Abstract
AIM: To investigate the dynamic features of insulin-like growth factor-I receptor (IGF-IR) expression in rat hepatocarcinogenesis, and the relationship between IGF-IR and hepatocytes malignant transformation at mRNA or protein level.
METHODS: Hepatoma models were made by inducing with 2-fluorenylacetamide (2-FAA) on male Sprague-Dawley rats. Morphological changes of hepatocytes were observed by pathological Hematoxylin and eosin staining, the dynamic expressions of liver and serum IGF-IR were quantitatively analyzed by an enzyme-linked immunosorbent assay. The distribution of hepatic IGF-IR was located by immunohistochemistry. The fragments of IGF-IR gene were amplified by reverse transcription-polymerase chain reaction, and confirmed by sequencing.
RESULTS: Rat hepatocytes after induced by 2-FAA were changed dynamically from granule-like degeneration, precancerous to hepatoma formation with the progressing increasing of hepatic mRNA or IGF-IR expression. The incidences of liver IGF-IR, IGF-IR mRNA, specific IGF-IR concentration (ng/mg wet liver), and serum IGF-IR level (ng/mL) were 0.0%, 0.0%, 0.63 ± 0.17, and 1.33 ± 0.47 in the control; 50.0%, 61.1%, 0.65 ± 0.2, and 1.51 ± 0.46 in the degeneration; 88.9%, 100%, 0.66 ± 0.14, and 1.92 ± 0.29 in the precancerosis; and 100%, 100%, 0.96 ± 0.09, and 2.43 ± 0.57 in the cancerous group, respectively. IGF-IR expression in the cancerous group was significantly higher (P < 0.01) than that in any of other groups at mRNA or protein level. The closely positive IGF-IR relationship was found between livers and sera (r = 0.91, t = 14.222, P < 0.01), respectively.
CONCLUSION: IGF-IR expression may participate in rat hepatocarcinogenesis and its abnormality should be an early marker for hepatocytes malignant transformation.
Keywords: Hepatoma, Insulin-like growth factor-I receptor, Immunohistochemistry, Gene amplification, Sequencing, Rat hepatoma model
Core tip: The abnormality of insulin-like growth factor-I receptor (IGF-IR) expression was progressively increased in hepatocarcinogenesis at mRNA or protein level, and there was a positive correlation between circulating blood and liver IGF-IR, suggesting that the over-expression of liver IGF-IR release into blood, and serum IGF-IR should be an early useful marker for monitoring malignant transformation of rat hepatocytes.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death, especially in the inshore area of the Yangtze River[1-3]. Multiple pathogenic factors, including infection with hepatitis B virus or hepatitis C virus, chronic hepatitis and cirrhosis with the subsequent multistage pathogenesis, have been extensively studied[4-6]. Recent advances in molecular genetics have identified a large number of activated or suppressed genes may play significant roles in HCC[7-9]. Insulin-like growth factor (IGF) signaling is specifically required for hepatoma progression[10], especially in IGF-II and IGF-I receptor (IGF-IR) which are the very important signaling contributors. Surgical liver transplantation or resection remains the mainstay of curative therapy for patients in the early stage of HCC[11,12].
Carcinoembryonic IGF-II was reported to be overexpressed at different stages of HCC development[12]. Integrative genomic analysis showed enrichment of IGF activation in the proliferation subclass of HCC. Effective blockage of IGF signaling with A12 provides the rationale for testing this therapy in clinical trials[13] and confirmed that it plays the biologic role primarily through IGF-IR[14]. Although the IGF-IR expression is distributed widely in multiple tissues, it is high in fetal liver but not in adult liver[10]. IGF-IR is an important regulatory factor necessary for cell growth and differentiation, provides mitogenic signal for tumor growth[15], exhibits a very potent anti-apoptotic activity[16], induces vascular endothelial growth factor expression, and able to maintain the cell phenotype transformation in carcinogenesis[17,18]. However, the features of IGF-IR expression during hepatocytes malignant transformation have not yet been reported. Thus, the objectives of the present study were to investigate the dynamic expression features of IGF-IR in rat hepatocarcinogenesis, and the relationship between IGF-IR expression and hepatocyte malignant transformation at mRNA or protein level.
