Abstract
Background
Fetal alcohol exposure produces multi-organ defects, making it difficult to identify underlying etiological mechanisms. However, recent evidence for ethanol sensitivity of the miRNA miR-9, suggests one mechanism whereby ethanol broadly influences development. We hypothesized that loss of miR-9 function recapitulates aspects of ethanol teratology.
Methods
Zebrafish embryos were exposed to ethanol during gastrulation, or injected with anti-miR-9 or nonsense control morpholinos during the 2-cell stage of development and collected between 24 and 72 hours post fertilization (hpf). We also assessed the expression of developmentally important, and known miR-9 targets, FGFR-1, FOXP2, and the non-targeted transcript, MECP2. Methylation at CpG islands of mammalian miR-9 genes was assessed in fetal murine neural stem cells (mNSCs) by methylation-specific PCR, and miRNA processing assessed by qRT-PCR for pre-miR-9 transcripts.
Results
Ethanol treatment and miR-9 knockdown resulted in similar cranial defects including microcephaly. Additionally, ethanol transiently suppressed miR-9, as well as FGFR-1 and FOXP2, and alterations in miR-9 expression were correlated with severity of ethanol-induced teratology. In mNSCs, ethanol increased CpG dinucleotide methylation at the miR-9-2 locus and accumulation of pre-miR-9-3.
Conclusions
Ethanol exerts regulatory control at multiple levels of miR-9 biogenesis. Moreover, early embryonic loss of miR-9 function recapitulated the severe range of teratology associated with developmental ethanol exposure. Ethanol also disrupts the relationship between miR-9 and target gene expression, suggesting a nuanced relationship between ethanol and miRNA regulatory networks in the developing embryo. The implications of these data for the expression and function of mature miR-9 warrant further investigation.
Keywords: miR-9, zebrafish, embryonic development, ethanol, methylation
I. Introduction
Maternal ethanol consumption during pregnancy can result in a spectrum of developmental defects collectively known as Fetal Alcohol Spectrum Disorders (FASD) (Riley and McGee, 2005). This spectrum of ethanol effects is observed in both clinical (Defendi, 2010), and preclinical studies (Driscoll et al., 1990), and suggests that mechanisms underlying FASD are complex. However, recent studies, identifying microRNAs (miRNAs) as novel developmental targets of ethanol (Balaraman et al., 2012; Sathyan et al., 2007; Tal et al., 2012), suggest that miRNAs may mediate some teratogenic effects of maternal ethanol consumption.
MiRNAs are small, non-protein coding RNAs that control the expression of target gene networks through silencing (for review, see (Miranda et al., 2010)). We previously showed that ethanol suppressed several miRNAs, including miR-9, in mammalian (murine) neural stem cells (mNSCs) (Balaraman et al., 2012; Sathyan et al., 2007), and that the ethanol sensitivity of these miRNAs partly accounted for ethanol’s effects on mNSC maturation (Camarillo and Miranda, 2008; Prock and Miranda, 2007; Santillano et al., 2005). MiR-9 is an evolutionarily conserved miRNA (Christodoulou et al., 2010), that is sensitive to ethanol both during development (Sathyan et al., 2007; Tal et al., 2012; Wang et al., 2009) and in the adult (Pietrzykowski et al., 2008). Because of gene duplication events, the identical mature miRNA is coded by three genes in mammals (miR-9-1, 9-2 and 9-3), and by seven genes in zebrafish. MiR-9 has recently been found to specify the midbrain-hindbrain boundary in zebrafish (Leucht et al., 2008), and control neurobehavioral development (Tal et al., 2012). MiR-9-2/3 knockout mice exhibit fetal growth retardation, microencephaly, and other features that are also common to FASD (Shibata et al., 2011). Such data, in conjunction with studies showing miR-9 is important for cell growth in mesoderm (Bazzoni et al., 2009) and endoderm (Plaisance et al., 2006), suggest this miRNA may broadly mediate ethanol effects during development. Since mature miR-9 exhibits absolute sequence identity in vertebrates, we used the experimentally tractable zebrafish embryo to model mammalian prenatal ethanol exposure during gastrulation and to assess whether manipulation of miR-9 expression mimicked the effects of ethanol exposure. We also assessed the effects of ethanol on methylation at miR-9 gene loci and on the regulation of pre-miRNA transcripts. Because of evolutionary divergence in miR-9 gene number between teleosts and mammals, assessment of these upstream events in teleost miR-9 regulation has limited implications for understanding the susceptibility of mammalian miR-9 loci. For this reason, and because the mammalian brain is an important target of both ethanol and miR-9, we employed mNSCs to study the effects of ethanol on mammalian miR-9 gene methylation and the expression of pre-miRNA transcripts.
Our data indicate that the early embryonic loss of miR-9 in zebrafish recapitulates many anatomical features associated with developmental ethanol exposure, and that ethanol disrupts the predicted relationship between miR-9 and its identified mRNA targets FGFR-1 and FOXP2. Moreover, in mNSCs, ethanol increases CpG methylation associated with the mammalian miR-9-2 locus, and interferes with the processing of pre-miR-9 transcripts.
Methods
Zebrafish and mice (C57Bl/6/SJ) were maintained in AAALAC-approved housing facilities, and all experimental procedures were approved by the Institutional Animal Care and Use Committee.
Zebrafish embryo model
Zebrafish (Danio rerio) are a well-established model for studying teratology and developmental genetics (Ton et al., 2006). Though zebrafish have seven miR-9 genes compared to three in the mammal, the mature miR-9 sequence is identical to mammalian miR-9 (Christodoulou et al., 2010). Therefore, zebrafish make a good model for the study of both ethanol teratology and for miR-9 loss of function studies. Adult wild-type Danio rerio male and female breeders were maintained in separate tanks with filtered, recirculating artificial seawater (60mg Instant Ocean® [Spectrum Brands, Cincinnati, OH] per liter distilled water) at a temperature of 26°–28°C, on a light/dark cycle of 14L:10D. Fish were fed both freshly hatched brine shrimp (artemia) and commercial tropical fish food daily. For breeding purposes, two males and two females were placed in a false bottom breeding container with a divider to separate the sexes, prior to the start of the dark cycle. The divider was removed immediately prior to the onset of the light cycle of the following day, allowing for fertilization to occur. Approximately 20 minutes after the onset of the light phase, embryos were collected from the bottom of the breeding container, rinsed, and maintained at 28°C.
Zebrafish embryo ethanol exposure paradigm
At 3.5 hours post fertilization (hpf) embryos were randomly assigned to either treatment or control group (maintained under identical environmental conditions, at the same developmental stages as the ethanol-treated group). Ethanol at a concentration of 430mM (2% w/v or 2,000 mg/dL) was added to artificial sea water (Bradfield et al., 2006). Based on published data showing that under these conditions, ~29% of ethanol reaches the embryo (Reimers et al., 2004), the ethanol exposure level to the developing embryo is estimated at 130mM. Embryos were exposed for 4 hours during gastrulation (3.5 to 7.5 hpf), after which point they were rinsed with fresh artificial seawater and allowed to develop until 24, 48, or 72 hpf (roughly corresponding to the end of the 1st, and the middle of the 2nd, and 3rd trimesters of human development (Kimmel et al., 1995)) and then sacrificed.
