Abstract
Reward-related memories are essential for adaptive behavior and evolutionary fitness, but are also a core component of maladaptive brain diseases such as addiction. Reward learning requires dopamine neurons located in the ventral tegmental area (VTA), which encode relationships between predictive cues and future rewards. Recent evidence suggests that epigenetic mechanisms, including DNA methylation, are essential regulators of neuronal plasticity and experience-driven behavioral change. However, the role of epigenetic mechanisms in reward learning is poorly understood. Here, we reveal that the formation of reward-related associative memories in rats upregulates key plasticity genes in the VTA, which are correlated with memory strength and associated with gene-specific changes in DNA methylation. Moreover, DNA methylation in the VTA is required for the formation of stimulus-reward associations. These results provide the first evidence that that activity-dependent methylation and demethylation of DNA is an essential substrate for the behavioral and neuronal plasticity driven by reward-related experiences.
Keywords: behavioral epigenetics, reward, associative learning, DNA methylation, ventral tegmental area, dopamine, decision making
The ability to form memories about events that predict desired outcomes is a fundamental aspect of adaptive behavior. Associative reward learning in animals, which is a useful model for key aspects of human decision making, impulsivity, and addiction1–4, is mediated by a network of brain nuclei that includes dopamine neurons located in the ventral tegmental area (VTA). These neurons release dopamine in striatal and cortical projection targets that regulate motivated behavior and decision making, and receive inputs from a number of brain regions that process information related to environmental cues and rewarding events5. Dopamine neurons undergo plastic changes during the formation of reward-related memories6, such that cues signaling future rewards acquire the ability to increase dopamine neuron firing and dopamine release in terminal regions6–9. As a result, these cues are attributed with incentive salience and able are to evoke conditioned approach responses, which are blocked by dopamine antagonists in target areas such as the nucleus accumbens (NAc) 8, 10.
While these changes are necessary and sufficient for reward-directed behaviors10–12, the molecular mechanisms that underlie this neuroplasticity remain elusive. Emerging evidence suggests that epigenetic mechanisms, including DNA methylation, are essential regulators of synaptic plasticity and experience-dependent behavioral change13–17, providing a candidate mechanism by which the environment and genetic landscape interact to support memory formation and maintenance18. Although DNA methylation was once viewed as an inherently stable mark incapable of rapid change, recent evidence conclusively demonstrates that this is not the case14, 15, 19, 20. In the brain, DNA undergoes rapid methylation and demethylation via distinct mechanisms, and is controlled in an activity-dependent fashion by the same receptor and intracellular signaling cascades that regulate memory formation21–23. Indeed, recent studies have revealed a central role for DNA methylation in various types of aversive learning, adaptive behaviors, and synaptic plasticity 13, 14, 16, 17. However, despite the importance of reward-related learning to critical issues such as decision making and addiction, nothing is known about the role of DNA methylation in this process. To investigate the molecular and epigenetic changes in the VTA that mediate reward learning, we employed a Pavlovian reward conditioning paradigm in which animals form an association between reward-paired cues and future rewards. Our results indicate that reward learning produced changes in DNA methylation at genes that are upregulated in dopamine neurons following learning, and that blocking DNA methylation in the VTA prevents memory formation.
RESULTS
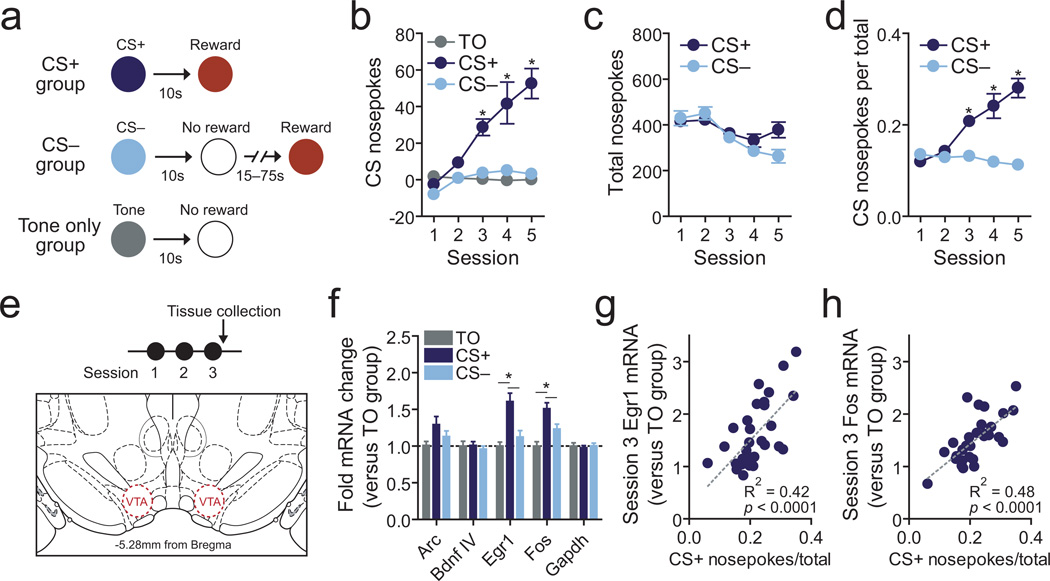
To enable dissociation of learning-related changes from those arising due to reward or environmental experiences alone, we trained rats in three distinct behavioral designs (Fig. 1a). “CS+” animals received exposure to an audio cue (the CS+) that was fully predictive of sucrose delivery to a centrally located food port in a standard conditioning chamber. The “CS–” group received identical exposure to the same audio cue and sucrose rewards, but these stimuli were explicitly unpaired. Finally, a tone-only (TO) group received exposure to the conditioning chamber and audio cue, but did not receive sucrose rewards. All behavioral sessions consisted of 25 audio stimulus presentations (all groups) and 25 reward presentations (CS+ and CS– groups only). As shown in Figure 1b, rats that received repeated pairings between the CS+ and reward exhibited increased reward-port nosepoke responses during the 10s CS presentation period as compared to CS– and TO animals, indicating selective experience-dependent associative memory formation (also see Supplementary Fig. 1a–b for representative examples). Importantly, there was no difference in the total number of nosepoke responses between CS+ and CS– animals in any behavioral session, suggesting that the emergence of cue-evoked conditioned responding could not be explained by a general difference in motor activity or motivational arousal (Fig. 1b–d). Additionally, in agreement with previous studies6, our results suggest that animals acquire the CS+/reward association gradually across training, emerging initially during the third training session. Critically, previous studies suggest that reward-paired cues first begin to increase dopamine release at this time point6, 8, which relies upon transient NMDA-receptor dependent synaptic plasticity (increased AMPA/NMDA ratio)6. Therefore, to determine what molecular and epigenetic changes may contribute to the neuronal plasticity underlying this form of reward learning, we focused our molecular experiments on this behavioral time point (Fig. 1e). Given that a wealth of research indicates that ongoing gene expression is critical not only for behavioral memory but also the maintenance of synaptic plasticity, we began by investigating changes in gene expression with RT-qPCR. We limited our focus to genes previously shown to regulate learning, memory, and synaptic plasticity in other brain regions, including Arc, Bdnf exon IV, Egr1, and Fos22, 24–26. As shown in Figure 1f, reward-related memory formation selectively increased expression of the immediate early genes Egr1 and Fos (genes shown to be essential for memory formation and long-term potentiation in other brain regions24, 26) as compared to TO and CS– controls.
Figure 1.
Reward learning causes experience dependent changes in VTA IEG expression that are correlated with memory strength. a, Pavlovian training scheme. For the CS+ group, audio cues predicted rewards whereas cues for the CS– and TO group were not associated with reward delivery. However, CS– animals received unpaired rewards. b, Animals formed associations between the CS+ and reward delivery, as determined by cue-specific nosepoke responses (37, 42, and 36 animals per group for sessions 1𠄳, 12–13 animals per group for sessions 4𠄵 two-way ANOVA: interaction between training and behavioral session, F(8,404) = 12.96, P < 0.0001; Bonferroni post-hoc tests indicated significantly elevated CS+ nosepokes at sessions 3, 4, and 5 (*P < 0.001)). c, Experience with a reward-predictive CS+ does not alter total nosepokes responses (two-way ANOVA: main effect of training group, F(1,273) = 2.088, P = 0.15; Bonferroni post hoc tests P > 0.05 for all within-session comparisons). d, Even when correcting for total nosepokes, CS+ animals made significantly more nosepoke responses during the cue presentation than CS– animals (two-way ANOVA: interaction between training and behavioral session, F(1,273) = 31.75, P < 0.0001; Bonferroni post-hoc test indicates significantly elevated CS+ nosepokes at sessions 3, 4, and 5 (*P < 0.001)). e, For biochemical experiments, VTA tissue was harvested after 3 training sessions, when memory formation occurs. f, CS+ training, but not CS– training, increases expression of Egr1 and Fos as compared to TO controls (n = 23, 30, and 24 per group; one-way ANOVA: main effect of training for Egr1 mRNA (F(2,76) = 12.32, P < 0.0001) and Fos mRNA (F(2,76) = 13.27, P < 0.0001); Tukey post-hoc tests indicate significant difference between TO and CS– groups as compared to CS+ group (*P < 0.05). g-h, Induction of Egr1 (g) and Fos (h) in CS+ animals was significantly correlated with strength of memory formation. Error bars represent s.e.m.
