIMP2 induced dopaminergic toxicity.
(a) Stereological assessment of tyrosine hydroxylase positive and Nissl positive dopaminergic neurons in the SNpc of control and TetP-AIMP2 mice 25 days following AAV-tTA injection with or without treatment with the PARP inhibitor, AG014699 (Vehicle, 50% DMSO in saline; 0.125 mg/kg or 1.25 mg/kg AG-14699, bid, i.p.), −3 days through 25 days after the viral injections (n = 3 per group).
(b) AIMP2, and PAR conjugated proteins in the ventral midbrain of control and TetP-AIMP2 mice 16 days following intranigral injection of AAV-tTA with the treatment of AG014699 monitored by western blot.
(c) Quantification of PAR conjugated protein levels in panel (b) normalized to β-actin (n = 3 for 1.25 mg/kg AG-14699 treatment group, and n = 4 for the other groups).
(d) PAR conjugated proteins, and AIMP2 in the substantia nigra of postmortem brain tissues from Parkinson’s disease (PD) patients and controls monitored by western blot. Extent of dopamine neurodegeneration was determined by anti-tyrosine hydroxylase immunoreactivity.
(e) Quantification of PAR conjugated protein levels in panel (d) normalized to β-actin.
Quantified data (a, c, e) are expressed as mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, ANOVA test followed by Student-Newman-Keuls post-hoc analysis (a), Kruskal-Wallis ANOVA test (c), unpaired two-tailed Student’s t-test (e). Full length blots are presented in Supplementary Figure 14 (b, d).
DISCUSSION
We report the discovery that transgenic expression of the parkin substrate, AIMP2, leads to age dependent selective degeneration of dopaminergic neurons in the substantia nigra. Mutations in Parkin are a common cause of autosomal recessive Parkinson’s disease 1,2. Postmortem studies from sporadic Parkinson’s disease reveal protein modifications of parkin that lead to its inactivation and accumulation of the K48 ubiquitinated proteins6–13,18,31,32 such as, PARIS, FBP-1 and AIMP2. Thus parkin inactivation could be a common pathogenic feature in both familial and sporadic Parkinson’s disease. Expression of either PARIS or AIMP2 leads to neuronal degeneration18,32. Here we show that expression of AIMP2 at levels similar to those in observed in Parkinson’s disease 7,9,18 leads to an age dependent selective neurodegeneration of dopaminergic neurons, which models the human disease. The progressive degeneration of dopaminergic neurons is a characteristic and unique pathological feature of Parkinson’s disease33. However, an extensive investigation on the molecular mechanisms underlying neurodegeneration in Parkinson’s disease has been hindered by the limited availability of animal models with age-dependent neurodegeneration34. These conditional AIMP2 transgenic mouse models with sustained AIMP2 expression at levels in dopaminergic neurons found in sporadic Parkinson’s disease reveal that the canonical nigrostriatal degeneration of human Parkinson’s disease can be successfully recapitulated in rodents. The characteristic age-dependent loss of SNpc dopaminergic neurons reaches up to 60% at 20 months of age. VTA dopaminergic neurons also degenerate, but to a lesser extent than the SNpc dopaminergic neurons mirroring human Parkinson’s disease 26 Moreover, striatal dopamine content and behavior of the transgenic mice corresponds with the deficits in nigral dopamine-producing neurons. The neuronal degeneration in AIMP2 transgenic mice is confined to dopamine neurons in the substantia nigra and VTA when compared to other brain regions such as cortex, which did not develop signs of age-dependent degeneration despite much higher levels of AIMP2. These results taken together suggest that overexpression of AIMP2 leads to selective degeneration of dopamine neurons.
Our cell and mouse models provided a unique platform to understand the cell death signaling events for AIMP2 mediated neurodegeneration. In the AIMP2 inducible PC12 cells, toxicity is independent of protein translation efficiency, implicating non-canonical functions of AIMP2 in the execution of the cell death. Pharmacological screening following transient expression of AIMP2 in SH-SY5Y cells identified that AIMP2 toxicity is caspase-independent and is mediated by PARP1. Activation of PARP1 typically involves DNA strand nicks and breaks21,35, but AIMP2 seems to activate PARP1 in the nucleus through a direct protein-protein association providing a novel mode of PARP1 activation. The dose-dependent protection of dopaminergic neurons against AIMP2 toxicity in mice treated with a PARP1 inhibitor and the complete protection of dopaminergic neurons and prevention of amphetamine-induced stereotypic rotations in PARP1−/− mice indicate that the suppression of PARP1’s enzymatic activity is sufficient to halt the dopaminergic neuronal loss due to AIMP2 overexpression. Given that the amphetamine-induced behavior is rescued in PARP1−/− mice even with AIMP2 induction, AIMP2 accumulation itself does not seem to impair nigrostriatal dopamine release, but rather affects the viability of dopaminergic neurons via PARP1 activation and cell death.