MiR-9 knockdown
Zebrafish embryos were generated as described above. At the 1–2 cell stage (confirmed by microscopic evaluation), embryos were injected with miR-9 morpholino (ACTCATACAGCTAGATAACCAAAGA), or 5-base mismatch control morpholino (ACTCATAGACCTACATAACGAAACA) (Gene Tools, Philomath, OR) at a concentration of 2mM, selected based on previous publications (Leucht et al., 2008) and preliminary testing for efficacy of miR-9 knockdown. Morpholinos were suspended in an injection medium (58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5.0 mM HEPES buffer pH 7.6) along with 3% sterile-filtered green food-grade coloring, to visualize location of injection, and delivered using a beveled, glass pipette, at the juncture of the developing blastomere and yolk sac (Phillips et al., 2001). A third group of non-manipulated controls was also acquired, in which embryos were generated in the absence of any experiment manipulation and allowed to develop.
Real-time PCR
Real-time reverse transcription (RT)-PCR analyses was performed for miR-9 with the Exiqon LNA® miRNA amplification system, and for mRNA utilizing a SYBR green-based amplification detection, using a MyiQ single-color real-time PCR detection system (Bio-Rad, Hercules, CA). For mature miR-9, reverse transcriptase was used to synthesize miR-9 specific or control specific (U6 snRNA) cDNA using the Exiqon miRCURY® LNA® microRNA PCR detection system (Exiqon, Woburn, MA) as per manufacturer’s instructions. For quantification of the premature miRNAs (pre-miR-9-1, pre-miR-9-2, and pre-miR-9-3), cDNA was synthesized using qScript cDNA supermix reverse transcription reaction according to manufacturer’s protocol, at an average concentration of 12.5ng RNA/μL total cDNA (Quanta Biosciences, Gaithersburg, MD). We also assessed the expression of three mRNAs that are highly expressed during embryogenesis across species (Hendrich and Tweedie, 2003; Ota et al., 2010; Shu et al., 2007) and are implicated in pervasive developmental disorders in humans (Allan et al., 2008; Haesler et al., 2004; Hagberg, 2002; Poot et al., 2009; Riley et al., 2007). These include a previously identified miR-9 target FGFR-1 (Leucht et al., 2008; Rodriguez-Otero et al., 2011), a predicted vertebrate conserved miR-9 target, FOXP2 (www.targetscan.org), and an mRNA not predicted to be targeted by miR-9 in either mouse or zebrafish, MECP2. U6 snRNA served as a normalization control for miRNA, and 18s for mRNA. Each sample was analyzed in triplicate. Thermal-stability analysis was performed to confirm the amplification of a single PCR product. RNA expression was quantified by calculating Delta (Δ) CT, the difference between the cycle threshold (CT value) of the miRNA, or mRNA of interest, and the normalization control. ΔΔCT, the difference between ΔCT of the control and the ΔCT of the ethanol-treated samples, and fold-change (2ΔΔCT), were calculated according to previously published protocols (Balaraman et al., 2012; Sathyan et al., 2007).
In situ hybridization
MiR-9 expression was detected in zebrafish and in GD12.5 mouse brain sections using in situ hybridization with biotinylated LNA® probes (Exiqon, Woburn, MA) according to the manufacturer’s instructions. Whole embryos were permeabilized prior to in situ by incubation in 10 μg/ml Proteinase K at 38°C for 30 minutes. Briefly, tissue sections or whole embryos were incubated in pre-hybridization blocking cocktail (50% formamide, 1 M NaCl, 1x Denharts, 0.5mg/ml yeast tRNA, 0.3% Triton x-100) for 2 hours at 52°C. Tissue sections and whole embryos were hybridized at 52°C overnight (>16 hours) in hybridization solution (including anti-miR-9 probes diluted to a concentration of 25ng/ml in prehybridization solution). Tissue sections and whole embryos were then processed through a series of wash steps; in 5x SSC for 5 minutes, followed by 50% formamide, 1xSSC and 0.1% Tween-20 at 52°C for 30 minutes, in 0.2xSSC for 30 minutes, followed with 3 washes in PBS each for 5 minutes. Tissues were then incubated in streptavidin conjugated rhodamine (1:500, Vector Labs, Burlingame, CA) or anti-biotin antibody conjugated to Alkaline Phosphatase (1:500, Abcam, Cambridge, MA) for 1 hour and processed for immunofluorescence or enzyme-linked histochemistry. Fluorescent-labeled sections were mounted in Vectashield with DAPI (4′,6-diamidino-2-phenylindole, Vector Labs, Burlingame, CA). All solutions included RNAse Inhibitor (Promega Inc, Madison, WI).
The neurosphere model of fetal murine cortical neuroepithelium
mNSCs were derived from the dorsal portion of the gestational day (GD) 12.5 fetal mouse telencephalon according to previously published protocols (Camarillo et al., 2007; Prock and Miranda, 2007; Santillano et al., 2005; Sathyan et al., 2007). Cortical neurosphere cultures were assigned to either a control (0 mg/dL ethanol), or 70mM (0.32%, or 320 mg/dL) ethanol-treated group, for either 24 hours (acute ethanol exposure) or for 5 days (chronic ethanol exposure). This concentration mimics levels of ethanol reached during chronic alcoholism (Adachi et al., 1991).
Bisulfite conversion of genomic DNA and Methylation-specific PCR for mammalian Mir-9 genes
Genomic DNA was isolated from mNSCs using the ZR Genomic DNA II kit™ (Zymo Research, CA, USA). The EZ DNA methylation kit (Zymo research, CA, USA) was used to detect cytosine methylation. Briefly, two micrograms of genomic DNA per sample were subject to sodium bisulfite treatment (Zhang et al., 2007). DNA was denatured with a dilution buffer containing 2 M NaOH and incubated overnight at 50°C with CT conversion reagent, followed by a purification, desulfonation, and elution. The bisulfite-modified DNA was used immediately for PCR or stored at −80°C. After bisulfite treatment, the methylation status of specific CpG sites in genomic DNA was analyzed by methylation-specific PCR. In brief, the bisulfite-converted genomic DNA was amplified by PCR using two sets of locus-specific Methylation Specific Primers (MSP), which recognize methylated or unmethylated DNA, respectively. MSPs within predicted CpG di-nucleotide containing regions associated with mammalian 5′-end transcription factor/RNA polymerase II binding regions identified by the Encode project (UCSC Genome Browser, http://genome.ucsc.edu, (ENCODE Project Consortium, 2011)), were designed using Methyl Primer Express ® software version 1.0 (Applied Biosystems, CA, USA). Each forward and reverse MSP primer was designed to cover between two to four CpG di-nucleotide repeats (Table 1). Universal methylated mouse DNA standard and control primers (Zymo research, CA, USA) were used to access the efficiency of bisulfite – mediated conversion of DNA. The PCR conditions were as follows: 1 cycle at 95°C for 5 min; 40 cycles at 95°C for 30 s, 30 s at the desired annealing temperature (48°C and 49°C for methylated and unmethylated primers for mir-9-1, mir-9-2 and mir-9-3 respectively), 72°C for 1 min; and 1 cycle at 72°C for 7 min. PCR samples were resolved by electrophoresis in 2% agarose gel and visualized with ethidium bromide.
Table 1.