MATERIALS AND METHODS
Hepatoma model and pathological analysis
Rat hepatoma models were made as described previously[13]. Briefly, 54 male Sprague-Dawley rats (4-wk-old, body weight 120-130 g) from the Experimental Center of Medical Animals, Nantong University, China were used to induce HCC development by fed foods with 0.05% 2-fluorenylacetamide (2-FAA, Sigma). Rats were monitored daily for survival and weight loss, recorded their clinical signs and sacrificed at different days. All procedures performed on the animals were conducted in accordance with the guidelines for experimental animals approved by the Animal Care and Use Committee of Nantong University, China. According to the plan, 5 mL of blood was drawn from rat hearts and serum was separated for IGF-IR test, and livers were used for pathology, immunohistochemistry, total RNA extraction, and quantitative analysis of IGF-IR expression. Rat liver tissues were fixed in 10% formaldehyde solution, sectioned after dehydration, and paraffin embedding and then performed with HE staining to observe morphological changes under microscope.
Total RNA extraction
Liver tissue (20 mg) was homogenized on ice after adding 1.0 mL of TRIzol reagent in a glass homogenizer which was made RNase-free by 0.1% DEPC water. One milliliter slurry was transferred into the tubes and centrifuged at 15000 rpm for 10 min at 4 °C. 0.8 mL of the supernatant was put into another tube, and then 0.2 mL of chloroform was added into the tube, mixed by vortexing 2 min, centrifuged at 10000 rpm for 10 min at 4 °C, 0.3 mL of the supernatant was collected into another RNase-free tube, equivalent 100% isopropanol and then another 0.5 mL of 50% isopropanol were added, mixed gently for 30 s, centrifuged at 15000 rpm for 5 min at 4 °C. The supernatant was removed, the RNA pellets were washed with 1.0 mL of 75% ethanol, mixed, and centrifuged at 15000 rpm for 5 min at 4 °C. The RNA pellets were air dried 3 min at room temperature, reconstituted in 20 μL of 0.1% lithium chloride, and incubated at 50 °C for 5 min. The purity and concentration of total RNA was estimated, and the ratio was calculated according to absorbance (A) readings at A260/A280 nm.
IGF-IR cDNA synthesis
In a 0.2 mL tube, 2 μg of RNA was combined with 1 μL of random primer(Oligo-dT18, 0.5 μg/μL) and adjusted volume to 12 μL with dH2O, centrifuged for 5 s. Denature RNA and primer were incubated at 70 °C for 5 min and placed on ice for 30 s, then centrifuged for 5 s, placed on ice and added the reagents as follows: 4 μL of 5 X cDNA Synthesis Buffer, 1 μL of RNase OUT (20 U/μL), 2 μL dNTP Mix (10 mmol/L), centrifuged for 5 s, incubated at 37 °C for 5 min, placed on ice for 30 s, then added 1 μL of RevertAid M-MuLV Reverse Transcriptase (20 U/μL). Lastly, the reaction was terminated by heating at 70 °C for 5 min after incubating for 60 min at 37 °C.
Primers and nested polymerase chain reaction
According to rat IGF-IR mRNA full-length sequence (NM052807), and the primers were designed by Premier Primer 5.0 software. External primer P1: 5’-CTGCGGCGA TGAAGAAAAGA-3’(nt 1038-1057) and P2: 5’-TGGAGGTGAAACGGAGAAC A-3’ (nt 1521-1540), 503 bp; Internal primer P3: 5’-ATGCCTTGGTCTCCTTGTC CT T-3’ (nt 1229-1250) and P4: 5’-TTTGCTCTGCCGTCCCTTTGTT-3’ (nt 1449-1470), 242 bp. IGF-IR cDNA was amplified by a nested Polymerase chain reaction (PCR), with the 1st stage was performed in 2 μL of cDNA (0.1 μg/μL), 2.0 μL of the mix primers (P1, P2), 12.5 μL of Priemix Taq, and adjusted volume to 25 μL with dH2O under the following conditions: 94 °C for 5 min followed by 35 cycles of 94 °C for 10 s, 55 °C for 30 s, 72 °C for 1 min and a final extension step of 10 min at 72 °C; the 2nd stage was performed according to above same condition except for 1 μL of the 1st PCR solution, and 2 μL of the mix primers (P3, P4), and finally, the PCR product was stored at 4 °C for analysis.
Electrophoresis and sequencing
Agarose (0.375 g) was added to 25 mL of TAE buffer, superheated for 3 times. When the agarose cooled to about 50 °C, 1.5 μL of ethidium bromide (10 mg/mL) was added, poured into taped gel tray, placed the combs. 300 mL of TAE buffer was poured into the electrophoresis box. Mixed 8 μL of PCR product with 1 μL of loading dye and put into a well, DNA ladder into another gel well, than electrophoresized on 1.5% agarose gel at 100 V for 30 min, observed with UV Transilluminator at 320 nm, Cut and purified the DNA fragments from the gels for sequencing in MegaBACE DNA sequencer, and the alignment of nucleotide sequences was compared with the original IGF-IR gene sequence.