Notably we found that AAV1-tTA mediated overexpression of AIMP2 at levels equivalent to CamKII-tTA mediated overexpression in the ventral midbrain of the transgenic mice dramatically accelerated the pathogenesis of AIMP2-mediated neurodegeneration. The mechanisms underlying the acceleration of the pathogenesis with viral delivery of tTA are not clear. One possibility is the acuteness of the virally induced model, which bypasses compensatory mechanisms, which in many cases are strongly induced during embryonic development (see 4). Consistent with this notion, virally induced adult onset deletion of Parkin causes age-dependent degeneration of dopamine neurons32, which is absent in germ-line Parkin knockouts36. Moreover, viral induction of other Parkinson’s disease genes such as LRRK237 or α-synuclein38 leads to degeneration of dopamine neurons, whereas expression of LRRK2 or α-synuclein via conventional approaches does not cause degeneration of dopamine neurons (see39,40). Conditional overexpression of α-synuclein via a CamKII-tTA/TetP-α-synuclein expression cassette takes months to cause degeneration of dopamine neurons whereas viral expression of α-synuclein occurs under a much shorter time frame similar to our observations41. Since adeno-associated virus serotype 1 transduces both neurons and glia cells42, whereas CamKII-tTA/TetP-AIMP2 expression is restricted to neurons, it is possible that the glial expression of AIMP2 accelerates the phenotype.
AIMP2 upregulation is not likely to be the sole mechanism by which inactivation of parkin contributes to loss of dopamine neurons. How and whether AIMP2 contributes to other touted mechanisms of loss of dopamine neurons due to parkin inactivation is not known. These additional mechanisms will need to be evaluated in future studies to determine their relative contributions to dopamine neuron degeneration due to parkin inactivation. In particular, it will be important to determine whether AIMP2 and PARIS or other mechanisms of parkin-induced dopamine neurodegeneration intersect in a common pathway or whether they are separate pathways. Since these other mechanisms may involve perturbations in mitochondrial control either through PARIS impairment of mitochondrial biogenesis32 or mitochondrial degradation via mitophagy43, it is conceivable that AIMP2 acts downstream of these mitochondrial perturbations. Indeed under physiological conditions AIMP2 is trapped in the aminoacyl-tRNA synthetase (ARS) complex as a structural cofactor in the cytosol and functions to maintain the stability of this complex44,45. Thus, AIMP2 without any stress is only found in the cytosol as a complex with ARS. Dopamine neurons in Parkinson’s disease may be particularly vulnerable to oxidative stress46,47, which could account for the AIMP2 elevation and PARP1 activation observed in the substantia nigra of patients with Parkinson’s disease as well as the selective vulnerability of dopamine neurons. Consistent with this notion is the selective degeneration of dopamine neurons in the AIMP2 transgenic mice since AIMP2 only interacts with and activates PARP1 in dopamine neurons.
In postmortem brain tissue from control and Parkinson’s disease patients there are increased levels of PAR-conjugated proteins in total substantia nigra lysates implicating PARP1 activation in Parkinson’s disease pathogenesis. In previous reports, AIMP2 accumulation was noted in familial and sporadic forms of Parkinson’s disease in which mutations of Parkin or loss of function of this E3 ligase have been observed9,17,18. The presence of AIMP2 accumulation and elevated PARP1 activation in Parkinson’s disease patient samples supports the pathological relevance of AIMP2-PARP1 signaling in Parkinson’s disease. Consistent with the notion that PARP1 may play a role in Parkinson’s disease is the observation that knockout or inhibition of PARP1 is protective against the MPTP model of Parkinson’s disease, supporting a role for parthanatos in Parkinson’s disease 48,49. PARP1 is a drugable target and there are many inhibitors of PARP in clinical trial for oncologic indications33. Some of these drugs cross the blood brain barrier and thus may potentially be useful in treating Parkinson’s disease.
ONLINE METHODS
Plasmid constructions
The Tet off transgenic construct (C-terminal FLAG human AIMP2) was cloned into the XhoI site of pPrP.TetP vector 50 (9 kb). pCMV-Tag2A-AIMP2 18, EGFP-PARP1 (provided by Dr. Guy G Poirier), and MYC-AIMP2 18 and its deletion mutants have been previously described. Deletion mutants of EGFP-PARP1(aa 1-372, and aa 1-555) were cloned into the restriction sites (XhoI/XmaI) of EGFP-N1 vector. The GST-AIMP2-FLAG plasmid was generated by using the XhoI restriction site of pGEX-6p1 vector. 044 AM/CBA-pl-WPRE-bGH (5374bp) was used to clone the rAAV1-tTA-IRES2-zsGreen viral construct into BamHI/EcoRV sites. The integrity of the cloned constructs was verified by sequencing.