To further implicate experience-dependent changes in these genes within the VTA in the acquisition of stimulus-reward associations, we took advantage of the fact that CS+ animals exhibited considerable variability in memory formation in session 3 (Supplementary Fig. 1c). Although some animals exhibited robust learned nosepoke behavior, other animals did not differ substantially from CS– animals in terms of cued nosepoke responses, suggesting poor memory formation. Notably, a regression analysis revealed that the degree of learned approach responding in CS+ animals was highly correlated with the induction of Egr1 and Fos expression (Fig. 1g, h), suggesting that the level of gene expression might influence the strength of learning. Further, neither Egr1 or Fos mRNA were altered after exposure to reward-paired cues when the stimulus-reward association had stabilized (following behavioral session 5), indicating specificity for the acquisition phase of reward learning (Supplementary Fig. 1d).
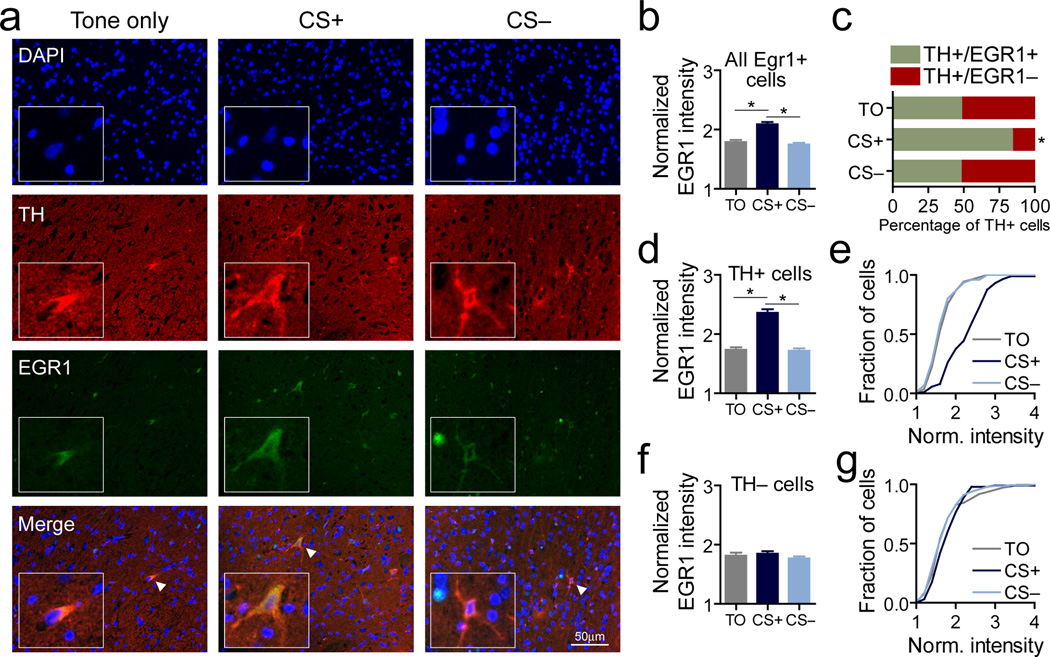
Although these results suggest that transcriptional changes occurring during reward learning are related to memory formation, the VTA is a heterogeneous structure with numerous cell types5, 27, 28 that have distinct functional roles29. To examine if these changes are limited to dopamine neurons, paraffin-embedded brain sections from trained animals (sacrificed after session 3) were subjected to immunohistochemical labeling of EGR1 protein and tyrosine hydroxylase (TH, a common marker for dopamine neurons; see Figure 2a for representative examples), which allowed us to examine colocalization of EGR1 changes within dopamine neurons. As with gene transcript levels, we found that reward learning increased total EGR1 fluorescence in all EGR1+ cells (Fig. 2b). Next, we sorted TH+ cells based on the presence of EGR1, which revealed that the proportion of TH cells that contained EGR1 was selectively increased in the CS+ group (Fig. 2c). Likewise, even when cells that did not contain EGR1 were excluded from analysis, we found that average EGR1 fluorescence was elevated in cells that also contained TH in the CS+ group (Fig. 2c–d), but was not altered in cells that lacked TH immunofluorescence (Fig. 2e–f). Thus, these results suggest that EGR1 protein is selectively induced in dopamine neurons in response to reward learning.
Figure 2.
Reward learning increases EGR1 specifically in dopamine neurons. a, Representative examples of DAPI, tyrosine hydroxylase, and EGR1 fluorescence following reward learning. Inset panels highlight TH+ neurons, marked by white triangles in merged panel. Scale, 50µm. b, Quantification of EGR1 fluorescence in all EGR1+ cells reveals increased EGR1 protein following reward learning (n = 5–6 animals per group; one-way ANOVA: main effect of training on EGR1 fluorescence (F(2,566) = 27.96, P < 0.0001); Tukey post-hoc tests indicate significant difference between TO and CS– groups as compared to CS+ group (*P < 0.001) c, Percentage of TH+ cells containing EGR1 fluorescence increased following reward learning (TO group, 49.6%, CS+ group, 85.4%; CS– group, 49.3%; Chi-square test for frequency, X2 = 8.779, *P = 0.0124). d, Quantification of EGR1+, TH+ cells revealed increased EGR1 in CS+ group (n = 53–88 cells per group; one-way ANOVA: main effect of training on EGR1 fluorescence (F(2,215) = 49.27, P < 0.0001); Tukey post-hoc tests indicate significant difference between TO and CS– groups as compared to CS+ group (*P < 0.001). e, Cumulative distributions of cell-by-cell EGR1 immunofluorescence for TH+, EGR1+ cells. f, Quantification of EGR1+, TH- cells revealed no changes in EGR1 in non-dopaminergic cells (n = 98–144 cells per group; one-way ANOVA: main effect of training on EGR1 fluorescence (F(2,350) = 0.94, P = 0.39). g, Cumulative distributions of cell-by-cell EGR1 immunofluorescence for TH-, EGR1+ cells. Error bars represent s.e.m.
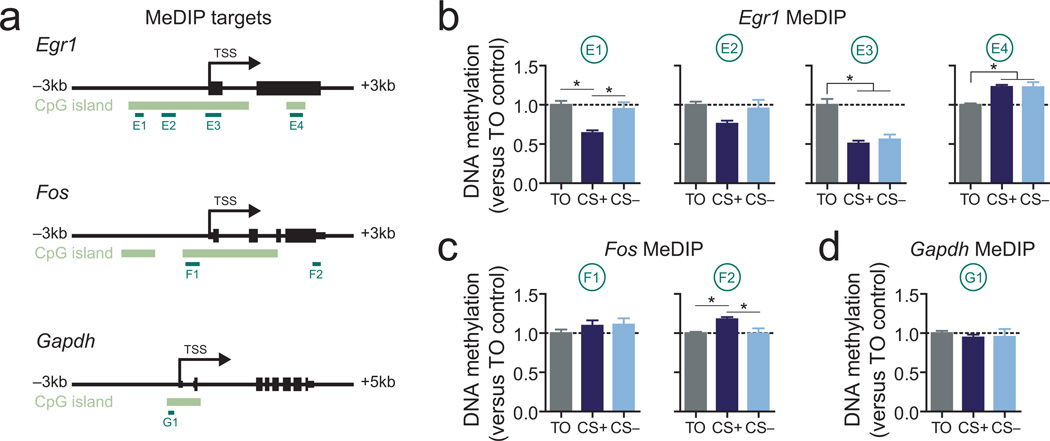
Activity- and experience-dependent changes in DNA methylation have recently been discovered to be important processes for neuronal function and plasticity19, 22, 30. To investigate whether reward learning-induced gene expression is associated with altered DNA methylation patterns, we next performed methylated DNA immunoprecipitation (MeDIP; see Methods and Supplementary Fig. 2), a technique capable of detecting site-and gene-specific changes in methylation status. Gene-specific primers were designed to assay methylation levels at various promoter and intragenic targets for the Egr1, Fos, and Gapdh loci, each of which contain a robust CpG island flanking the transcription start site (Fig. 3a). Intriguingly, reward experience induced bidirectional site- and gene-specific changes in DNA methylation (Fig. 3b–d). Thus, two different sites in the promoter region of Egr1 were demethylated by reward experience (decreased methylation vs. TO controls), with one site being specific to CS+/reward associations. Conversely, the 3’ end of the gene body for both Egr1 and Fos underwent active methylation in response to rewards (and was specific to the CS+ group for Fos), whereas promoter methylation at the housekeeping gene Gapdh was unaltered. Overall, these results reveal that reward learning induces gene-specific changes in DNA methylation patterns that correlate with transcriptional changes, and furthermore selectively implicate 3’ gene body methylation as a potential mechanism for experience-dependent gene activation.
Figure 3.
Reward-related memory formation alters VTA DNA methylation profiles at Egr1 and Fosa, Gene targets for MeDIP assay. Annotated lines below genes illustrate loci for gDNA primer pairs. b-d, MeDIP reveals site-specific learning and reward-related changes in DNA methylation at Egr1 and Fos (n = 8 per group; one-way ANOVA: main effect of training for E1, E3, E4, F2 sites, P < 0.008 for each comparison; Tukey post-hoc tests revealed significant group differences (*P < 0.05)). Error bars represent s.e.m.