Immunohistochemistry
Rat livers were fixed in 10% neutral formalin, embeded in paraffin and 4 μm of section was cut, dewaxed, hydrated, and treated with 3% H2O2 to quench endogenous peroxidase; tissue antigens were retrieved with microwave; and then the sections were blocked by normal animal serum. The first antigen of IGF-IR was dropped on tissue section overnight at 4 °C and thoroughly washed by PBS. Subsequently, the slides were washed in PBS again after adding biotinylated secondary antibody and incubating for 10 min, then added drops of streptomycin avidin-peroxidase (S-P), incubated for 10 min at room temperature, rinsed by dH2O and added DAB. After immunostaining, the slides were restrained with hematoxylin, dehydrated in a series of ethanol solutions, covered with neutral gum, and observed under the microscope (Olympus BX 50). The negative control included 0.01 mol/L PBS instead of the primary and secondary antibodies and S-P agent. Breast-cancerous tissues with positive IGF-IR expression served as positive control. Sensitive gamma-glutamyl-transferase (GGT) is used as a control at the same stage[19]. As IGF-IR-positive expression was evident as brown particles in tissues. IGF-IR expression in livers was divided in weakly positive (+, 10%-25% positive cells); moderately positive (++, 26%-75%); and strongly positive (+++, > 75%).
Extraction and measurement of total protein
Liver tissue (50 mg) was homogenized for 3 min after the addition of 1.0 mL of PBS. The slurry was transferred into tubes (1.5 mL) and centrifuged at 5000 rpm for 4 min at 4 °C. The supernatant was put into another tube and stored at 4 °C. The concentration of total protein was determined by BCA protein measuring kit. Working Reagent was prepared by mixing BCA Reagents (50:1, Reagent A:B), 10 μL of each protein standard was pipetted into a 96-well plate and diluted to 100 μL with 0.9% normal saline. Besides, the standards were added into the 96-well plate as follows: 0, 1, 2, 4, 8, 12 and 20 μL, and each well was added up to 20 μL with diluent. Then 5 μL of tissue proteins were pipetted into the wells and adjusted volume to 20 μL with 0.9% normal saline. In the end, added 200 μL BCA Working Reagent, incubated at 37 °C for 30 min and the absorbance was measured at 562 nm on a plate reader.
Detection of serum and liver IGF-IR levels
The levels of liver IGF-IR were detected using a rat IGF-IR enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer’s instructions. For the study, 100 μL of standard or tissue protein were separately put into each well of a 96-well ELISA plate, mixed gently for 30 s, incubated for 120 min at 37 °C. 100 μL of biotinylated anti-rat-IGF-IR was added, and incubated for 60 min at 37 °C. And then 100 μL of HRP-conjugate was added to the wells, and incubated for 60 min at 37 °C. Next, 100 μL of enzyme substrate working fluid was added to each well in dark for 5-10 min at 37 °C. Then, 100 μL of stop solution was put into each well, and absorbance was read at 492 nm. During the procedure, washing the plate was according to the ELISA kit. The levels of serum IGF-IR were detected in the same way.
Statistical analysis
Data were expressed as mean ± SD and subjected to analysis of variance. Differences between groups were assessed by using Newman-Keuls test, the Fisher’s exact test or Rank-Sum test. Statistical significance was accepted at the level of P value less than 0.05 by using the Stata 7.0 software.
RESULTS
Pathological morphological alteration of livers
The morphological changes of rat livers in hepatocarcinogenesis induced with 2-FAA are shown in Table 1 and Figure 1. The morphological alteration of liver in rat hepatocarcinogenesis (Figure 1A-C) was confirmed by HE staining (Table 1; Figure 1D-F), and the model rats were divided into 4 groups: the control group (n = 12), the degeneration group (n = 18 Figure 1D), the precancerous group (n = 9, Figure 1E), and the HCC group (n = 9, Figure 1F). The granule-like degeneration appeared in the cytoplasm and a large heterogeneous nucleus was seen occasional (the degeneration group, Figure 1A and D) at an early stage. At the intermediate stage, some areas had the trend to form nodules, hepatic plate cell layers increased, focal cell layers surpassed three, the nuclear chromatin was denser, and the ratio of nucleus to cytoplasm increased (the precancerous group, Figure 1B and E). The nodular hyperplasia at the later stage was observed in many areas, the structure of hepatic tissue disappeared, the hepatic cells arranged into nest or funicular form, the medium large and the nuclear chromatin was more dense, and the ratio of nucleus to cytoplasm increased, and all were confirmed as well differentiated HCC (the HCC group, Figure 1C and F), indicated that histological changes in hepatocytes from granule-like degeneration to precancerous and HCC, and confirmed by the immunohistochemistry of corresponding stage hepatic GGT (Figure 1G-I).