Methylation-specific PCR primer sequence
| Primer set | Forward primer, 5′→3′ | CpG di-nucleotides | Reverse primer, 5′→3′ | CpG di-nucleotides |
|---|---|---|---|---|
| MSP-9-1-M | CGT TTT TTT GAG GTT TCG TC | 3 | GTA ATC TCG AAT CGC TCG AC | 2 |
| MSP-9-1-U | TTT TGT TTT TTT GAG GTT TTG TT | TCC ATA ATC TCA AAT CAC TCA AC | ||
| MSP-9-2-M | GCG TTA AGG AGG TAA AAG GTC | 2 | TAA CGA CGA CGA CAA CAA C | 3 |
| MSP-9-2-U | TTA GTG TTA AGG AGG TAA AAG GTT | TAA CAA CAA CAA CAA CAA CAA C | ||
| MSP-9-3-M | TAT TCG TCG GTA TCG TTG TC | 4 | CTC TAA CTC TCC GCG TAC G | 3 |
| MSP-9-3-U | TTT TAT TTG TTG GTA TTG TTG TT | CCC CTC TAA CTC TCC ACA TAC A |
Statistical Analyses
For data obtained from real-time RT-PCR, statistical analyses were performed on ΔCT values. The data were analyzed using one– or two-way ANOVA; and planned, pair-wise comparisons were conducted using t-tests when applicable (GraphPad Prism version 5.0 for Windows, San Diego, CA; p-value *<0.05, **<0.01, ***<0.001). Data were expressed both as ΔCT to represent the original data, and as fold-change (2ΔΔCT), to visually represent the relationship between exposure condition and gene expression.
Results
Embryonic ethanol exposure suppresses the expression of MiR-9 in vivo (zebrafish)
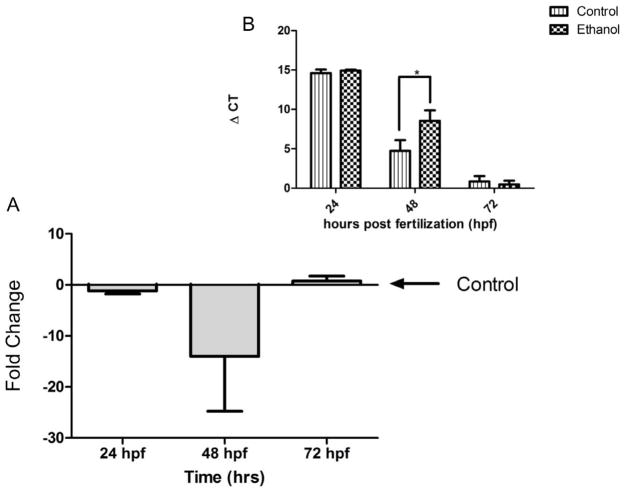
We (Balaraman et al., 2012; Sathyan et al., 2007), and others (Tal et al., 2012; Wang et al., 2009), showed that ethanol suppresses miR-9 expression during development. In the in vivo zebrafish model, we validated this finding. Following a four-hour period of ethanol exposure during gastrulation (3.5–7.5 hpf), zebrafish embryos were collected at 24, 48, or 72 hpf to assay miR-9 expression. The analysis showed a significant main effect of age of the embryo (24 vs. 48 vs. 72 hpf) on miR-9 expression, [F(2, 46)=16.29, p < 0.001]. With regard to ethanol’s effects, planned comparisons showed that ethanol significantly down-regulated miR-9 at 48 hpf [t(16)=3.46, p < 0.005], but not at other embryonic ages. The data suggest that early exposure to ethanol during gastrulation (~3.5 to 7.5 hpf) results in a delayed decrease in miR-9 expression at 48 hpf (Figure 1A shows fold-change and inset, 1B, shows the ΔCT).
Figure 1.
MiR-9 Expression is down-regulated at 48 hpf following early Ethanol exposure in zebrafish embryos. A) Ethanol exposure resulted in a ~14-fold decrease in miR-9 expression in zebrafish embryos, compared to controls at 48 hpf. No changes to miR-9 expression observed at 24 or 72 hpf. B) ΔCT value comparisons (increased ΔCT indicates lower miR-9 expression). Data expressed as mean±SEM, n=7–9. Asterisks indicate statistically significant comparisons, p<0.05.
In situ hybridization (Figure 2a) shows that at 48hpf miR-9 is expressed in the developing zebrafish, in brain and yolk sac (blue reaction product, Figure 2 a and c). No hybridization was observed with the nonsense control probe (Figure 2b). In control zebrafish, within the cephalic region specifically, miR-9 expression was observed throughout the forebrain, with a ridge of hybridization observed adjacent and posterior to the midbrain/hindbrain boundary (MHB, Figure 2c, white arrow), similar to that observed elsewhere (Leucht et al., 2008). Following ethanol exposure, zebrafish that exhibited the least anatomical defects nevertheless exhibited a disorganized pattern of miR-9 expression including the appearance of multiple boundary-like ridges of miR-9 (red arrows), anterior to the MHB. This mis-expression of miR-9 suggests defects in spatial patterning of the developing brain. As observed previously in a variety of animal models, and in humans, ethanol exposure also resulted in varying degrees of microcephaly accompanied by either microphthalmia or anophthalmia. No neural miR-9 expression was observed in any of these fetuses (Figure 2 e–h), and the ridge of miR-9 expression adjacent to the MHB was similarly abrogated. Similarly, ethanol exposure resulted in loss of miR-9 hybridization product in the yolk sac compared to unexposed controls (Figure 2 i vs. j and k, arrows point to posterior yolk sac specifically)..
Figure 2.
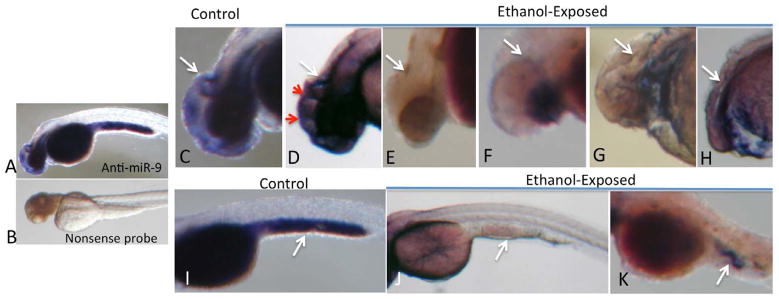
In situ hybridization for miR-9 at 48 hpf. Specific hybridization, visualized by alkaline-phosphates linked histochemistry (blue color product), was detected with an anti-miR-9 probe (A) but not with a nonsense control probe (B). In control zebrafish, specific hybridization was localized to the brain (D) and yolk sac (I). Within the brain (C), hybridization was observed in telencephalon and diencephalon, with a prominent hybridization ridge (white arrow) adjacent to the midbrain/hindbrain boundary. Ethanol exposure resulted aberrant miR-9 expression along ectopic ridges (D, red arrows) in zebrafish exhibiting the least dysmorphology. Ethanol exposure also resulted in a range of microcephaly, microphthalmia and anophthalmia (D–H), with a complete loss in neural miR-9 expression, including loss of hybridization adjacent to the midbrain/hindbrain boundary in microcephalic zebrafish. MiR-9 expression was also observed in yolk sac (I, white arrow points to hybridization in the posterior yolk sac). Ethanol exposure results in loss of miR-9 expression (J,K).