Antibodies
The following antibodies were used with dilution factors for each application according to the manufacturer’s instructions. Primary antibodies include: rabbit anti-AIMP2 (Cat#: 10424-1-1AP, ProteinTech Group, Inc), rabbit anti-eGFP (Cat#: ab290, Abcam), mouse anti-MYC (Cat#: 11 667 149 001, Roche Diagnostics), mouse anti-FLAG (M2, Sigma-Aldrich), mouse and rabbit anti-poly(ADP-ribose) antibodies (Cat#: 4335-MC-100 and 4336-BPC-100, Trevigen), anti-poly(ADP-ribose) polymerase (Cat#: 556494, mouse, BD bioscience; Cat#: 9542, rabbit, Cell signaling), rabbit anti-tyrosine hydroxylase (NB300-109, Novus Biologicals), mouse anti-GFAP (GA5, Cell Signaling Technology), rabbit anti-MnSOD (generated as described previously51), rabbit anti-MEF2C (Cat#: 5030, Cell signaling). Secondary antibodies include: horse radish peroxidase (HRP) conjugated mouse anti-β-actin (AC15, Sigma-Aldrich), HRP conjugated mouse anti-FLAG (Cat#: A8592, Sigma-Aldrich), HRP conjugated sheep anti-mouse IgG (GE Healthcare), HRP conjugated donkey anti-rabbit IgG (GE healthcare), Biotin conjugated goat anti-mouse IgG (Vector laboratories), Biotin conjugated goat anti-rabbit IgG (Vector laboratories), Alexa Fluor 488 conjugated donkey anti-mouse or rabbit IgG (H+L) (Invitrogen), Alexa Fluor 568 conjugated donkey anti-mouse or rabbit IgG (Invitrogen)
Conditional AIMP2 transgenic mouse generation
The linearized transgenic constructs (NotI digestion, 7 kbp) were microinjected into the embryos of the B6C3F2 strain and the one- or two-cell embryos were transferred into B6D2F1 pseudopregnant female mice (Transgenic Animal Core of National Cancer Institute, NIH). Using genomic DNA prepared from tail snip (Proteinase K, Roche diagnostics; direct PCR (tail) Lysis, Viagen), pups were genotyped by PCR (GoTaq Green Master Mix, Promega) using TetP-AIMP2 primers (F: CGG GTC GAG TAG GCG TGT AC; R: TCT AGA TGA TCC CCG GGT ACC GAG, PCR product = 173 bp) to select positive founders. Positive founders were further subjected to semi-quantitative PCR and normalized by GAPDH PCR (F: AAA CCC ATC ACC ATC TTC CAG; R: AGG GGC CAT CCA CAG TCT TCT, PCR product = 300 bp) to screen for high copy number founders. The top three high copy founders were mated with C57/BL6 for two~three generations to establish the transgenic lines. The following primer sets were used for genotyping of CamKIIα-tTA (F: TGA AAG TGG GTC CGC GTA C; R: TAC TCG TCA ATT CCA AGG GC, PCR product = 391 bp) or PARP1 (primer 1: AGG TGA GAT GAC AGG AGA TC; primer 2: CCA GCG CAG CTC AGA GAA GCC A; primer 3: CAT GTT CGA TGG GAA AGT CCC, PCR products: Null=350bp, WT=112 bp). AIMP2 induction in conditional Tg mice was suppressed by feeding the mice with doxycycline (Dox) containing food (Dox Diet-Sterile, 200 mg per kg doxycycline, Bio-Serv). All procedures involving animals were approved by and conformed to the guidelines of the Institutional Animal Care Committee of The Johns Hopkins University. Mice were kept in a 12 hour dark, 12 hour light cycle.