Table 1.
Histopathological changes of liver in hepatocarcinogenesis (hematoxylin and eosin staining)
| Group | n |
Histopathological changes in rat liver tissue |
|||
| Control | Degeneration | Precancerosis | HCC | ||
| Control | 12 | 12 | 0 | 0 | 0 |
| Experimental | |||||
| 2nd wk | 6 | 0 | 6 | 0 | 0 |
| 4th wk | 6 | 0 | 6 | 0 | 0 |
| 6th wk | 6 | 0 | 5 | 1 | 0 |
| 8th wk | 6 | 0 | 1 | 4 | 1 |
| 10th wk | 6 | 0 | 0 | 3 | 3 |
| 12th wk | 6 | 0 | 0 | 1 | 5 |
| Total | 48 | 12 | 18 | 9 | 9 |
HCC: Hepatocellular carcinoma.
Figure 1.
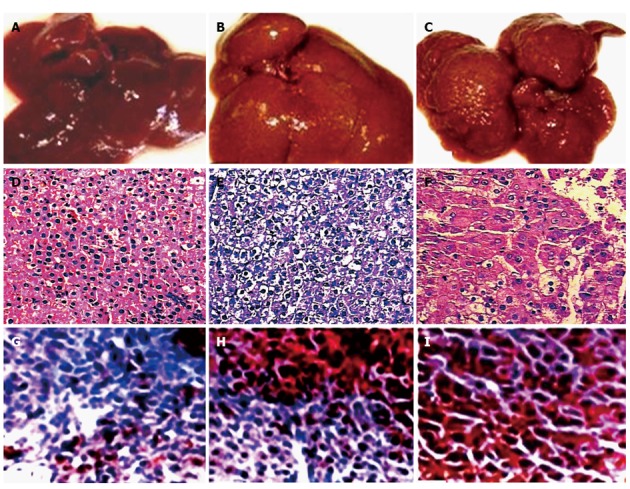
Rat livers and the histopathological alterations of rat tissues during malignant transformation of hepatocytes. A-C: The appearances of rat livers at different stages at early stage (A), at intermediate stage (B), and at later stage (C); D-F: The histopathology alteration (hematoxylin and eosin staining) of the corresponding rat liver section (× 200); G-I: The immunohistochemical confirmation of corresponding rat hepatic gamma-glutamyl transferase at the same stage (× 200).
Expression of hepatic IGF-IR mRNA
The quantitative analysis of total liver RNA from the different stages and the positive fragments of liver IGF-IR mRNA amplification in hepatocarcinogenesis are shown in Table 2 and Figure 2. The metabolic levels of hepatic nucleic acid were vigorous and the expression of hepatic total RNA was progressively increased from the control to HCC. The specific concentration of total RNA (μg/mg wet liver) was significantly higher in the HCC group than that in the control group (F = 6.840, P < 0.001, Table 2). The amplified fragments of hepatic IGF-IR mRNA could be detected clearly in the HCC, precancerous, or part of degeneration group (Figure 2A) and the IGF-IR gene fragments were confirmed by sequencing (Figure 2B). The expression of IGF-IR mRNA was progressively increased in hepatocarcinogenesis and the incidence was 0% in the control, 61.1% in the degeneration, 100% in the precancerous, and 100% in the HCC group (Table 2), respectively.
Table 2.
Total RNA and insulin-like growth factor-I receptor mRNA expression in hepatocarcinogenesis n (%)
| Group | n |
Total RNA level |
IGF-IR mRNA |
|||
| (μg/mg wet liver) | q | P value | Positive | P value | ||
| Control | 12 | 1.58 ± 0.94 | 0 (0.0) | |||
| Degeneration | 18 | 1.91 ± 0.601 | 1.177 | 0.249 | 11 (61.1)1 | < 0.001 |
| Precancerosis | 9 | 2.00 ± 0.211 | 1.308 | 0.206 | 9 (100)1 | < 0.001 |
| HCC | 9 | 2.86 ± 0.601 | 3.843 | 0.001 | 9 (100)1 | < 0.001 |
Compared with the control group. IGF-IR: Insulin-like growth factor-I receptor; HCC: Hepatocellular carcinoma.