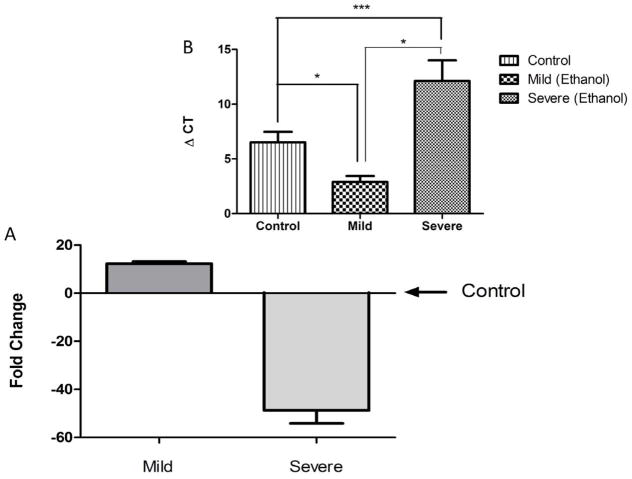
We next verified the observed association between severity of ethanol-induced defects and miR-9 expression by real-time RT-PCR. Ethanol exposed embryos were assessed for the presence of six readily scored anatomical defects (cyclopia, pericadial/yolk sac edema, truncation of tail, vertebral curvature, and cephalic/MHB defects) at 48 hpf, and sorted into two groups, unaffected to mildly affected (<2 defects), and severely affected (>3 defects). MiR-9 expression in each group, as determined by RT-PCR, was then compared to untreated controls. Our data indicate that ethanol exposure, whether resulting in mildly affected or severely affected embryos, resulted in a statistically significant alteration in miR-9 expression relative to control animals (ΔCT, F(2,11)=21.77, p<0.0001). Surprisingly, the mildly affected embryos exhibited a 12-fold increase in miR-9 expression (p<0.02, Figure 3), whereas the severely affected animals, with >3 scored defects, exhibited a 48-fold suppression of miR-9 expression relative to untreated controls (p<0.004). The group classified as unaffected to mildly affected was also statistically different from the heavily affected group (p<0.00004).
Figure 3.
Real-time RT-PCR analysis indicates that at 48 hpf, ethanol exposed zebrafish that show mild anatomical defects (< 2 scored defects) exhibit a significant increase (fold change relative to the mean of controls) in miR9 expression, whereas severely affected animals (>3 scored defects) exhibited significantly decreased miR-9 expression compared to non exposed controls. Inset graph shows ΔCT relative to U6 snRNA. Lower ΔCTs indicate higher levels of miRNA expression compared to higher ΔCTs. Data expressed as mean ± SEM, n = 3–7/group. *, p<0.05; ***, p<0.0001.
Loss of miR-9 recapitulates anatomical phenotypes associated with embryo exposure to ethanol
Since ethanol exposure during gastrulation significantly suppressed miR-9, and because miR-9 is expressed in neural and non-neural tissues during embryonic development, we hypothesized that suppression of miR-9 would mimic the gross anatomical defects observed in the severely affected embryos following ethanol exposure. Antisense, anti-miR-9 morpholinos injected at the one- to two-cell stage of zebrafish development resulted in significant 10-fold decrease in miR-9 expression at 48 hpf compared to nonsense control morpholino ([t(10) = 2.25, p < 0.05]). The suppression of miR-9 in zebrafish embryos in vivo resulted in anatomical defects including microcephaly and anophthalmia at 24 and 48 hpf (Figure 4 a,b, arrows), similar to those observed following exposure to ethanol during the gastrulation (Figure 2f–h). Other defects that overlapped with ethanol exposure phenotypes included tail truncation, pericardial edema and vertebral curvature (data not shown). No gross structural changes were observed in control embryos injected with a mismatched control oligonucleotide when compared with non-manipulated controls (Figure 4 c–f).
Figure 4.
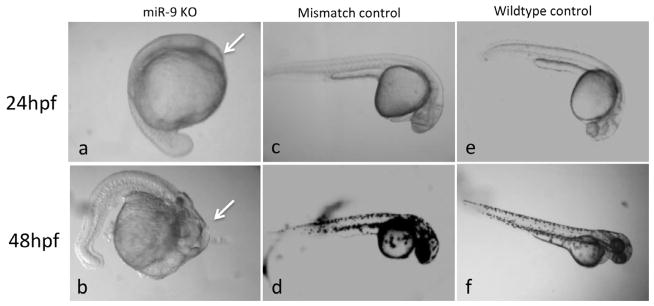
MiR-9 knockdown mimics ethanol phenotype at 24 and 48 hpf. (a,b), sample miR-9 knockdown phenotypes at 24 and 48 hpf showing microcephaly and disorganization of cranial structures (arrows). Embryos injected with 2 mM mismatched morpholinos (c,d) were not different from wildtype controls (e,f) at the corresponding developmental stages.
MiR-9 target gene regulation by ethanol
MiRNAs silence mRNAs through transcript degradation or translational repression. Since ethanol suppressed miR-9, we predicted that target mRNAs would be induced following ethanol exposure. We assessed the expression of three important developmental genes, FOXP2, FGFR-1 and MECP2 (Haesler et al., 2004; Hendrich and Tweedie, 2003; Ota et al., 2010; Riley et al., 2007; Shu et al., 2007). FOXP2 is predicted to be a strong, vertebrate-conserved target of miR-9, with two 7mer-m8 sites in the proximate 3′UTR (www.targetscan.org), while FGFR-1 has been shown to be a miR-9 target in the zebrafish (Leucht et al., 2008). The 3′UTR of MECP2 in contrast, exhibits a presumptive miR-9 binding site in the human, but is not predicted to be a miR-9 target in zebrafish, or mouse, and was therefore used to compare developmentally regulated miR-9 targets, to a non-targeted, developmentally important gene. Contrary to our hypothesis, ethanol exposure did not lead to an increase in FOXP2, or FGFR-1 transcripts in zebrafish embryos. Rather, planned comparisons showed that ethanol transiently decreased the expression of FGFR-1 [t(6) = 4.03, p < 0.01], as well as FOXP2 [t(8) =2.80, p < 0.05] mRNA transcripts at 48 hpf. Levels of mRNA for the non-miR-9 target MECP2 were unchanged at all developmental stages regardless of ethanol treatment (Figure 5).
Figure 5.
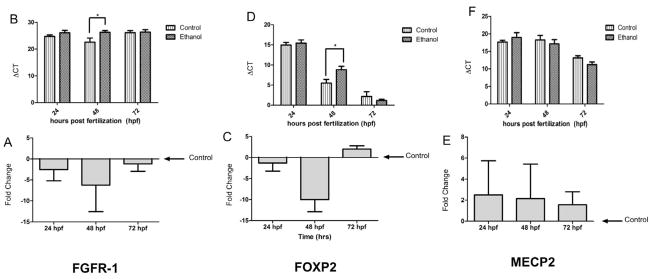
FGFR-1 and FOXP2 are downregulated following ethanol exposure in zebrafish embryos. A, C and E show fold-change relative to control and B, D and F show ΔCT (higher ΔCTs indicate lower mRNA expression). Following ethanol exposure, FGFR-1 (A and B) and FOXP2 (C and D) are downregulated at 48hpf while MECP2 (E and F) remains unchanged. All mRNA levels remain unchanged at 24 and 72hpf. Data expressed as mean±SEM, n=4–5/group. Asterisks indicate statistically significant comparisons of ΔCT, p<0.05.