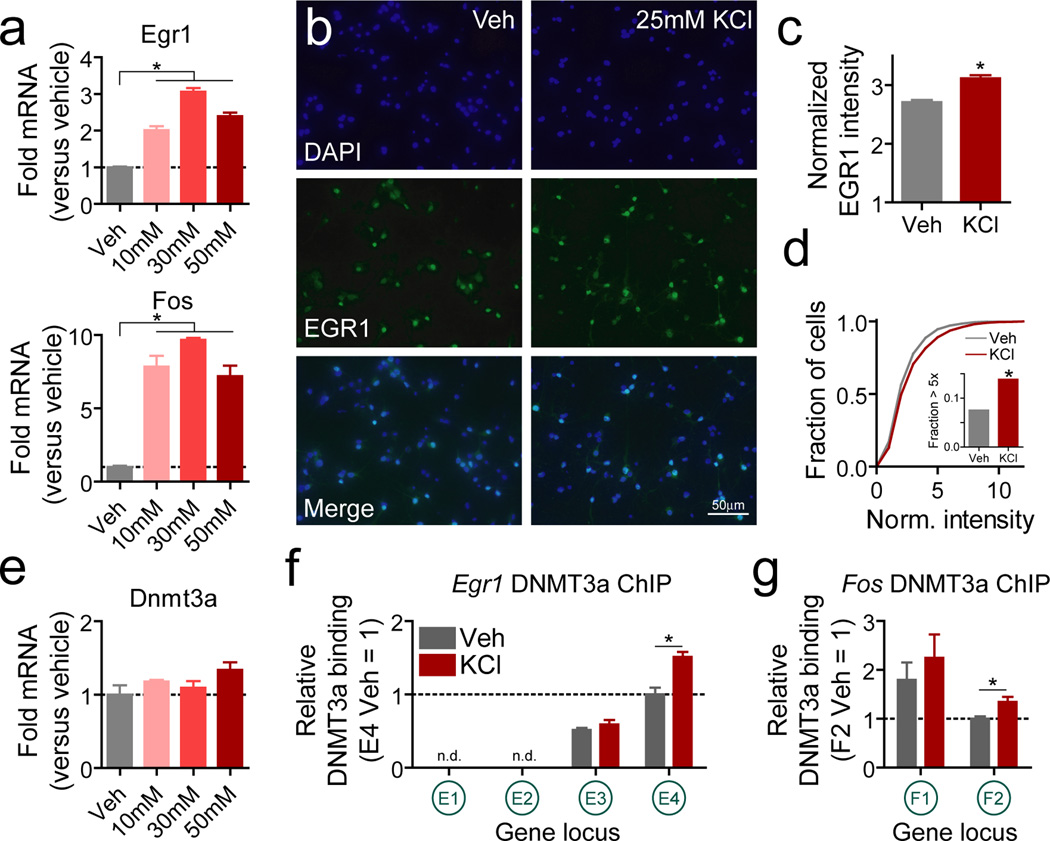
We next sought to explore the relationship between DNA methylation and transcription of these gene targets in more depth. Given that the cellular heterogeneity, small structure volume, and difficulty in access to the VTA in vivo can impose technical limitations that impact mechanistic interpretations, we employed a well-studied31, 32, fully controllable in vitro culture system with a defined neuronal population as a model system for this phase of our studies. Stimulation-dependent changes in gene expression and protein levels were first verified using potassium chloride (KCl) treatment of neurons in vitro, which causes membrane depolarization and increases neuronal activity31. mRNA levels of both Egr1 and Fos were significantly elevated following 1hr KCl stimulation (Fig. 4a). EGR1 immunofluorescence was also elevated at this time point (Fig. 4b–d), due mainly to a significant increase in the proportion of neurons with high levels of EGR1 protein (Fig. 4d; Supplementary Fig. 3a). We were surprised to find that KCl depolarization did not alter mRNA for the major de novo DNA methyltransferases (DNMT3a and DNMT3b; Fig. 4e and Supplementary Figure 3b),especially in light of recent findings that DNMT3a transcript levels can be altered by aversive memory formation and cocaine experience 15, 33. Nevertheless, DNA methylation can be altered by activity-dependent binding of DNMTs. Given that DNMT3a is the most highly expressed DNMT isoform in both neuronal culture and VTA tissue in adult animals (Supplementary Fig. 3c–d), we performed a chromatin immunoprecipitation (ChIP) experiment with a DNMT3a antibody to determine if DNMT3a binding patterns at Egr1 and Fos are altered by neuronal stimulation (Fig. 4f–g). Although some promoter elements for Egr1 contained no detectable DNMT3a binding, we found that binding at intragenic sites for both genes was significantly elevated by KCl stimulation, whereas proximal promoter binding was unaltered. Intriguingly, the sites with increased DNMT3a binding were also the sites that underwent de novo methylation in reward learning experiments, suggesting a shared mechanistic link in activation of these genes.
Figure 4.
Neuronal activity alters IEG expression and gene body DNMT binding in vitroa, Neuronal depolarization with multiple KCl concentrations increases mRNA levels of Egr1 and Fos (n = 3 per group; one-way ANOVA: main effect of treatment for Egr1 mRNA (F(3,11) = 92.99, P < 0.0001) and Fos mRNA (F(3,11) = 52.50, P < 0.0001; Tukey post-hoc tests revealed significant differences between vehicle treatment and all KCl concentrations (*P < 0.001)). b, EGR1 protein immunofluorescence in vehicle and KCl (25mM) treated neuronal cultures. c, Quantification of immunofluorescence revealed increased EGR1 levels following KCl stimulation (n = 1283 cells for vehicle treatment, 1138 cells for KCl treatment; Student’s t-test t2419 = 6.12, *P < 0.0001). d, Cumulative distribution of cell-by-cell EGR1 immunofluorescence revealed that KCl stimulation selectively increased the proportion of highly-expressing (> 5-fold over baseline) EGR1 positive cells (7.56% of vehicle treated cells vs. 13.88% of KCl treated cells; z-test for proportions, z = −5.0588, *P < 0.0001). e, 1hr KCl stimulation did not alter DNMT3a levels (n=3 per group; one-way ANOVA: main effect of treatment, F(3,11) = 2.28, P = 0.157). f-g, Chromatin immunoprecipitation for DNMT3a at Egr1 and Fos. KCl stimulation increased DNMT3a binding at intragenic locations for Egr1 (E4; n = 4 per group; Student’s t-test t6 = 4.463, *P = 0.0043) and Fos (F2; Student’s t-test t6 = 3.002, *P = 0.0239). DNMT3a binding at E1 and E2 Egr1 loci was below the detectable threshold. There were no differences in binding at promoter (E3 and F1) sites following KCl stimulation, indicating selective activity-dependent gene body methylation. Error bars represent s.e.m. Gene loci conventions follow from Figure 1i.
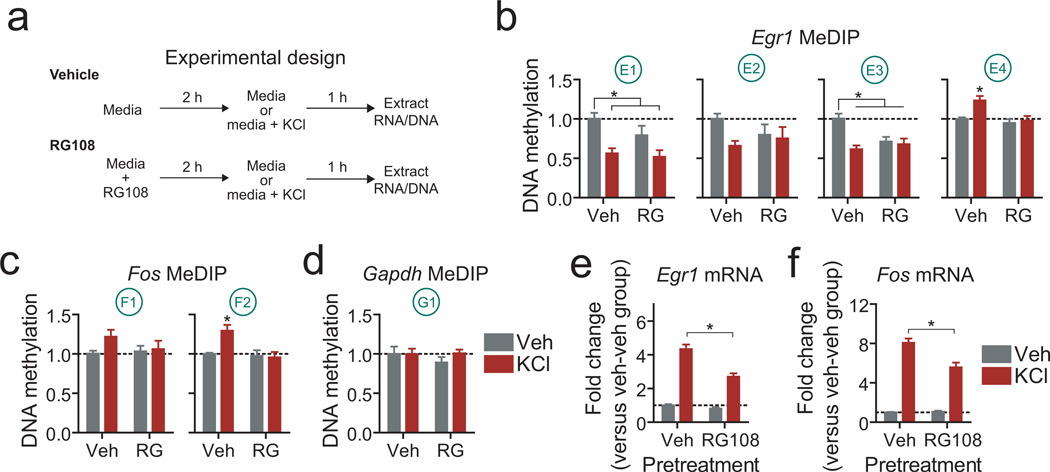
Next, we sought to determine the necessity of DNA methylation and DNMT activity on stimulation-induced transcription of Egr1 and Fos. DNA methylation was blocked in vitro with RG108 for 2hrs prior to KCl-induced depolarization (Fig. 5a). DNA extracted from these cultures was subject to MeDIP at the same target sites assayed in reward learning experiments. Strikingly, activity-dependent demethylation and de novo methylation was observed at the same promoter and intragenic sites at Egr1 and Fos (Fig. 5b–d). Thus, two of three sites assayed in the Egr1 promoter region were demethylated by KCl stimulation alone. This demethylation was not altered by pre-treatment with RG108, which is expected given that DNMT activity is not required for activity-dependent DNA demethylation19. However, 3’ intragenic sites at Egr1 and Fos were hypermethylated following neuronal activation, and this increase was completely blocked by DNMT inhibition. Together, these results reveal that intragenic DNA methylation at key plasticity genes is induced by neuronal activity. However, it is possible that these changes are merely the byproduct of gene activation by other transcriptional elements, and do not serve a dynamic role in transcriptional activation. To determine whether this was the case, we analyzed mRNA levels following KCl stimulation both in the presence and absence of DNMT inhibition (Fig. 5e–f). Remarkably, DNMT inhibition prior to KCl stimulation significantly attenuated depolarization-induced expression of both genes without altering basal transcript levels in the absence of KCl stimulation, revealing that activity-dependent changes in DNA methylation represent a core component of the transcriptional response to neuronal stimulation.
Figure 5.