Immunohistochemistry (tyrosine hydroxylase, AIMP2, and GFAP) and stereological assessment of the number of tyrosine hydroxylase and Nissl positive cells
Mice were intracardially perfused with ice-cold PBS and 4% paraformaldehyde/PBS (pH 7.4) after deep anesthesia by intraperitoneal (i.p.) injection of Nembutal sodium solution (50 μl of two fold dilution in PBS of pentobarbital sodium 50 mg/ml, Lundbeck Inc.). Brains were removed and postfixed 16 hrs in the same fixative. After cryoprotection in 30% sucrose/PBS (pH 7.4), brains were frozen on dry ice, and serial coronal sections (40 μm sections) were cut with a microtome and the striatum and ventral midbrain regions were saved. Every four sections were collected for subsequent procedures. Free-floating sections in one group were blocked with 4% goat serum (Sigma-Aldrich)/PBS plus 0.2% Triton X-100 and incubated with antibodies against tyrosine hydroxylase or AIMP2 or GFAP followed by incubation with biotin conjugated anti-rabbit (tyrosine hydroxylase and AIMP2 IHC) or anti-mouse (GFAP IHC) antibodies, ABC reagents (Vector Laboratories), and Sigmafast 3,3′-diaminobenzidine (DAB) tablets (Sigma-Aldrich). Sections were counterstained with Nissl (0.09% thionin) after tyrosine hydroxylase staining as described previously 32. Sections were dehydrated in 100% ethanol and cleared in Xylene (Fisher Scientific) followed by mounting with DPX (Sigma-Aldrich) before imaging under a microscope. Cell counting was performed with the aid of the optical fractionator software, which is an unbiased method for the counting of tyrosine hydroxylase positive or Nissl positive cells in the substantia nigra region of the left hemisphere of AIMP2 transgenic mice (Experimenters were blinded for genotypes of mice during stereological counting) or in the injected and non-injected sides of animals used in the virus-injection procedure. This unbiased stereological counting was done with a computer-assisted image analysis system, consisting of an Axiophot photomicroscope (Carl Zeiss Vision) equipped with a computer-controlled motorized stage (Ludl Electronics), a Hitachi HV C20 video camera, and the Stereo Investigator software (MicroBrightField). The total number of tyrosine hydroxylase- and Nissl-stained neurons was calculated as previously described52.
Immuofluorescence
4% paraformaldehyde/PBS (pH 7.4)-fixed coronal brain sections or SH-SY5Y cells were blocked with 4% donkey serum (Sigma-Aldrich)/PBS plus 0.2% Triton X-100 and incubated with antibodies against tyrosine hydroxylase or AIMP2 or PARP1 or MEF2C. After brief washes with PBS containing 0.2% Triton X-100, floating brain sections or cells were incubated with corresponding secondary antibodies conjugated with fluorescent dyes (Alexa Fluor 488 conjugated donkey anti-mouse IgG for tyrosine hydroxylase and PARP1, Alexa Fluor 568 conjugated donkey anti-rabbit IgG for AIMP2). For immunostaining of endogenous and transgenic AIMP2 in mouse brain sections, TSA Fluorescence Systems Tyramide signal amplification (Streptavidin-HRP, Blocking reagent, amplification diluent, and Cyanine 3 tyramide, PerkinElmer Life Sciences, Inc.) was used to amplify the signal following the manufacturer’s instructions. Images were obtained using fluorescent microscope (Zeiss Axiovert 200M) or confocal microscope (Zeiss Confocal LASM 710).
Co-immunoprecipitation (Co-IP) and immunoblot analysis
Human brain tissue or mouse brains or cells were homogenized and prepared in lysis buffer (for tissue, 10 mM Tris-HCl, pH 7.3, 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 1% TritonX-100, 0.5% Na-deoxycholic acid; for cells, PBS, 1% Nonidet P-40 supplemented with Phosphatase Inhibitor Cocktail I and II (P2850 (I), P5726 (II), Sigma-Aldrich), and Complete Protease Inhibitor Mixture). Protein levels were quantified using a BCA protein assay kit (Pierce). Co-immunoprecipitation was performed with antibodies and Protein G Sepharose and Glutathione Sepharose 4B (GE Healthcare Sciences). The immunocomplexes were then washed with immunoprecipitation buffer for four times and samples were prepared by adding 2 × sample loading buffer (Bio-Rad). Brain tissue lysates (40 μg) or IP’ed samples were electrophoresed on SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were blocked with 5% skim milk in TBS-T and incubated with primary antibodies. After HRP-conjugated secondary antibodies incubation, the immunoblot signal was detected using chemiluminescent substrates (Thermo scientific).
Subcellular fractionation
Ventral midbrain tissues from mice or ice cold PBS-washed SH-SY5Y cells were homogenized in a hypotonic buffer (10 mM HEPES, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and 0.1% NP-40) supplemented with a complete protease inhibitor cocktail and passed through 26G syringe needle for 3 times. After incubation on ice for 20 minutes with 10 second vortexing two times, the lysates were centrifuged at 720 g for 5 minutes. The supernatant was used as a postnuclear fraction and the pellet was washed with the same buffer twice and used as a nuclear fraction for subsequent western blot analysis (Loading for western blot: 2% of PN fraction and 8% of Nu fraction).
Measurement of neurotransmitters in the striatum
Biogenic amine concentrations were measured by High-performance liquid chromatography (HPLC) with electrochemical detection (HPLC-ECD). Briefly, mice were sacrificed by decapitation and the striatum was quickly removed. Striatal tissue was weighed and sonicated in 0.2 ml ice cold 0.01 mM perchloric acid containing 0.01% EDTA and 60 nM 3,4-dihydroxybenzylamine (DHBA) as an internal standard. After centrifugation (15,000 × g, 30 min, 4°C), the supernatant was passed through a 0.2 μm filter. Twenty microliters of the supernatant were analyzed in the HPLC column (4.6 mm × 150mm C-18 reverse phase column, MC Medical, Tokyo, Japan) with detection by a dual channel Coulchem III electrochemical detector (Model 5300, ESA, Inc Chelmsford, MA, USA). The protein concentrations of tissue homogenates were measured using the BCA protein assay kit (Pierce, Rockford, IL, USA). Data were normalized to protein concentrations (ng neurotransmitters per mg tissue).