Ethanol selectively promotes increased methylation of CpG dinucleotides in mammalian miR-9 genes
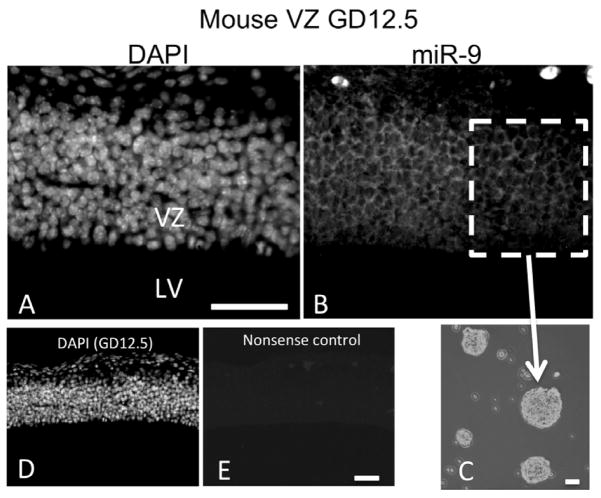
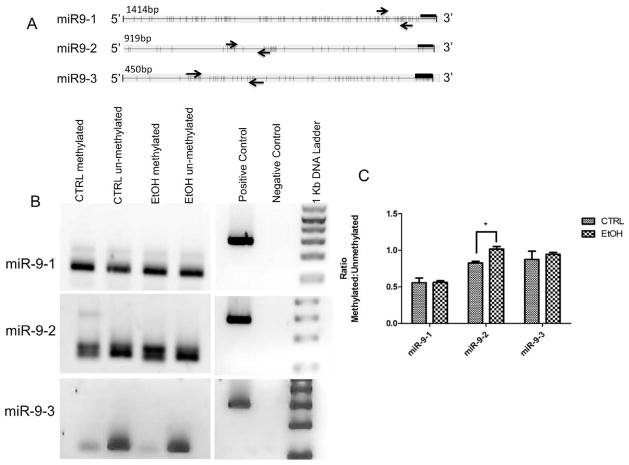
We hypothesized that ethanol suppression of mature miR-9 coincides with changes in methylation at one or more miR-9 genes. Though mature zebrafish miR-9 is identical to mammalian miR-9, mammalian genomes contain only three miR-9 genes, while zebrafish contain seven. Consequently, epigenetic modifications of zebrafish miR-9 genes have limited implications for the regulation of mammalian miR-9 transcription. We therefore used a mammalian model to study miR-9 gene methylation. In situ hybridization (Figure 6) shows that miR-9 is expressed in ventricular zone (VZ) neural progenitors in the GD12.5 fetal murine brain. Moreover, we previously showed in two independently replicated experiments, that ethanol at 70 mM significantly decreased miR-9 expression in mNSCs derived from the fetal VZ (Balaraman et al., 2012; Sathyan et al., 2007). We therefore used mNSCs (Figure 5c), to study methylation of the mammalian genomic miR-9 loci. In silico analyses (UCSC genome browser; genome.ucsc.edu) revealed presumptive CpG dinucleotide repeats associated with identified mammalian transcription factor binding domains (ENCODE Project Consortium, 2011) at the 5′-end of each of the three pre-miR-9 genes. Methylation-specific PCR analysis indicated that under control conditions, the miR-9-3 locus exhibits lower levels of methylation compared to the miR-9-1 and miR-9-2 loci (Figure 7A). Following a 5-day exposure to 70mM ethanol, methylation was specifically and significantly increased at CpG dinucleotides associated with the miR-9-2 (t(5) = 3.92, p < 0.05), but not the miR-9-1 or miR-9-3 loci (Figure 7B).
Figure 6.
MiR-9 is expressed in the fetal telencephalon during mammalian development, and in neural and non-neural tissue during zebrafish development. A) DAPI stain localizes to nuclei. B) MiR-9 is expressed in the ventricular zone of the fetal murine brain at gestational day 12.5. C) Image of cultured neurospheres derived from the GD 12.5 mouse ventricular zone and cultured ex vivo. D–E) DAPI staining to visualize nuclei (D) and nonsense control (E) and in the fetal murine brain at GD 12.5, scale bar indicates 40 um. Abbreviations, LV, Lateral ventricle; VZ, ventricular zone. Scale bars, 40 um;
Figure 7.
Ethanol effects on methylation at miR-9 loci changes with ethanol exposure. A) Maps of mammalian miR9 gene loci depicting the proximal 5′ sequence with predicted CpG dinucleotides represented as vertical bars. Each 5′ region coincides with assessed RNA polymerase II and transcription factor binding sites (UCSC genome browser, genome.ucsc.edu). Arrows indicate forward and reverse methylation specific primers Methyl Primer Express ® software version 1.0 (Applied Biosystems, CA, USA). Black bars indicate location of the miR-9 gene. B) Size fractionation of PCR products shows that the 5′ regions of miR-9 genes exhibit evidence for differential methylation. MSP primers detect high levels of methylation in miR-9-1 and miR-9-2 promoter regions compared to the miR-9-3 locus. C) Quantitative analysis of miR-9 loci methylation. Graph represents ratio of methylated to unmethylated CpG dinucleotides shown in A (n=3–4/group). Ethanol exposure leads to a significant increase in methylation at the miR-9-2 locus.
Ethanol promotes accumulation of specific mammalian pre-miR-9 transcripts
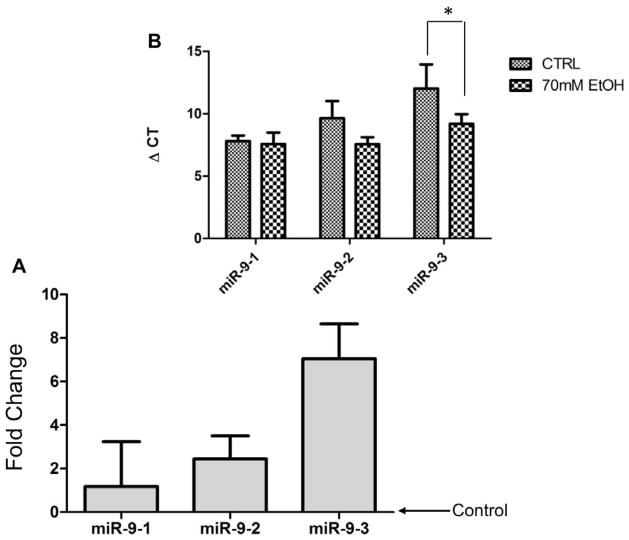
Pre-miRNAs are short-lived, intermediate transcripts that are generated by Drosha/DGCR8 processing and are in turn, rapidly processed by Dicer complexes into mature miRNAs (for a review, see (Miranda et al., 2010)). Normally, pre-miRNA transcripts are present in low abundance in cells (Kim, 2005) and the accumulation of intermediate species is indicative of disruptions in miRNA processing (Tomari and Zamore, 2005). We therefore tested the hypothesis that a decrease in mature miR-9 was associated with disrupted miRNA processing leading to accumulation of pre-miR-9 transcripts. Since the identical mature miR-9 is processed from three independent genes in mammals, we examined the expression of pre-miR-9-1, -9-2 and -9-3 following ethanol exposure. mNSCs were treated with 70 mM ethanol, a dose previously shown to suppress miR-9 expression (Balaraman et al., 2012; Sathyan et al., 2007), for 1 or 5 days. The results showed that under normal conditions pre-miR-9-1 transcripts were 18-fold more abundant than pre-miR-9-2 transcripts, which were in turn, 47-fold more abundant than pre-miR-9-3 transcripts. Ethanol exposure resulted in a significant increase in pre-miR-9-3 transcript levels compared to control, (F(1, 17) =7.44, p < 0.05, Figure 8). There was a trend towards an increase, though not statistically significant (F(1, 17) =3.13, p<0.095), in pre-miR-9-2 transcript levels. Pre-miR-9-1 was not altered by ethanol. Moreover, there was no significant change in the expression of any of the pre-miR-9 transcript isoforms with length of exposure to ethanol (i.e. 1 vs. 5 days).