DNA methylation is required for normal activity-induced IEG expression in vitroa, Experimental design. Neuronal cultures were pre-treated with RG108 to block DNMT activity prior to depolarization with 25mM KCl. b, Neuronal activity induced promoter demethylation and intragenic hypermethylation as measured by MeDIP at Egr1 loci (n = 7 per group; one-way ANOVA: main effect of treatment on Egr1 methylation (E1, F(3,27) = 6.526, P = 0.002; E2, F(3,27) = 1.819, P = 0.17; E3, F(3,27) = 8.239, P = 0.0006; E4, F(3,27) = 7.398, P = 0.0011; Tukey post-hoc tests revealed significant differences between treatment groups (*P < 0.05)). Importantly, intragenic hypermethylation was completely blocked by RG108 treatment (E4 locus; P < 0.01 for comparison between vehicle + KCl and RG108 + KCl group). c, KCl treatment produced intragenic hypermethylation, which was completely prevented by DNMT inhibition (n = 7 per group; one-way ANOVA: main effect of treatment on Fos methylation (F2, F(3,27) = 6.595, P = 0.0021; Tukey post-hoc tests revealed significant differences between vehicle + KCl treatment and all other groups (*P < 0.05)). Fos promoter methylation was unaltered by KCl stimulation. d, Methylation at the housekeeping gene Gapdh was not altered by neuronal activity or RG108 treatment (analysis performed as above, P = 0.6). e-f, DNMT inhibition impaired KCl-induced Egr1 and Fos mRNA expression (n = 13–15 per group; one-way ANOVA: main effect of treatment on Egr1 mRNA (F(3,53) = 94.77, P < 0.0001) and Fos mRNA (F(3,53) = 122.8, P < 0.0001). Tukey post-hoc tests revealed significant differences between vehicle + KCl treatment and RG108 + KCl treatment for both transcripts (*P < 0.001)). Error bars represent s.e.m. Gene loci conventions follow from Figure 3a.
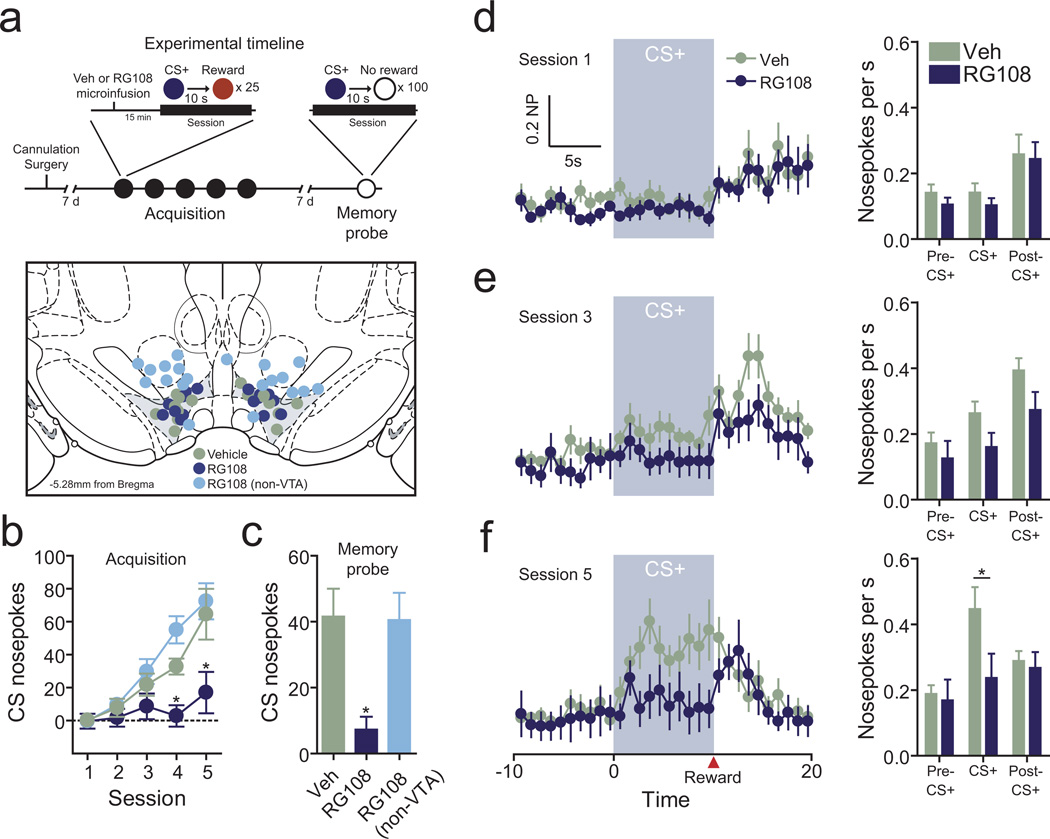
Given that Egr1 and Fos expression were correlated with reward learning, that learning induced changes in methylation status at these genes, and that DNA methylation is required for normal activity-induced expression of these genes, we hypothesized that DNMT activity is an essential mechanism for the acquisition of reward-related memories. To examine the functional role of DNA methylation in the VTA during reward learning, we surgically implanted bilateral cannulae to deliver RG108 directly to the VTA before conditioning sessions (Fig. 6a). Vehicle or RG108 infusions were made 15min prior to each of 5 conditioning sessions, in which CS+ cues were paired with rewards as before. DNMT inhibition in the VTA significantly reduced conditioned nosepoke responding in sessions four and five as compared to vehicle infused controls (Fig. 6b). However, intra-VTA RG108 did not significantly alter reward consumption (all animals consumed all sucrose rewards in each session), and did not alter total nosepokes made during any behavioral session (data not shown). Moreover, DNMT inhibition at sites outside of the VTA (Fig. 6a–b) did not alter reward learning. Thus, these results reveal for the first time that DNA methylation in the VTA is required for acquisition of a learned appetitive response. To ensure that DNMT inhibition selectively induced memory deficits (instead of creating more general deficits in motivation), we tested the same animals one week after the initial conditioning period in a “memory probe” session. This session consisted of 100 CS+ presentations without reward delivery, and was designed to measure memory for the previous conditioning in the absence of any new memory formation. If RG108-treated animals actually learned the cue/reward association but were motivationally impaired in the presence of RG108, we might predict that they would resume conditioned nosepokes in the drug free state. In contrast, we found that animals treated with the DNMT inhibitor only during conditioning did not exhibit selective learned nosepoke responses during this memory probe (Fig. 6c), suggesting that they did not form the cue/reward association. Consistent with this evidence, we observed that RG108 selectively impaired cue-related nosepoke responses, without significantly affecting nosepoke behavior during reward procurement/consumption (10s after reward delivery) in any behavioral session (Fig. 6d–f). Thus, together these results suggest that DNMT inhibition in the VTA impaired reward-related memory formation but not general motivational processes.
Figure 6.
Reward learning requires DNA methylation in the VTA. a, Upper panel, behavioral design. During the acquisition phase of learning, vehicle or RG108 (200µM) was bilaterally infused into the VTA 15 minutes prior to each of 5 CS+ conditioning sessions. Animals were returned to the conditioning context 7 days later to probe memory for the previously formed association. Lower panel, VTA infusion sites. Vehicle and RG108 infusion sites are shown in relation to the VTA (shaded region; n = 8 and 9 animals per group). RG108 infusions made at sites outside of the VTA are also noted (light blue circles). b, DNMT inhibition in the VTA significantly impaired reward learning. RG108 infusions made into the VTA (dark blue circles) prevented acquisition of conditioned nosepoke responses as compared to vehicle infusions (two-way ANOVA: interaction between treatment and behavioral session on CS+ nosepoke responses, F(8,120) = 2.854, P = 0.0061; Bonferroni post-hoc tests indicated significantly diminished CS+ nosepoke responses in the RG108 group in training sessions 4 and 5 (P < 0.05 and P < 0.001, respectively)). There was no difference in reward learning between vehicle treated group and RG108 infused group when infusions were made to sites outside the VTA, indicating site-specific action. Intra-VTA RG108 did not alter total nosepokes (data not shown; two-way ANOVA: interaction between treatment and behavioral session on overall nosepoke responses, F(8,120) = 0.46, P = 0.88) or reward consumption in any behavioral session. c, Animals infused with RG108 during memory acquisition phase exhibit no memory for the CS+/reward association 7 days after training. Cued nosepokes were significantly diminished in VTA-RG108 infused animals as compared to vehicle treated and RG108 (non VTA) animals (one-way ANOVA: main effect of treatment on cue-specific nosepokes (F(2,26) = 6.341, P = 0.0062; Tukey post-hoc tests revealed significant differences between VTA-RG108 animals and vehicle/non-VTA RG108 groups (*P < 0.05). d-f, Nosepoke responses during the 30s CS+ presentation window in acquisition sessions 1, 3, and 5 reveal DNA methylation is only required for the development of learned cue-evoked nosepoke responses. Although nosepoke responding did not differ at any time point (pre-CS+ presentation, CS+ presentation, or post-CS+/reward presentation) in sessions 1 or 3, VTA RG108 infusions blocked cue-related nosepokes during session 5 (two-way repeated measures ANOVA: interaction between infusion and time bin (F(2,30) = 3.744, P = 0.0353; Bonferroni post-hoc tests revealed significant differences between VTA-RG108 animals and vehicle-infused animals during the cue presentation window alone (*P < 0.05)). Error bars represent s.e.m.