Stereotaxic intranigral virus injection
For stereotaxic injection of AAV1-IRES-zsGreen overexpressing tTA and zsGreen, eight-week-old mice of indicated genotypes were anesthetized with pentobarbital (60 mg per kg). An injection cannula (26.5 gauge) was applied stereotaxically into the SNpc (anteroposterior, 3.2 mm from bregma; mediolateral, 1.3 mm; dorsoventral, 4.3 mm) or the cortex (anteroposterior, 3.2 mm; mediolateral, 2 mm; dorsoventral, 0.5 mm) unilaterally (applied into the right hemisphere). The infusion was performed at a rate of 0.2 μl per min, and 2 μl or 1 μl of a high titer AAV1-tTA-IRES-zsGreen (3.5 × 10^13 VG per ml in PBS) was injected into each mouse. After the final injection, the injection cannula was maintained in the substantia nigra for an additional 5 minutes for a complete absorption of the virus and then slowly removed from the mouse brain. The head skin was closed by suturing and wound healing and recovery were monitored following surgery. For western blot analysis, brains were removed 17 days (nigral injection) or 25 days (cortical injection) after viral injection respectively and protein samples were prepared as noted above. For stereological analysis, animals were perfused and fixed intracardially with ice-cold PBS followed by 4% paraformaldehyde 25 days after intranigral viral injection. The brain was removed and processed for immunohistochemistry or immunofluorescence. Amphetamine-induced stereotypic rotation was performed 3 weeks after the unilateral intranigral virus injection. PARP1 inhibition was accomplished by AG014699 (PF-01367338, Selleck Chemicals) treatment (no randomization was used in the drug treatment protocol) that was initiated 3 days before intranigral AAV-tTA injection and continued until perfusion.
Behavioral tests
All behavior tests were performed in the Behavioral Core Facility at the Johns Hopkins University School of Medicine. Since behavioral data for open field and rotarod tests were obtained automatically using instruments detailed below, no particular blinding strategy was employed.
Open field test
General locomotor and exploratory activities were assessed in open field square-shaped chambers equipped with an automated beam tracking system. Unless otherwise noted, the tests were performed in the afternoon. Briefly, a mouse was placed in the center of the open field arena and allowed to explore the area for a total of 30 minutes. The activities of a mouse were recorded over 30 minutes, which consists of six 5 minute sessions, by photobeam activity system (PAS) software installed in a computer connected to the open field equipment. Before and after each testing, the surface of the arena was cleaned with 70% ethanol. The total number of beam breaks during the 30 minute period was used to determine gross locomotor activity of a mouse. These mice were also used for rotarod testing.
Rotarod test
Motor coordination of mice was measured as the retention time on an accelerating rotarod of the rotamex V instrument equipped with photobeams and a sensor to automatically detect mice that fall from the rotarod. Prior to the actual rotarod test, animals were trained on the rotarod at 4.0 rpm for 5 min and allowed to rest for at least 30 minutes. After training and resting, four mice were placed on separate rods and the durations on the accelerating rods were recorded automatically by the software installed on a computer connected to the instrument. The setting of the rotamex was as follows and remained constant throughout all trials. Start speed, 4.0 rpm; maximum speed, 40 rpm; Acceleration interval, 30 sec; acceleration step, 4 rpm. The tests were repeated three times and the average retention time and end speed were recorded for each mouse. The retention time was used to determine the motor coordination of the animal.
Amphetamine induced stereotypic rotation
25 days after mice received the AAV1-tTA (2 μl) intranigral injection into the right hemisphere, 5 mg per kg amphetamine (Sigma-Aldrich) was intraperitoneally administered into mice. Mice were placed into a white paper cylinder of 20 cm diameter and monitored for 30 minutes. The behavior of mice was filmed at three one-minute intervals between 20 and 30 minutes following amphetamine administration. Full body ipsilateral rotations (clockwise) during one minute session were counted for each mouse from the video recordings.
In vitro binding assay of recombinant AIMP2
The GST fusion protein, GST-AIMP2, was prepared following a standard protocol and GST was cleaved by prescission protease (GE Healthcare). For in vitro binding assays, recombinant AIMP2 was incubated with immunoprecipitated human PARP1 from SH-SY5Y using a PARP1 specific antibody and protein G beads. After washing three times with a binding buffer (1 % triton-X in PBS), the bound proteins were separated by SDS PAGE and immunoblotted with the indicated antibodies. Anti-FLAG antibody pull down was performed by incubating recombinant AIMP2-FLAG and recombinant PARP1 with protein G sepharose beads overnight at 4°C using a similar procedure as described for the anti-PARP1 pull down.