Figure 8.
Pre-mir-9 transcripts accumulate following chronic ethanol exposure in mammals. A and B) Following 70mM ethanol exposure, miR-9-3 exhibits significant accumulation, while miR-9-2 exhibits a near-significant increase in accumulation. In both paradigms, miR-9-1 levels remained unaffected by ethanol. Asterisks indicate statistically significant comparisons of ΔCT, p<0.05 (lower ΔCT indicates increased expression of pre-miRNA), n=4–5/group. Data from acute and chronic ethanol treatment conditions were combined because, while there was a main effect of ethanol exposure on Pre-miR-9 accumulation, there was not a statistically significant effect of time.
Discussion
MiRNAs may be key to understanding the detrimental effects of prenatal ethanol exposure. This current study confirms previous observations that ethanol suppresses miR-9 during development (Balaraman et al., 2012; Sathyan et al., 2007; Tal et al., 2012) and demonstrates that ethanol produces developmental defects in zebrafish that span a spectrum of severity similar to that observed in mouse models (Randall and Taylor, 1979), and in human populations (Roebuck et al., 1998). Our data demonstrate that loss of miR-9 is associated with severe ethanol-induced developmental defects. We also present novel data suggesting that, miR-9 loss of function in the zebrafish embryo recapitulates several morphological features associated with ethanol exposure, including microcephaly. This study also presents evidence that miR-9 suppression is associated with increased methylation of specific miR-9 genes and decreased processing of pre-miR-9 transcripts.
In our study, the timing of ethanol exposure, in the zebrafish embryo coincides with the initiation of the mid-blastula transition (MBT) and overlapping gastrulation, and results in delayed miR-9 suppression and morphological defects. During the MBT, maternal mRNAs are cleared by newly synthesized embryonic miRNAs (Giraldez et al., 2006), the zygote initiates de novo gene transcription to control subsequent embryo development (Kane and Kimmel, 1993), and asynchronous cell division is initiated. During subsequent gastrulation, the epithelial-to-mesenchymal transition (EMT, (Nakaya and Sheng, 2008)) results in the emergence of a distinct ectoderm, mesoderm and endoderm. The underlying complexity of the MBT and gastrulation, including emerging desynchronization in the timing of mitotic events among neighboring progenitor cells, may contribute to the varying severity of structural abnormalities observed following ethanol exposure.
MiR-9 is a neural-enriched miRNA that is also expressed in mesoderm and endoderm derived tissues (Saunders et al., 2010; Zhang et al., 2011). Like ethanol (Mooney et al., 2004; Santillano et al., 2005), miR-9 influences neural progenitor cell migration and neurogenesis (Delaloy et al., 2010). Our observation that ethanol exposure during gastrulation resulted in a delayed, transient suppression of miR-9 at 48 hpf, a period associated with neural tube closure and neural crest migration (Kimmel et al., 1995), suggests that ethanol exerts temporally displaced control over miRNA expression, to perhaps influence important downstream neurodevelopmental programs. MiR-9 expression was lost only in severely ethanol-affected animals, i.e., those that also exhibited microcephaly and varying degrees of microphthalmia. In zebrafish with few or no scored morphological defects, miR-9 expression was actually significantly increased, though in situ hybridization showed that in these instances, miR-9 was still mis-expressed, perhaps defining additional, aberrant molecular boundaries in the developing brain. The loss of miR-9 expression adjacent to the MHB in microcephalic ethanol-exposed animals warrants further investigation, because of the importance of the MHB as an organizer for hindbrain development. Recent evidence suggests that miR-9 maintains the organizer function of the MHB (Leucht et al., 2008), and consequently, loss of miR-9 may facilitate aberrant development of known ethanol-sensitive hindbrain structures like the cerebellum. Conversely, the increase in miR-9 in animals that were less affected by alcohol may represent a neuroprotective adaptation.
Zebrafish, like humans, are genetically heterogeneous. While the source of variation in alcohol teratogenesis and miR-9 regulation in zebrafish is unknown, case reports of prenatal ethanol exposure in human monozygotic and dizygotic twins have previously documented a genetic contribution to variations in susceptibility (Streissguth and Dehaene, 1993). It is likely that common genetic susceptibility factors contribute to both miR-9 regulation and to alcohol-induced teratology. Moreover, our data showing that the miR-9 knockdown recapitulated the severe end of the range of alcohol-induced anatomical defects, including microcephaly, suggests that disruption of miR-9 may constitute part of the pathway to teratogenesis.
Emerging evidence suggests that mammalian miR-9 genes are regulated epigenetically by DNA methylation (Lehmann et al., 2007). Though mature miR-9 is identical in all vertebrates, the number of miR-9 genes varies between mammals and teleosts. We therefore used a mammalian fetal mNSC model, wherein ethanol also suppresses miR-9 expression (Balaraman et al., 2012; Sathyan et al., 2007), to assess evidence for differential methylation sensitivity of mammalian miR-9 genes in developing neural tissue. Our data show that ethanol exposure increases methylation only in the mammalian miR-9-2 gene, indicating that the miR-9-2 locus is specifically sensitive to ethanol. More comprehensive screens will be required to fully assess the vulnerability of the miR-9-2 locus. However, these data have significant developmental implications. In the fetal telencephalon, all miR-9 genes are expressed in neural precursors of the ventricular zone (the neurosphere model equivalent), whereas miR-9-2 alone is expressed in maturing neurons of the cerebral cortical plate (Shibata et al., 2011). Therefore, transcriptional control of miR-9-2 appears to be specifically critical for miR-9 function in differentiating neurons, whereas miR-9-1 and miR-9-3 might preferentially contribute to miR-9 function in neural stem cells. Deletion of miR-9-2/-9-3 leads to growth retardation and microencephaly (Shibata et al., 2011), features associated with fetal ethanol exposure. Collectively, these data suggest that ethanol-induced methylation at the miR-9-2 gene may interfere with mature miR-9 regulated neuronal maturation.