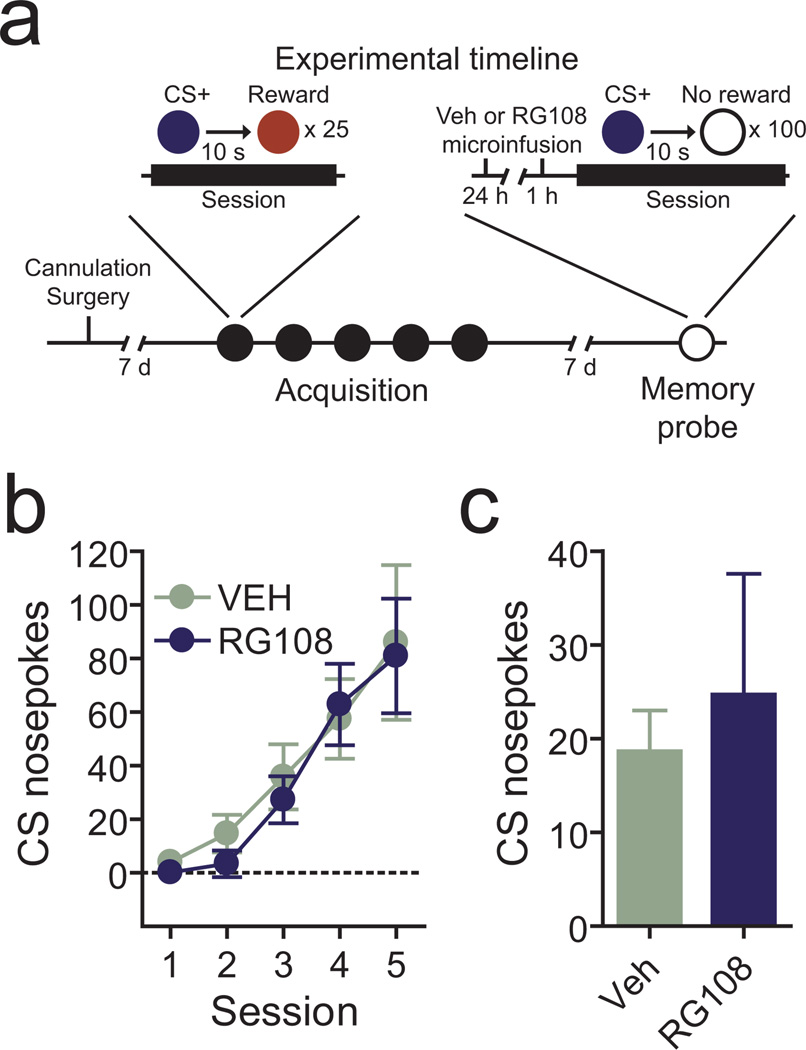
In addition to its involvement in memory formation, DNA methylation has also been implicated in the storage/retrieval of long term “remote” memories14. To dissociate the possible effects of DNA methylation on reward-related memory formation and memory storage, we next examined the ability of DNMT inhibition to alter previously formed associations. Animals were implanted with bilateral cannulae aimed at the VTA, and trained in an identical reward learning task without any drug infusions (Fig. 7a). After normal acquisition of cue-reward associations in the absence of DNMT inhibition, animals were divided into two groups, with half receiving two RG108 infusions into the VTA 24hrs and 1hr prior to a memory probe session and the other half receiving vehicle infusions. Critically, DNMT inhibition did not alter conditioned nosepoke responding evoked by the reward-paired cue from a previously learned association (Fig. 7b–c), indicating that ongoing DNA methylation in the VTA is not necessary to maintain or retrieve remote reward memories.
Figure 7.
Active DNA methylation in the VTA is not required for remote memory storage or retrieval. a, Behavioral design. Animals were implanted with bilateral cannulae aimed at the VTA, and trained in the CS+ task without any drug infusions. Animals were then separated into vehicle or RG108 infusion groups. Microinfusions were made directly into the VTA both 24hrs and 1 hr before the memory probe session 7 days after initial training. This design was used to target the storage and/or retrieval of long-term reward related memories. b, Acquisition of stimulus reward associations occurred normally in a drug-free state. Animals were later separated into the labeled groups. There were no pre-existing differences in reward learning between animals that ended up receiving vehicle and RG108 after memory formation (n = 6 and 7 per group; two-way ANOVA: interaction between future treatment and behavioral session on CS+ nosepoke responses, F(4,44) = 0.17, P = 0.95). c, DNMT inhibition prior to the memory probe session did not alter memory recall. There was no difference in learned nosepoke responses between vehicle and RG108 treated animals (Student’s t-test t11 = 0.4157, P = 0.6856). Error bars represent s.e.m.
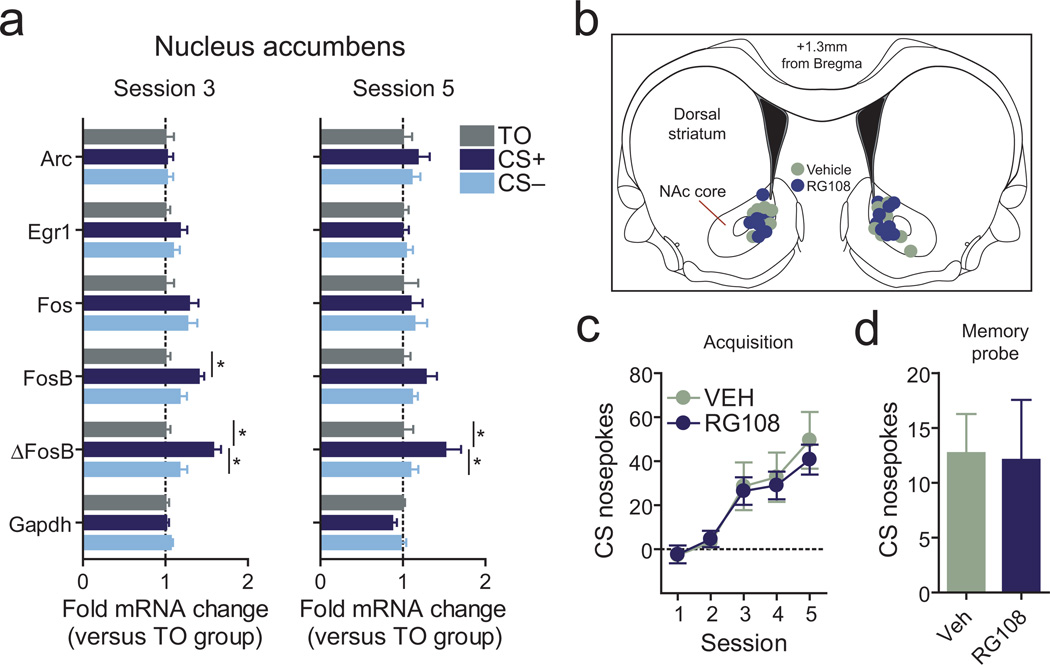
Reward learning also requires neuronal activity and dopamine release within the nucleus accumbens (NAc)8, 10, a downstream target of dopaminergic neurons projecting from the VTA. Intriguingly, we found that reward learning increased mRNA expression of FosB and its splice variant ΔFosB in the NAc core (but not other activity-regulated genes; Fig. 8a), which are downstream of dopamine receptor activation and important functional regulators of behavioral responses to drugs of abuse such as cocaine. Given that psychostimulant-related neuronal and behavioral adaptations are regulated by DNA methylation in the NAc 33–35, we next examined a role for NAc DNA methylation in reward learning. As above, we microinfused RG108 bilaterally into the NAc core to block DNA methylation prior to reward conditioning (Fig. 8b–d). In contrast to the previously published role of DNA methylation in cocaine related responses, we found no alterations in normal reward memory acquisition or long-term memory following NAc DNMT inhibition, suggesting a specific role for DNA methylation in the VTA in natural reward learning. This finding also represents an intriguing distinction for circuit-specific (VTA vs. NAc) epigenetic changes in normal reward memory as compared to psychostimulant-driven behavioral change.
Figure 8.
Reward learning alters gene expression in the nucleus accumbens, but does not require NAc DNA methylation. aFosB and ΔFosB mRNA levels were significantly elevated only in CS+ animals 1hr following behavioral training session 3 (n = 15–18 per group; FosB one-way ANOVA: main effect of group, F(2,47) = 4.656, P = 0.0145; ΔFosB one-way ANOVA: main effect of group, F(2,47) = 10.93, P = 0.0001; Tukey post-hoc test for between-group differences; *P < 0.05). ΔFosB levels remained elevated in CS+ animals 1hr after behavioral session 5 (n = 8 per group; ΔFosB one-way ANOVA: main effect of group, F(2,23) = 3.657, P = 0.0434; Tukey post-hoc test for between-group differences; *P < 0.05). Expression of other IEGs, including Egr1 and Fos, was not altered by learning or reward experience. b, NAc infusion sites. c, Using an identical infusion paradigm and drug concentration from VTA microinfusion experiments, bilateral RG108 infusions into the NAc did not impair the acquisition or expression of associative reward memories. Vehicle and RG108 infused animals exhibited similar increases in cue-evoked nosepoke responses across training (n = 8 per group; two-way ANOVA: interaction between treatment and behavioral session on CS+ nosepoke responses, F(4,75) = 0.15, P = 0.96). d, NAc RG108 infusions during acquisition did not alter total nosepoke responses (data not shown) or long-term memory performance (j). Error bars represent s.e.m.