Cell culture and transfection
SH-SY5Y cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10 % fetal bovine serum and 100 U per ml of penicillin/streptomycin antibiotics and were grown at 37 °C, 7 % CO2, and 90 % humidity. AIMP2 inducible tet-off PC12 cell lines were maintained in DMEM supplemented with 10 % horse serum, 5 % tet-approved fetal bovine serum (Clontech) and the following antibiotics: 100 U per ml penicillin/streptomycin (Invitrogen), 100 μg per ml G418 (Invitrogen), 200 μg per ml hygromycin (Invitrogen), and 200 ng per ml doxycycline (Invitrogen) for suppression of AIMP2 induction. For differentiation, PC12 cells were passed onto collagen-coated plates and provided with 100 ng/ml nerve growth factor (NGF, Roche) in low serum media (1% horse serum in DMEM). NGF was replenished every other day during differentiation. All transient transfections of SH-SY5Y cells were performed using Fugene transfection reagent (Roche).
Cell viability analysis
AIMP2 inducible PC12 cells or SH-SY5Y cells were plated in a 6-well plate (collagen coating for PC12 cells) at a seeding density of 0.5 × 106 cells per well. 7 days following differentiation and AIMP2 induction (AIMP2 inducible PC12 cells) or 2 days after transient transfection (SH-SY5Y cells) with the treatment of indicated chemicals (Z-VAD-FMK (N-CBZ-Val-Aal-Asp(O-Me) fluoromethyl ketone, Sigma-Aldrich; DPQ, sc-202755, Santa Cruz Biotechnology), trypan blue exclusion was performed to assess cell toxicity by counting the number of dead (blue) and live cells. After the indicated days of differentiation and induction, cultures were briefly washed with PBS, followed by trypsinization. Resuspended cells were mixed with equal volume of 0.4 % trypan blue. Live and dead cells were counted under light microscope using a hemacytometer.
35S – Pulse methionine incorporation assay
Differentiated AIMP2 inducible PC12 cells grown in regular media were pulsed incubated in media containing 35S-labelled methionine together with appropriate chemicals such as 1-methyl-4-phenylpyridinium (MPP+), H2O2, and thapsigargin (all purchased from Sigma-Aldrich). After 10 minutes incubation, total lysates were prepared and separated in SDS-PAGE followed by transferring to nitrocellulose membrane and taking signals in X-ray film to visualize newly synthesized proteins with 35S-methionine incorporation.
In vitro PARP1 activity assay
In vitro PARP1 activity assay was performed following the manufacturer’s instructions (Trevigen, Universal PARP Colorimetric Assay Kit). As a modification, activated DNA was left out of the reactions to prevent PARP1 activation saturation. Briefly, GST and GST-tagged recombinant AIMP2 were added to different units of recombinant PARP1 in histone coated wells. After reaction for 60 minutes at room temperature, the levels of PARsylation of the PARP1 substrate histone were measured using colorimetric assay (absorbance at 450 nm) and normalized as fold change compared to the level at 0.125 units of PARP1 with GST.
Statistics
Power analysis was performed by using G*Power 3.1 software to determine approximate sample sizes for behavior tests or TH stereological analysis of CamKII-tTA/TetP-AIMP2 mice and AAV-tTA virus injection mice. Based upon mean difference from our preliminary experiments with AIMP2 transgenic mice, 6 total sample size was calculated for behavior, tyrosine hydroxylase stereology in CamKII-tTA;TetP-AIMP2 mice (Effect size f = 3.33 for 20% mean difference, 0.2 standard deviation value within each group; α = 0.05). 4 total samples size was calculated for tyrosine hydroxylase stereology with virally induced AIMP2 transgenic mice (Effect size f = 22.5 for 90 % mean difference, 0.2 standard deviation within each group; α = 0.05). Normality of the data was tested with the Shapiro-Wilk test. The equality of variance was determined with Levene statistics. All quantitative data are expressed as the mean ± s.e.m. Statistical significance was determined either by an unpaired two-tailed Student’s t test and nonparametric Mann-Whitney U test for comparison of two groups (control and test) or a one-way ANOVA test and Student-Newman-Keuls post-hoc analysis and nonparametric Kruskal-Wallis ANOVA test for comparison among multiple groups of more than three as indicated in each figure legend. P values lower than 0.05 were considered to indicate significant difference among groups.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH NS38377 and the JPB Foundation. YL was supported by the Samsung Scholarship Foundation. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases. The authors acknowledge the joint participation by the Adrienne Helis Malvin Medical Research Foundation and the Diana Helis Henry Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with The Johns Hopkins Hospital and the Johns Hopkins University School of Medicine and the Foundation’s Parkinson’s Disease Programs. DS and LT were supported by the Intramural Research Program of the NCI, Center for Cancer Research, NIH.