Pre-miRNAs are processed by Dicer, into mature miRNAs, resulting in low steady-state levels of pre-miRNAs. Our data show that ethanol exposure led to a significant accumulation of pre-miR-9-3 and a marginal accumulation of pre-miR-9-2, suggesting altered processing of pre-miR-9 transcripts. Altered processing of some pre-miRNA transcripts has recently been found to predict glioma stage progression (Moore et al., 2012) and may well be a factor in the etiology of neurodevelopmental disorders as well. The mechanisms underlying the regulation of pre-miRNA processing are poorly understood. A small but important body of evidence indicates that RNA editing by adenosine deaminases selectively interferes with Dicer processing of some pre-miRNAs to mature miRNAs, leading to the accumulation of these pre-miRNAs (e.g., (Kawahara et al., 2007)). Additionally, miRNA-binding proteins like lin28 can selectively inhibit processing of the let7 miRNA family (Viswanathan et al., 2008) without interfering with processing of other miRNAs. The recruitment of such mechanisms by ethanol needs further investigation, and may ultimately explain the selective vulnerability of pre-miR-9-3 but not pre-miR-9-1 to ethanol. It is important to note that in the current study, the timing and dosing of ethanol exposure is not identical between the in vivo and ex vivo models, though in both models, our current and previous (Balaraman et al., 2012; Sathyan et al., 2007) data show that ethanol suppresses the expression of miR-9. Such differences necessitate caution when extrapolating the results from one paradigm to the other. It will be important to determine the effect of developmental timing and ethanol dosage on pre-miR accumulation, as well as the extent to which ethanol interferes with miRNA processing mechanisms. Together, the methylation and pre-miRNA accumulation data do advance a hypothesis that the altered epigenetic landscape of one miR-9 gene combined with disrupted miRNA processing from a second miR-9 gene promote a common outcome, i.e., suppression of mature miR-9 by ethanol.
We further tested in the zebrafish model, the extent to which ethanol regulated presumptive miR-9 targets FGFR-1 and FOXP2, compared to the non-targeted MECP2. These mRNAs were selected because of their association with pervasive developmental disorders in humans (Allan et al., 2008; Haesler et al., 2004; Hagberg, 2002; Poot et al., 2009; Riley et al., 2007). Because ethanol suppressed miR-9, we predicted ethanol would induce FGFR-1 and FOXP2, but not MECP2. Contrary to our prediction, ethanol significantly suppressed miR-9 mRNA targets FGFR-1 and FOXP2, while the non miR-9 target, MECP2, remained unaffected. These data suggest that ethanol specifically suppresses mRNAs that are also targeted by miR-9, and in doing so, uncouples the regulatory function of miR-9. This outcome is perhaps not surprising, since the stability of FGFR-1 and FOXP2 mRNAs are likely controlled by multiple miRNAs that may over-compensate for the loss of miR-9. FGFR-1 is important for stem cell differentiation and migration (Bansal et al., 2003) while FOXP2 promotes neuronal differentiation in mouse models, in part, by binding to promoters of important neuron-specific miRNAs like miR-124 (Vernes et al., 2011). These data collectively support the hypothesis that ethanol interferes with miR-9 dependent neural stem cell maturation and with genes that support the maturation of neurons and neural networks.
Mature miR-9 is evolutionarily conserved in vertebrates, suggesting that its developmental functions are also conserved. The vulnerability of miR-9 to ethanol in both teleosts and mammals indicates that miR-9 may mediate broader developmental vulnerability of vertebrate organisms to environmental disrupters other than ethanol, a concept supported by studies demonstrating that miR-9 suppression is associated with retinoic acid induced spina-bifida (Zhao et al., 2008), and induction, associated with nicotine exposure in fetal mNSCs (Balaraman et al., 2012). These mechanistic relationships between miR-9 and developmental disorders, further suggest that this miRNA may be a clinically relevant target for future studies on the prevention and treatment of teratogen mediated birth defects.
Acknowledgments
We would like to thank Drs. Bruce Riley and Mahesh Padanad, for their expertise and technical instruction regarding the zebrafish model. This research was funded by NIH Grant # AA013440 to RCM and by a fellowship from the Texas Brain and Spine Institute to DLP.
Abbreviations
- EMT
epithelial-mesenchymal transition
- BMT
mid-blastula transition
- MHB
midbrain-hindbrain boundary, mNSCs, mouse neurosphere cultures
References
- Adachi J, Mizoi Y, Fukunaga T, Ogawa Y, Ueno Y, Imamichi H. Degrees of alcohol intoxication in 117 hospitalized cases. J Stud Alcohol. 1991;52:448–453. doi: 10.15288/jsa.1991.52.448. [DOI] [PubMed] [Google Scholar]
- Allan AM, Liang X, Luo Y, Pak C, Li X, Szulwach KE, Chen D, Jin P, Zhao X. The loss of methyl-CpG binding protein 1 leads to autism-like behavioral deficits. Hum Mol Genet. 2008;17:2047–57. doi: 10.1093/hmg/ddn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaraman S, Winzer-Serhan U, Miranda RC. Opposing actions of ethanol and nicotine on microRNAs are mediated by nicotinic acetylcholine receptors in fetal cerebral cortical-derived neural progenitor cells. Alcohol Clin Exp Res. 2012;36:1669–1677. doi: 10.1111/j.1530-0277.2012.01793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal R, Lakhina V, Remedios R, Tole S. Expression of FGF receptors 1, 2, 3 in the embryonic and postnatal mouse brain compared with Pdgfralpha, Olig2 and Plp/dm20: implications for oligodendrocyte development. Dev Neurosci. 2003;25:83–95. doi: 10.1159/000072258. [DOI] [PubMed] [Google Scholar]
- Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A. 2009;106:5282–7. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradfield JY, West JR, Maier SE. Uptake and elimination of ethanol by young zebrafish embryos. Neurotoxicol Teratol. 2006;28:629–633. doi: 10.1016/j.ntt.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Camarillo C, Kumar LS, Bake S, Sohrabji F, Miranda RC. Ethanol regulates angiogenic cytokines during neural development: evidence from an in vitro model of mitogen-withdrawal-induced cerebral cortical neuroepithelial differentiation. Alcohol Clin Exp Res. 2007;31:324–335. doi: 10.1111/j.1530-0277.2006.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarillo C, Miranda RC. Ethanol exposure during neurogenesis induces persistent effects on neural maturation: evidence from an ex vivo model of fetal cerebral cortical neuroepithelial progenitor maturation. Gene Expr. 2008;14:159–171. [PMC free article] [PubMed] [Google Scholar]
- Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, Klaus S, Snyman H, Hannon GJ, Bork P, Arendt D. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463:1084–1088. doi: 10.1038/nature08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defendi GL. Fetal Alcohol Spectrum Disorder: How to Recognize the Various Manifestations. Consultant for Pediatricians. 2010;9:8. [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll CD, Streissguth AP, Riley EP. Prenatal alcohol exposure: comparability of effects in humans and animal models. Neurotoxicol Teratol. 1990;12:231–237. doi: 10.1016/0892-0362(90)90094-s. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium . A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Haesler S, Wada K, Nshdejan A, Morrisey EE, Lints T, Jarvis ED, Scharff C. FoxP2 expression in avian vocal learners and non-learners. J Neurosci. 2004;24:3164–3175. doi: 10.1523/JNEUROSCI.4369-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg B. Clinical manifestations and stages of Rett syndrome. Ment Retard Dev Disabil Res Rev. 2002;8:61–65. doi: 10.1002/mrdd.10020. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–77. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The Zebrafish Midblastula Transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lehmann U, Hasemeier B, Romermann D, Muller M, Langer F, Kreipe H. Epigenetic inactivation of microRNA genes in mammary carcinoma. Verh Dtsch Ges Pathol. 2007;91:214–220. [PubMed] [Google Scholar]
- Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RC, Pietrzykowski AZ, Tang Y, Sathyan P, Mayfield D, Keshavarzian A, Sampson W, Hereld D. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcohol Clin Exp Res. 2010;34:575–587. doi: 10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney SM, Siegenthaler JA, Miller MW. Ethanol Induces Heterotopias in Organotypic Cultures of Rat Cerebral Cortex. Cereb Cortex. 2004;14:1071–1080. doi: 10.1093/cercor/bhh066. [DOI] [PubMed] [Google Scholar]
- Moore LM, Kivinen V, Liu Y, Annala M, Cogdell D, Liu X, Liu CG, Sawaya R, Yli-Harja O, Shmulevich I, Fuller GN, Zhang W, Nykter M. Transcriptome and Small RNA Deep Sequencing Reveals Deregulation of miRNA Biogenesis in Human Glioma. J Pathol. 2012 doi: 10.1002/path.4109. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya Y, Sheng G. Epithelial to mesenchymal transition during gastrulation: an embryological view. Dev Growth Differ. 2008;50:755–766. doi: 10.1111/j.1440-169X.2008.01070.x. [DOI] [PubMed] [Google Scholar]
- Ota S, Tonou-Fujimori N, Nakayama Y, Ito Y, Kawamura A, Yamasu K. FGF receptor gene expression and its regulation by FGF signaling during early zebrafish development. Genesis. 2010;48:707–716. doi: 10.1002/dvg.20682. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Bolding K, Riley BB. Zebrafish fgf3 and fgf8 encode redundant functions required for otic placode induction. Dev Biol. 2001;235:351–365. doi: 10.1006/dbio.2001.0297. [DOI] [PubMed] [Google Scholar]
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, Siegelmann HT, Treistman SN. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59:274–287. doi: 10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, Regazzi R. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J Biol Chem. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- Poot M, Beyer V, Schwaab I, Damatova N, Van’t Slot R, Prothero J, Holder SE, Haaf T. Disruption of CNTNAP2 and additional structural genome changes in a boy with speech delay and autism spectrum disorder. Neurogenetics. 2009;11:81–89. doi: 10.1007/s10048-009-0205-1. [DOI] [PubMed] [Google Scholar]
- Prock TL, Miranda RC. Embryonic cerebral cortical progenitors are resistant to apoptosis, but increase expression of suicide receptor DISC-complex genes and suppress autophagy following ethanol exposure. Alcohol Clin Exp Res. 2007;31:694–703. doi: 10.1111/j.1530-0277.2007.00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall CL, Taylor WJ. Prenatal ethanol exposure in mice: teratogenic effects. Teratology. 1979;19:305–311. doi: 10.1002/tera.1420190305. [DOI] [PubMed] [Google Scholar]
- Reimers MJ, Flockton AR, Tanguay RL. Ethanol- and acetaldehyde-mediated developmental toxicity in zebrafish. Neurotoxicol Teratol. 2004;26:769–781. doi: 10.1016/j.ntt.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Riley BM, Mansilla MA, Ma J, Daack-Hirsch S, Maher BS, Raffensperger LM, Russo ET, Vieira AR, Dode C, Mohammadi M, Marazita ML, Murray JC. Impaired FGF signaling contributes to cleft lip and palate. Proc Natl Acad Sci U S A. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley EP, McGee CL. Fetal alcohol spectrum disorders: an overview with emphasis on changes in brain and behavior. Exp Biol Med (Maywood) 2005;230:357–365. doi: 10.1177/15353702-0323006-03. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Otero P, Roman-Gomez J, Vilas-Zornoza A, Jose-Eneriz ES, Martin-Palanco V, Rifon J, Torres A, Calasanz MJ, Agirre X, Prosper F. Deregulation of FGFR1 and CDK6 oncogenic pathways in acute lymphoblastic leukaemia harbouring epigenetic modifications of the MIR9 family. Br J Haematol. 2011;155:73–83. doi: 10.1111/j.1365-2141.2011.08812.x. [DOI] [PubMed] [Google Scholar]
- Roebuck TM, Mattson SN, Riley EP. A review of the neuroanatomical findings in children with fetal alcohol syndrome or prenatal exposure to alcohol. Alcohol Clin Exp Res. 1998;22:339–344. doi: 10.1111/j.1530-0277.1998.tb03658.x. [DOI] [PubMed] [Google Scholar]
- Santillano DR, Kumar LS, Prock TL, Camarillo C, Tingling JD, Miranda RC. Ethanol induces cell-cycle activity and reduces stem cell diversity to alter both regenerative capacity and differentiation potential of cerebral cortical neuroepithelial precursors. BMC Neurosci. 2005;6:59. doi: 10.1186/1471-2202-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyan P, Golden HB, Miranda RC. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci. 2007;27:8546–8557. doi: 10.1523/JNEUROSCI.1269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, Sharma AD, Tawney J, Nakagawa M, Okita K, Yamanaka S, Willenbring H, Verdin E. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY) 2010;2:415–431. doi: 10.18632/aging.100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W, Lu MM, Zhang Y, Tucker PW, Zhou D, Morrisey EE. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development. 2007;134:1991–2000. doi: 10.1242/dev.02846. [DOI] [PubMed] [Google Scholar]
- Streissguth AP, Dehaene P. Fetal alcohol syndrome in twins of alcoholic mothers: concordance of diagnosis and IQ. Am J Med Genet. 1993;47:857–861. doi: 10.1002/ajmg.1320470612. [DOI] [PubMed] [Google Scholar]
- Tal TL, Franzosa JA, Tilton SC, Philbrick KA, Iwaniec UT, Turner RT, Waters KM, Tanguay RL. MicroRNAs control neurobehavioral development and function in zebrafish. FASEB J. 2012;26:1452–1461. doi: 10.1096/fj.11-194464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. MicroRNA biogenesis: drosha can’t cut it without a partner. Curr Biol. 2005;15:R61–64. doi: 10.1016/j.cub.2004.12.057. [DOI] [PubMed] [Google Scholar]
- Ton C, Lin Y, Willett C. Zebrafish as a model for developmental neurotoxicity testing. Birth Defects Res A Clin Mol Teratol. 2006;76:553–567. doi: 10.1002/bdra.20281. [DOI] [PubMed] [Google Scholar]
- Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, Taylor JM, Ho J, Mombereau C, Brewer A, Lowy E, Nicod J, Groszer M, Baban D, Sahgal N, Cazier JB, Ragoussis J, Davies KE, Geschwind DH, Fisher SE. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 2011;7:e1002145. doi: 10.1371/journal.pgen.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Zhang Z, Li Q, Yang R, Pei X, Xu Y, Wang J, Zhou SF, Li Y. Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Hum Reprod. 2009;24:562–579. doi: 10.1093/humrep/den439. [DOI] [PubMed] [Google Scholar]
- Zhang F, Pomerantz JH, Sen G, Palermo AT, Blau HM. Active tissue-specific DNA demethylation conferred by somatic cell nuclei in stable heterokaryons. Proc Natl Acad Sci U S A. 2007;104:4395–4400. doi: 10.1073/pnas.0700181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chintalgattu V, Shih T, Ai D, Xia Y, Khakoo AY. MicroRNA-9 is an activation-induced regulator of PDGFR-beta expression in cardiomyocytes. J Mol Cell Cardiol. 2011;51:337–346. doi: 10.1016/j.yjmcc.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Sun DG, Wang J, Liu SR, Zhang CY, Zhu MX, Ma X. Retinoic acid downregulates microRNAs to induce abnormal development of spinal cord in spina bifida rat model. Childs Nerv Syst. 2008;24:485–492. doi: 10.1007/s00381-007-0520-5. [DOI] [PubMed] [Google Scholar]