DISCUSSION
Taken together, these findings establish that activity-dependent DNA methylation is critical for basic reward learning. Moreover, these findings highlight the region-selective manner in which epigenetic processes regulate memory formation. Thus, although DNA methylation in the VTA is required for the formation of reward-related memories, it is not required in the NAc despite the essential role of this structure in reward processing and behavioral responses to rewards. As such, these findings also draw an important distinction between the neuroepigenetic regulation of natural reward responses and those that occur downstream of drugs of abuse or psychiatric illness33–35. For example, cocaine experience alters DNMT levels and DNA methylation in the NAc, regulates the structural plasticity at NAc synapses that is thought to regulate addiction-related behaviors, and controls the formation of cocaine-associated memories (conditioned place preference)33.
A central hypothesis in the drug addiction field is that substance use co-opts normal reward pathways to generate maladaptive behaviors. Our findings suggest the possibility that epigenetic mechanisms may contribute differently to addictive mechanisms and normal reward processes in terms of NAc function. Overall, then, while drug experiences may co-opt normal reward mechanisms to some extent, it is also possible that they engage distinct epigenetic mechanisms (including but not limited to DNA methylation) that contribute to addiction yet are not involved in normal reward learning. A critical step going forward will be to determine the precise differences between epigenetic regulation induced by drugs of abuse and natural rewards, as well as the contribution of epigenetic mechanisms in other reward-processing structures such as the basolateral amygdala36 and prefrontal cortex37 to reward-directed behaviors. Furthermore, given that reward learning phenotypes have been linked to drug responses1, it is intriguing to speculate that epigenetic differences between animals are contributing factors to the unique vulnerability of a specific subset of the population to addictive behaviors 38, 39.
The present findings also build upon the long-appreciated role for DNA methylation in transcriptional regulation and suggest that methylation patterns in terminally differentiated neurons may represent a final common pathway for alterations in neuronal function. In addition to supporting the much-appreciated role of gene promoter methylation or demethylation in silencing or activating memory-related genes14–17, this report is the first to suggest the distinct transcriptional and behavioral relevance of active gene body methylation with respect to learning mechanisms. Thus, despite the fact that the promoters of memory enhancing genes can remain demethylated following DNMT inhibition, stimulation or learning-related gene transcription may be significantly diminished without activity-dependent intragenic methylation. Methylation in gene bodies is an evolutionarily ancient form of DNA methylation that, unlike gene promoter methylation, is not strictly correlated with gene expression levels, as many actively transcribed genes possess enrichment of DNA methylation40. The present results suggest that activity and learning-dependent forms of gene body methylation may serve a critical role in determining the final transcriptional potential of a given gene, thereby representing an essential step in the biochemical pathways underlying the neural regulation of reward-dependent learning and memory.
Finally, we observed that even in the absence of meaningful gene expression changes, experience with rewards in our CS– group was capable of inducing some of the same alterations in DNA methylation that were observed in our CS+ group. Of course, this is not necessarily surprising given that unexpected rewards evoke increases in dopamine neuron signaling7, 9. An intriguing interpretation of these results is that multiple signal transduction cascades simultaneously impinge on epigenetic regulation of gene transcription, and that these changes work together in a combinatorial fashion to determine the ultimate transcriptional profile of a given activity-dependent gene41. Indeed, while some experiences (such as exposure to rewards) may be capable of inducing epigenetic changes alone, they may require the epigenetic changes induced by other signals (such as exposure to reward-paired cues) to result in meaningful changes in gene activity. Thus, a “methylation code” may arise in which modifications at different gene areas (proximal and distal promoters, gene body) work together to regulate whether a gene will undergo activity-dependent transcription. Future research will be required to parse the precise mechanisms by which these modifications interact and how they ultimately regulate gene readout and memory function in the adult brain.
Online Methods
Animals
Male Sprague-Dawley rats, approximately 90–120 days old and weighing 300–400 grams, were housed individually in plastic cages in an AAALAC approved animal care facility on a 12 h light/dark cycle with water available ad libitum. During all behavioral experiments, animals were food deprived to 85% of their original bodyweight by restricting food access to ~15g of lab chow per day. During the week following surgical procedures (described below), food was available ad libitum. All procedures were performed in accordance with the University of Alabama at Birmingham Institutional Animal Care and Use Committee. No formal statistical method was used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications 14, 15, 22. All animals were randomly assigned to respective groups.
Conditioning Procedures
Pavlovian reward conditioning occurred in standard experimental chambers (30 cm × 23 cm × 23 cm; Med Associates, Albans, VT). Each chamber was equipped with a single house light, a floor with metal bars, and a speaker that delivers a 3 kHz, 80 db tone. A food cup was mounted on the front wall of the testing chamber, and was attached to a food dispenser that released a single 45 mg sucrose pellet (Research Diets) when activated. An infrared photobeam source and detector were mounted on either side of the food cup, enabling millisecond recording of food cup-approach behavior. For each conditioning session, animals were transported to the conditioning chamber, and the session began with houselight illumination and initiation of background white noise. For the CS+ groups, conditioning trials (25 per session) consisted of a 10s tone followed immediately by the delivery of a sucrose pellet to the food receptacle. Individual trials were initiated on a variable schedule every 30−90s with an average inter-trial interval of 60s. Cue-reward learning was assayed by the development of conditioned approach behavior, in which rats make goal-directed nose-pokes into the sucrose pellet receptacle during presentation of the CS+ 6. All nose-pokes were recorded and available for analysis. However, the primary measure of learning was the number of nose-pokes occurring during the CS period subtracted by the number of nose-pokes in the 10s prior. A second measure of learning was the total number of nosepokes made during the CS period divided by the total number of nosepokes. The CS– group was exposed to delivery of the same tone according to an identical inter-trial interval (30–90s, average 60s), and the same number of sucrose rewards. However, for these animals, delivery of sucrose rewards was timed to occur sporadically between CS– tone presentations, such that rewards were never delivered within 15s of tone onset or offset. The result is that while the tone predicts the future reward for the CS+ group, it does not predict the reward for the CS– group. Finally, another group of animals (tone-only controls; TO) underwent the same exposure to the audio tone as CS+ and CS– animals, but no rewards were delivered. To examine long-term memory retention, animals were trained in the CS+ task as described above, returned to home cages for seven days, and then placed back in the conditioning environment for a single session which consisted of 100 CS+ presentations without reward delivery.
Isolation of the VTA and NAc
For RT-PCR, ChIP, and MeDIP experiments, animals were sacrificed by rapid decapitation 1 hour after completion of the final conditioning session. Brains were immediately removed and flash-frozen in isopentane chilled on dry ice. Brains were sectioned with a 1mm brain block on ice and sections were placed on microscope slides and immediately frozen in chilled isopentane and stored at −80°C. VTA and NAc core sections were removed with a 1mm punch tool and processed for downstream analysis.
Measuring mRNA levels by real-time, reverse transcriptase PCR
VTA and NAc punches were processed for mRNA quantification. Total RNA was extracted using the RNeasy Mini kit (Qiagen) following the manufacturer’s instructions. mRNA was reverse transcribed using the iScript RT-PCR kit (Bio-Rad). Specific intron-spanning primers were used to amplify cDNA regions for transcripts of interest (Arc, Bdnf exon IV, Egr1, Fos, Fosb, ΔFosb; see Supplementary Table 1 for primer sequences). q-PCR amplifications were performed in triplicate using an iQ5 real-time PCR system (Bio-Rad) at 95°C for 5 min, followed by 45 cycles of 95°C for 15s and 58°C for 60s, and then incubation at 72°C for 10 min followed by real-time melt analysis to verify product specificity. Gapdh was used as an internal control for normalization using the ΔΔCt method 42.
Methylated DNA immunoprecipitation (MeDIP)
Methylated DNA immunoprecipitation was performed using a 5-methylcytosine (5mC) antibody (5-methylcytosine monoclonal mouse antibody from Epigentek) as described previously with minor modifications14, 43. Genomic DNA was extracted (DNeasy Blood and Tissue Kit, Qiagen), treated with RNase A, and quantified (Quant-IT HS dsDNA kit, Invitrogen) using the manufacturer’s recommended protocols. 200ng (in vivo experiments) or 400ng (in vitro experiments) of DNA per sample was removed and sonicated (Fisher Sonic Dismembrator 120) to 200–1000bp fragments for methylation analysis. To increase yields of methylated DNA, purified genomic DNA was pooled across 2 biological samples per IP for in vivo experiments. Sonicated DNA was incubated for 1 hour with 4µl mC antibody and then methylated DNA was collected with protein A coated magnetic beads (Invitrogen), washed (1× Bind Wash Buffer, Epimark kit, New England Biolabs), extracted for 2 hrs at 60° with proteinase K in TE buffer with 1% SDS, and purified (Qiagen DNA micro kit). Methylation levels at selected DNA regions were assayed via qPCR on an iQ5 real-time PCR system (Bio-Rad) at 95°C for 5 min, followed by 45 cycles of 95°C for 15s and 58°C for 60s, and then incubation at 72°C for 10 min followed by real-time melt analysis to verify product specificity. Ct values for IP samples were normalized to unprocessed (input) DNA. Gapdh, which did not change across samples, was used as an internal normalization control.