Footnotes
Note: Supplementary information is available on the Nature Neuroscience website.
AUTHOR CONTRIBUTIONS
Y.L. designed and performed the in vitro and in vivo experiments. S.S.K. performed the HPLC analysis and behavior tests. J.-H.S. and Y.L. performed stereotaxic intranigral virus injection and stereological counting of TH neurons. Y.-I.L. performed subcellular fractionation and confocal microscopy. H.S.K., B.D.L., H.C.K., S.-U.K and D.K. provided materials and helped with the analysis of the results. H.J. performed immunofluorescence and cell counting for the intracortical virus injection samples. D.S. and L.T. performed pronuclear injections of the TetP-AIMP2 construct and provided founder mice. V.L.D. and T.M.D. formulated the hypothesis, initiated and organized the study and wrote the manuscript. Y.L., V.L.D. and T.M.D. contributed to the final manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Abbas N, et al. A wide variety of mutations in the parkin gene are responsible for autosomal recessive parkinsonism in Europe. French Parkinson’s Disease Genetics Study Group and the European Consortium on Genetic Susceptibility in Parkinson’s Disease. Hum Mol Genet. 1999;8:567–574. doi: 10.1093/hmg/8.4.567. [DOI] [PubMed] [Google Scholar]
- 2.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 3.Corti O, Lesage S, Brice A. What genetics tells us about the causes and mechanisms of Parkinson’s disease. Physiol Rev. 2011;91:1161–1218. doi: 10.1152/physrev.00022.2010. [DOI] [PubMed] [Google Scholar]
- 4.Dawson TM. Parkin and defective ubiquitination in Parkinson’s disease. Journal of neural transmission. Supplementum. 2006:209–213. doi: 10.1007/978-3-211-45295-0_32. [DOI] [PubMed] [Google Scholar]
- 5.Moore DJ. Parkin: a multifaceted ubiquitin ligase. Biochem Soc Trans. 2006;34:749–753. doi: 10.1042/BST0340749. [DOI] [PubMed] [Google Scholar]
- 6.Chung KK, et al. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 7.Imam SZ, et al. Novel regulation of parkin function through c-Abl-mediated tyrosine phosphorylation: implications for Parkinson’s disease. J Neurosci. 2011;31:157–163. doi: 10.1523/JNEUROSCI.1833-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Ko HS, et al. Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin’s ubiquitination and protective function. Proc Natl Acad Sci U S A. 2010;107:16691–16696. doi: 10.1073/pnas.1006083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ. Dopamine covalently modifies and functionally inactivates parkin. Nat Med. 2005;11:1214– 1221. doi: 10.1038/nm1314. [DOI] [PubMed] [Google Scholar]
- 11.Meng F, et al. Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation. Mol Neurodegener. 2011;6:34. doi: 10.1186/1750-1326-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C, et al. Stress-induced alterations in parkin solubility promote parkin aggregation and compromise parkin’s protective function. Hum Mol Genet. 2005;14:3885–3897. doi: 10.1093/hmg/ddi413. [DOI] [PubMed] [Google Scholar]
- 13.Yao D, et al. Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci U S A. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 15.Olzmann JA, Chin LS. Parkin-mediated K63-linked polyubiquitination: a signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy. 2008;4:85–87. doi: 10.4161/auto.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cookson MR. Parkin’s substrates and the pathways leading to neuronal damage. Neuromolecular medicine. 2003;3:1–13. doi: 10.1385/NMM:3:1:1. [DOI] [PubMed] [Google Scholar]
- 17.Corti O, et al. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum Mol Genet. 2003;12:1427–1437. doi: 10.1093/hmg/ddg159. [DOI] [PubMed] [Google Scholar]
- 18.Ko HS, et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci. 2005;25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kistner A, et al. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci U S A. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sprengel R, Hasan MT. Tetracycline-controlled genetic switches. Handb Exp Pharmacol. 2007:49–72. doi: 10.1007/978-3-540-35109-2_3. [DOI] [PubMed] [Google Scholar]
- 21.de Murcia G, et al. Structure and function of poly(ADP-ribose) polymerase. Mol Cell Biochem. 1994;138:15–24. doi: 10.1007/BF00928438. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrabi SA, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci Signal. 2011;4:ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayford M, et al. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- 26.German DC, Manaye K, Smith WK, Woodward DJ, Saper CB. Midbrain dopaminergic cell loss in Parkinson’s disease: computer visualization. Annals of neurology. 1989;26:507–514. doi: 10.1002/ana.410260403. [DOI] [PubMed] [Google Scholar]
- 27.Park SG, Choi EC, Kim S. Aminoacyl-tRNA synthetase-interacting multifunctional proteins (AIMPs): a triad for cellular homeostasis. IUBMB Life. 2010;62:296–302. doi: 10.1002/iub.324. [DOI] [PubMed] [Google Scholar]