Given that recent discoveries have identified the existence of additional modifications to cytosine bases in DNA (including conversion of methylated cytosines (mC) to hydroxymethylated cytosines (hmC)20, 44, 45), we also validated this technique to ensure specific binding to methylated DNA, linear enrichment of DNA based on input material, and specific capture of methylated DNA over non-methylated DNA (Supplementary Figure 2). To confirm mC-specific antibody binding, we performed an antibody dot blot with pure mC, hmC, or cytosine (C) PCR fragments (Zymo Research). Double-stranded DNA fragments (100ng per sample) were incubated at 95°C for 10 minutes to denature DNA, blotted onto a PVDF membrane (Bio-Rad), and then allowed to air-dry to bind DNA to the membrane. Membranes were then blocked with Odyssey blocking buffer (Li-Cor Biosciences) and incubated for 1hr in a mixture of primary antibodies for mC (mouse AB, Epigentek; 1:1000) and hmC (rabbit antibody, Active Motif; 1:1000), diluted in PBS. Membranes were then washed 3 times with PBS and incubated for 45 minutes in a mixture of goat anti-mouse (IR dye 800, Li-Cor Biosceinces) and goat anti-rabbit (IR dye 680, Li-Cor Biosceinces) secondary antibodies (both at 1:15,000 concentration in PBS) and imaged on an Odyssey infrared imaging system. Our results (Supplementary Fig. 2a–c) indicate specific (>25 fold) binding to the antibody target, and no cross-binding to unmodified cytosines. To confirm that methylated DNA could be immunoprecipitated in a linear range (which is required for proper identification of between-group differences), we performed a serial dilution (range, 125ng – 1000ng) with purified genomic DNA pooled from biological samples. Primers for the Fos promoter were used as a readout for methylation levels using qPCR, and indicated linear enrichment of DNA across sample concentrations, including the concentrations used for the experiments described here (Supplementary Fig. 2d). Finally, we also validated that MeDIP enriches methylated DNA fragments with little cross-enrichment of nearly identical non-methylated DNA fragments. Synthetic methylated and non methylated control DNA fragments (1pg each, Methyl Miner kit, Invitrogen) were spiked into biological samples prior to MeDIP, and levels of methylated and non-methylated DNA capture were assayed via qPCR with primers specific for each synthetic sequence. Although methylated DNA was actually observed to be less abundant in the input fraction, we found that MeDIP enriched the methylated DNA fragment over 200-fold as compared to the non-methylated fragment (Supplementary Fig. 2e–f). These experiments therefore validate the specificity and accuracy of this assay for enriching methylated DNA.
Cultured neuron experiments
Rat cortical neuronal cultures were generated from embryonic day 18 rat cortical tissue. Briefly, tissue culture wells were coated overnight at 37°C with poly-lysine (50µg/ml) and rinsed 3 times with diH2O. Dissected cortices were incubated with papain for 20 minutes at 37°C. After rinsing in Hank’s Balanced Salt Solution (HBSS), a single cell suspension of the tissue was re-suspended in Neurobasal media (Invitrogen) by trituration through a series of large to small fire-polished Pasteur pipets. Primary neuronal cells passed through a 70uM cell strainer were plated on poly-lysine coated culture wells. Cells were grown in Neurobasal media plus B-27 and L-glutamine supplement (complete Neurobasal media) for 10 days in vitro in a humidified CO2 (5%) incubator at 37°C.
At 10–12 days in vitro, neuronal cultures were treated as described in the text. For KCl stimulation experiments, KCl was added to complete neurobasal media at 2X the specified concentration, and half of the cell culture media (500µl) was replaced with KCl solution or vehicle (neurobasal media alone). Cells were incubated with KCl for 1hr prior to RNA or DNA extraction (RT-qPCR and MeDIP experiments), fixation with 1% paraformaldehyde (ChIP experiments), or immunostaining (EGR1 immunofluorescence experiments). For DNMT inhibition experiments, cultures were incubated for 2 hours with either vehicle solution (complete Neurobasal media) or 200µM RG108 solution (dissolved in complete Neurobasal media). After 2 hr pretreatment, cells were treated for 1hr with either vehicle (complete Neurobasal media) or KCl (dissolved in complete Neurobasal media; final concentration of 25mM) to induce neuronal depolarization. This combination of treatments yielded 4 experimental groups: 1) Vehicle pretreatment plus vehicle, 2) RG108 pretreatment plus vehicle, 3) Vehicle pretreatment plus KCl activation, and 4) RG108 pretreatment plus KCl activation. At a minimum, all cell culture experiments were performed in triplicate. Experiments involving RG108 incubation were repeated 3 times.
DNMT3a Chromatin Immunoprecipitation (ChIP)
DNMT3a binding at specific genomic loci was assayed using available ChIP protocols with minor modifications46, 47. Briefly, cultured cells were treated as described and immediately fixed in 1% paraformaldehyde, washed in PBS, lysed, and then sonicated (Fisher Sonic Dismembrator 120) to shear DNA to 200–1000bp fragments. Sheared, cross-linked DNA was incubated with 4µl DNMT3a antibody (#2160, Cell Signaling) and 25ul protein A coated magnetic beads (Invitrogen) overnight, washed sequentially in low salt, high salt, and LiCl buffers, and then incubated for 2hrs at 65°C in TE buffer containing 1% SDS and proteinase K solution (Qiagen) to reverse crosslinks. Following magnetic removal of protein A coated beads, extracted DNA was then purified (Qiagen DNA micro kit), and DNMT3a binding levels at selected DNA regions were assayed via qPCR as described above. Ct values for IP samples were normalized to unprocessed (input) DNA, which was not incubated with DNMT3a antibody. We found no difference in input DNA between samples (Student’s t-test, p > 0.21 for each comparison). Furthermore, use of a control rabbit IgG in place of the DNMT3a antibody led to no detectable binding (typical Ct value > 37 in qPCR amplification).
Egr1 Immunofluorescence in cell culture
To examine the effect of KCl stimulation on neuronal EGR1 protein levels, cell cultures grown on coverslips were treated as described and then washed briefly in sterile PBS (pH 7.4), incubated at room temperature for 20 minutes in freshly prepared 4% paraformaldehyde in PBS, washed 3 times with PBS, and then incubated with PBS containing 0.25% Triton X-100 to permeabilize membranes. Cells were then washed 3 times in PBS, blocked in 5% normal goat serum in PBS for 1hr, and incubated with an EGR1 antibody (Cell Signaling EGR1 (44D5) Rabbit mAb #4154; 1:200 in PBS) at 4°C overnight. Cells were then washed 3 times in PBS and incubated for 1hr at room temperature with a fluorescent secondary antibody (Alexa 488 goat anti-rabbit, Invitrogen; 1:500 in PBS with 1% BSA), washed again 3 times with PBS, and coverslips were mounted onto microscope slides with Prolong Gold anti-fade media containing DAPI stain (Invitrogen) as a marker for cell nuclei. Multidimensional image acquisition for DAPI (blue) and EGR1 (green) staining was performed using a Zeiss AxioImager widefield epifluorescence microscope at 20× magnification. A total of 6 images were collected for each culture well, for a total of 18 images per treatment group. EGR1 fluorescence was quantified in Image J software by first identifying DAPI-positive cells, and then measuring the EGR1 integrated density in a circular field around the cell body for each individual cell (1283 total cells from vehicle treatment, 1138 total cells from KCl stimulation). Integrated density measurements for each cell were then normalized to a background EGR1 fluorescence reading (mean integrated density measurement from 10 image locations that lacked both EGR1and DAPI staining).
Egr1/tyrosine hydroxylase double immunofluorescence
To determine whether changes in EGR1 protein in vivo following learning occurred in dopamine neurons within the VTA, animals underwent 3 behavioral training sessions (tone only, CS+, or CS–, as described above) and were perfused with saline 1hr after the session ended. Brains were removed and fixed in Buoin’s solution (Sigma) overnight, cleared with ethanol, and then embedded in paraffin wax. 7µm brain sections were cut on a microtome, baked on glass microscope slides, deparaffinized with xylene, washed in PBS, blocked in 10% normal goat serum/1% BSA for 1hr, and subjected to sequential immunostaining for tyrosine hydroxylase (Pel Freez sheep anti TH antibody, 1:500) and EGR1 (Cell Signaling EGR1 (44D5) Rabbit mAb #4154; 1:200) overnight. Slides were then washed, incubated in fluorescent secondary antibodies (Alexa 488 goat anti-rabbit and Alexa 647 donkey anti-sheep, Invitrogen; each at 1:250 in PBS with 10% normal goat serum/1% BSA), and coverslipped with Prolong Gold anti-fade media containing DAPI stain (Invitrogen). Multidimensional image acquisition for DAPI (blue), EGR1 (green), and tyrosine hydroxylase (red) staining was performed using a Zeiss AxioImager widefield epifluorescence microscope at 20× magnification. At least 3 images were collected for each animal, for a total of 14–15 images per treatment group. EGR1+ cells and TH+ cells were counted using Image J software, and EGR1 fluorescence for each cell was measured by obtaining the mean integrated density in a circular field around the cell body. Integrated density measurements for each cell were then normalized to a background EGR1 fluorescence reading (mean integrated density measurement from 10 image locations that lacked both EGR1and DAPI staining). Co-labeled cells (TH+, EGR1+) and single-labeled cells (either TH+ or EGR1+) were analyzed separately to determine if EGR1 changes occurred specifically in dopaminergic neurons.
Surgical procedures
Rats were anesthetized with 5% isoflurane and secured in a stereotaxic apparatus (Kopf Instruments). Under aseptic conditions, stainless steel guide cannulae (26G; Plastics One) were implanted bilaterally 2mm above the region of interest (NAc core stereotaxic coordinates: anteroposterior, +1.3 mm from bregma, ±1.3 mm lateral from midline, and −4.2 mm from dura; VTA stereotaxic coordinates: anteroposterior, −5.2 mm from bregma, ±2.2 mm lateral from midline, and −5.3 mm from dura). A stainless steel obdurator of the same length was placed in each cannula to ensure clearance. Animals were given at least 7 days of recovery during which they received buprenorphine for pain management. Following recovery, animals were habituated to dummy cannula removal prior to drug infusions.
RG108 infusions in vivo
The role of DNA methylation in reward-related memory formation and maintenance was examined by blocking DNMT activity with RG108, a potent small molecule DNMT inhibitor48, 49 with demonstrated efficacy in blocking DNA methylation in vivo 14, 19, 33 and in vitro48, 50, 51. All infusions were made using a 30 gauge stainless steel injection cannula that extended into the infusion site. Bilateral microinfusions of 1 µL solution were made over a 6 min period (2 min per side) using a syringe pump (Harvard Apparatus) at a rate of .5 µL/min. Injectors remained in place for 1 min following infusion to allow for diffusion. Drug concentrations were as follows: Vehicle, 0.2% DMSO in sterile saline; RG108, dissolved in DMSO and diluted to a concentration of 200µM in sterile saline. This concentration was chosen based on behavioral efficacy of doses in this range for in vivo microinfusion experiments14, 22, 33, 52, as well as the ability to block DNA methylation in vivo33 and in vitro51. Moreover, although we did not conduct a dose-effect curve to determine optimal RG108 concentration, this dose of RG108 was found to completely block activity-dependent methylation at intragenic loci of Egr1 and Fos where DNMT3a was observed to increase binding following KCl stimulation in neuronal cultures (Figure 4f–g and Figure 5b–c) For acquisition experiments, infusions were made 15min prior to each of the first 5 behavioral sessions. For memory maintenance experiments, infusions were made 24hr and 1hr prior to the memory probe session.
Histological verification of infusion sites
Upon completion of microinfusion experiments, cannula placement was verified histologically. Rats were deeply anesthetized with sodium pentobarbital and perfused transcardially with physiological saline and formalin, followed by brain removal. After post-fixing and freezing, 50 µm coronal brain sections were taken though the rostro-caudal extent of the region of interest (NAc core subregion or VTA). Sections were stained with cresyl violet and the placement of the cannulae were verified by microscopic inspection of the sections. Placement of an infusion tip within the region of interest was determined by examining the relative position of the cannula track to visual landmarks represented in a stereotaxic atlas53.
Statistical analysis
Differences in conditioned nose-poke responses were examined using a between-subjects group × session ANOVA. Tukey post hoc tests were employed to identify sessions in which approaches produced by the CS+ and CS− differed. Differences in gene expression and DNA methylation status between conditioning groups were determined using one-way ANOVAs, with Tukey post hoc tests for between-group comparisons. EGR1 immunofluorescence was compared with an unpaired two-way Student’s t-test or one-way ANOVA, as appropriate. Differences in the proportion of highly immunofluorescent cells between groups was compared with a z-test for proportions (cell culture experiment) or Chi-square test for multiple groups (in vivo experiments). Differences in reward learning produced by drug infusions were examined using a two-way treatment (vehicle and RG108) × session ANOVA with Tukey post hoc tests for within-session comparisons. Differences in memory maintenance were compared with one-way ANOVA with Tukey post-hoc tests, or t-tests where appropriate. Normality was formally tested and verified where appropriate. Statistical significance was designated at α = 0.05 for all analyses. Statistical and graphical analyses were performed with Graphpad software (Prism). Where necessary (immunofluorescence experiments), data analysis was performed blind to experimental condition.
Supplementary Material
Acknowledgments
We would like to thank all members of the Sweatt laboratory, particularly Garrett Kaas and Iva Zovkic, for comments and helpful suggestions during the completion of these studies. We would also like to thank the Intellectual and Developmental Disabilities Research Core at UAB for assistance with cell culture experiments. This work is supported by the National Institute on Drug Abuse (DA029419 to JJD), the National Institute on Mental Health (MH091122 and MH057014 to JDS), and the Evelyn F. McKnight Brain Research Foundation.
Footnotes
Author Contributions: J.J.D. and J.D.S. designed the experiments and wrote the manuscript with help from all authors. J.J.D. carried out behavioral and biochemical experiments with assistance from D.C., M.C.G-K., M. K., J.M., E.S., and A.T.
The authors declare no competing financial interests.
References
- 1.Flagel SB, et al. An animal model of genetic vulnerability to behavioral disinhibition and responsiveness to reward-related cues: implications for addiction. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2010;35:388–400. doi: 10.1038/npp.2009.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hyman SE, Malenka RC, Nestler EJ. Neural Mechanisms of Addiction: The Role of Reward-Related Learning and Memory. Annual review of neuroscience. 2006 doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- 3.Saunders BT, Robinson TE. Individual variation in resisting temptation: Implications for addiction. Neuroscience and biobehavioral reviews. 2013 doi: 10.1016/j.neubiorev.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saunders BT, Yager LM, Robinson TE. Preclinical studies shed light on individual variation in addiction vulnerability. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2013;38:249–250. doi: 10.1038/npp.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fields HL, Hjelmstad GO, Margolis EB, Nicola SM. Ventral tegmental area neurons in learned appetitive behavior and positive reinforcement. Annual review of neuroscience. 2007;30:289–316. doi: 10.1146/annurev.neuro.30.051606.094341. [DOI] [PubMed] [Google Scholar]
- 6.Stuber GD, et al. Reward-predictive cues enhance excitatory synaptic strength onto midbrain dopamine neurons. Science. 2008;321:1690–1692. doi: 10.1126/science.1160873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day JJ, Roitman MF, Wightman RM, Carelli RM. Associative learning mediates dynamic shifts in dopamine signaling in the nucleus accumbens. Nat Neurosci. 2007;10:1020–1028. doi: 10.1038/nn1923. [DOI] [PubMed] [Google Scholar]
- 8.Flagel SB, et al. A selective role for dopamine in stimulus-reward learning. Nature. 2011;469:53–57. doi: 10.1038/nature09588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz W, Dayan P, Montague PR. A neural substrate of prediction and reward. Science. 1997;275:1593–1599. doi: 10.1126/science.275.5306.1593. [DOI] [PubMed] [Google Scholar]
- 10.Di Ciano P, Cardinal RN, Cowell RA, Little SJ, Everitt BJ. Differential involvement of NMDA, AMPA/kainate, and dopamine receptors in the nucleus accumbens core in the acquisition and performance of pavlovian approach behavior. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:9471–9477. doi: 10.1523/JNEUROSCI.21-23-09471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai HC, et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yun IA, Wakabayashi KT, Fields HL, Nicola SM. The ventral tegmental area is required for the behavioral and nucleus accumbens neuronal firing responses to incentive cues. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:2923–2933. doi: 10.1523/JNEUROSCI.5282-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng J, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller CA, et al. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 16.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biological psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weaver IC, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 18.Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo JU, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 24.Jones MW, et al. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- 25.Bramham CR, Worley PF, Moore MJ, Guzowski JF. The immediate early gene arc/arg3.1: regulation, mechanisms, and function. J Neurosci. 2008;28:11760–11767. doi: 10.1523/JNEUROSCI.3864-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamprecht R, Dudai Y. Transient expression of c-Fos in rat amygdala during training is required for encoding conditioned taste aversion memory. Learn Mem. 1996;3:31–41. doi: 10.1101/lm.3.1.31. [DOI] [PubMed] [Google Scholar]
- 27.Margolis EB, Coker AR, Driscoll JR, Lemaitre AI, Fields HL. Reliability in the identification of midbrain dopamine neurons. PloS one. 2010;5:e15222. doi: 10.1371/journal.pone.0015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margolis EB, Toy B, Himmels P, Morales M, Fields HL. Identification of rat ventral tegmental area GABAergic neurons. PloS one. 2012;7:e42365. doi: 10.1371/journal.pone.0042365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Zessen R, Phillips JL, Budygin EA, Stuber GD. Activation of VTA GABA neurons disrupts reward consumption. Neuron. 2012;73:1184–1194. doi: 10.1016/j.neuron.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levenson JM, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 31.Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng JV, et al. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci. 2010;13:1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tye KM, Stuber GD, de Ridder B, Bonci A, Janak PH. Rapid strengthening of thalamo-amygdala synapses mediates cue-reward learning. Nature. 2008;453:1253–1257. doi: 10.1038/nature06963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parkinson JA, Willoughby PJ, Robbins TW, Everitt BJ. Disconnection of the anterior cingulate cortex and nucleus accumbens core impairs Pavlovian approach behavior: further evidence for limbic cortical-ventral striatopallidal systems. Behavioral neuroscience. 2000;114:42–63. [PubMed] [Google Scholar]
- 38.Deroche-Gamonet V, Belin D, Piazza PV. Evidence for addiction-like behavior in the rat. Science. 2004;305:1014–1017. doi: 10.1126/science.1099020. [DOI] [PubMed] [Google Scholar]
- 39.Kreek MJ, Nielsen DA, Butelman ER, LaForge KS. Genetic influences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nat Neurosci. 2005;8:1450–1457. doi: 10.1038/nn1583. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 41.Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 44.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 48.Brueckner B, et al. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- 49.Schirrmacher E, et al. Synthesis and in vitro evaluation of biotinylated RG108: a high affinity compound for studying binding interactions with human DNA methyltransferases. Bioconjugate chemistry. 2006;17:261–266. doi: 10.1021/bc050300b. [DOI] [PubMed] [Google Scholar]
- 50.Metivier R, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 51.Rajasethupathy P, et al. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maddox SA, Schafe GE. Epigenetic alterations in the lateral amygdala are required for reconsolidation of a Pavlovian fear memory. Learning & memory. 2011;18:579–593. doi: 10.1101/lm.2243411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paxinos G, Watson C. The rat brain in stereotaxic coordinates. New York: El Sevier; 2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.