- 28.MacCallum PR, et al. Cap-dependent and hepatitis C virus internal ribosome entry site- mediated translation are modulated by phosphorylation of eIF2alpha under oxidative stress. J Gen Virol. 2006;87:3251–3262. doi: 10.1099/vir.0.82051-0. [DOI] [PubMed] [Google Scholar]
- 29.Plummer R, et al. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin Cancer Res. 2008;14:7917–7923. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudow G, et al. Morphometry of the human substantia nigra in ageing and Parkinson’s disease. Acta Neuropathol. 2008;115:461–470. doi: 10.1007/s00401-008-0352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ko HS, Kim SW, Sriram SR, Dawson VL, Dawson TM. Identification of far upstream element-binding protein-1 as an authentic Parkin substrate. J Biol Chem. 2006;281:16193–16196. doi: 10.1074/jbc.C600041200. [DOI] [PubMed] [Google Scholar]
- 32.Shin JH, et al. PARIS (ZNF746) Repression of PGC-1alpha Contributes to Neurodegeneration in Parkinson’s Disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obeso JA, et al. Missing pieces in the Parkinson’s disease puzzle. Nat Med. 2010;16:653–661. doi: 10.1038/nm.2165. [DOI] [PubMed] [Google Scholar]
- 34.Dawson TM, Ko HS, Dawson VL. Genetic animal models of Parkinson’s disease. Neuron. 2010;66:646–661. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Von Coelln R, et al. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci U S A. 2004;101:10744–10749. doi: 10.1073/pnas.0401297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee BD, et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson’s disease. Nat Med. 2010;16:998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirik D, et al. Nigrostriatal alpha-synucleinopathy induced by viral vector-mediated overexpression of human alpha-synuclein: a new primate model of Parkinson’s disease. Proc Natl Acad Sci U S A. 2003;100:2884–2889. doi: 10.1073/pnas.0536383100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chesselet MF, Richter F. Modelling of Parkinson’s disease in mice. Lancet neurology. 2011;10:1108–1118. doi: 10.1016/S1474-4422(11)70227-7. [DOI] [PubMed] [Google Scholar]
- 40.Lee Y, Dawson VL, Dawson TM. Animal models of Parkinson’s disease: vertebrate genetics. Cold Spring Harbor perspectives in medicine. 2012;2 doi: 10.1101/cshperspect.a009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin X, et al. Conditional expression of Parkinson’s disease-related mutant alpha-synuclein in the midbrain dopaminergic neurons causes progressive neurodegeneration and degradation of transcription factor nuclear receptor related 1. J Neurosci. 2012;32:9248–9264. doi: 10.1523/JNEUROSCI.1731-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Royo NC, et al. Specific AAV serotypes stably transduce primary hippocampal and cortical cultures with high efficiency and low toxicity. Brain research. 2008;1190:15–22. doi: 10.1016/j.brainres.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corti O, Brice A. Mitochondrial quality control turns out to be the principal suspect in parkin and PINK1-related autosomal recessive Parkinson’s disease. Curr Opin Neurobiol. 2013;23:100–108. doi: 10.1016/j.conb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Han JM, et al. AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc Natl Acad Sci U S A. 2008;105:11206–11211. doi: 10.1073/pnas.0800297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JY, et al. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. Proc Natl Acad Sci U S A. 2002;99:7912–7916. doi: 10.1073/pnas.122110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan CS, et al. ‘Rejuvenation’ protects neurons in mouse models of Parkinson’s disease. Nature. 2007;447:1081–1086. doi: 10.1038/nature05865. [DOI] [PubMed] [Google Scholar]
- 47.Guzman JN, et al. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468:696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandir AS, et al. Poly(ADP-ribose) polymerase activation mediates 1-methyl-4-phenyl-1, 2,3,6-tetrahydropyridine (MPTP)-induced parkinsonism. Proc Natl Acad Sci U S A. 1999;96:5774–5779. doi: 10.1073/pnas.96.10.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Shimoji M, Yu SW, Dawson TM, Dawson VL. Apoptosis inducing factor and PARP-mediated injury in the MPTP mouse model of Parkinson’s disease. Ann N Y Acad Sci. 2003;991:132–139. doi: 10.1111/j.1749-6632.2003.tb07471.x. [DOI] [PubMed] [Google Scholar]
- 50.Lin X, et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson’s-disease-related mutant alpha-synuclein. Neuron. 2009;64:807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu SW, et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 52.West MJ. New stereological methods for counting neurons. Neurobiology of aging. 1993;14:275–285. doi: 10.1016/0197-4580(93)90112-o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.