Figure 2.
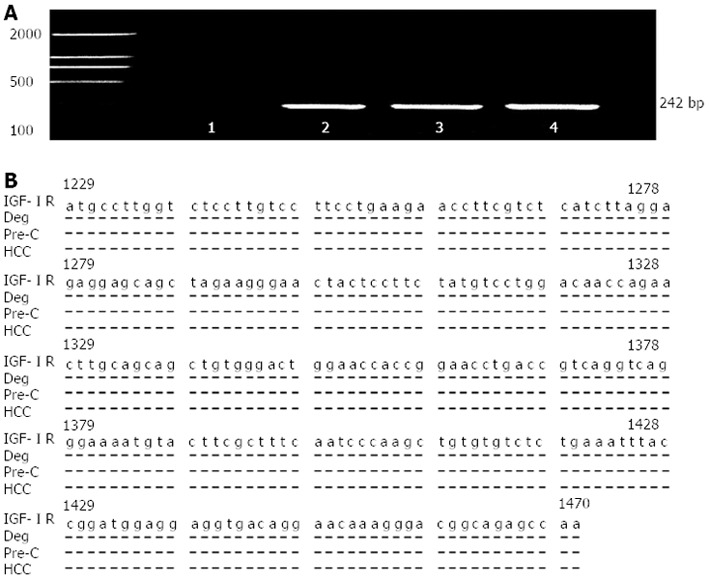
Gene amplification, sequencing and homological analysis of liver insulin-like growth factor I receptor. A: The amplified fragments of rat insulin-like growth factor I receptor (IGF-IR) mRNA by reverse transcription-nested polymerase chain reaction 1, no positive fragments from control rat liver 2, the positive fragments from degeneration rat (Deg) liver; 3, the positive fragments from precancerous rat (Pre-C) liver, and 4, the positive fragments from hepatocellular carcinoma (HCC) rat liver; B: The amplified fragment of rat IGF-IR mRNA was confirmed by sequencing and alignment of nucleotide sequences from rat liver.
Dynamic expression of hepatic IGF-IR
The positive with brown particles and cellular distribution of liver IGF-IR expression analyzed by immunohistochemistry is shown in Figure 3 and the result is summarized in Table 3. The immunohistochemistry evidences indicated the positive IGF-IR expression and hepatocyte distribution of IGF-IR positive expression increased gradually along with the occurrence (Figure 3) of hepatocyte malignant transformation. The nucleus was stained in some atypical hyperplasia areas; while near the area of necrosis, the cells were mainly stained in cytoplasm and nucleus, and the positive cells were always located in the edge of the portal area or central vein area (Figure 3C or D), and the IGF-IR levels in the precancerous or HCC group were significantly higher than that in the control group (Table 3, P < 0.001). Of the 18 cases with degeneration, the IGF-IR-expression was detected in 9 cases and 5 of them showed moderately or strongly positive; of the 9 cases with HCC, IGF-IR over - expression was detected in 9 cases and 8 of them showed moderately or strongly positive expression (Table 3).
Figure 3.
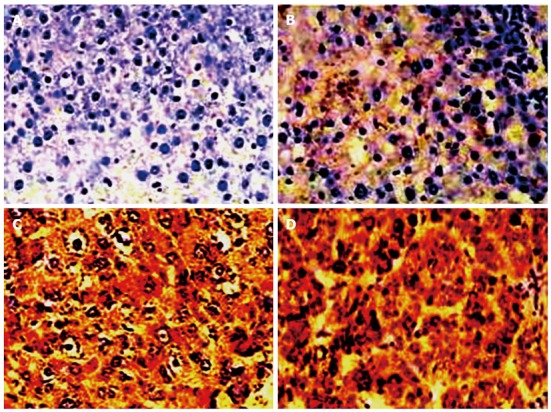
Immunohistochemical analysis of rat liver insulin-like growth factor I receptor at the different stage of rat hepatocyte malignant transformation (× 200). A: No positive staining in the liver from control rat; B: The weaker insulin-like growth factor I receptor (IGF-IR) expression in the liver from degeneration rat; C: The significantly IGF-IR expression in the liver from precancerous rat; D: The over-expression of liver IGF-IR in hepatoma rats.
Table 3.
Dynamic alteration of liver insulin-like growth factor-I receptor expression in hepatocarcinogenesis n (%)
| Group | n | Positive | P value |
IGF-IR intensity |
Nemenyi test | P value | |||
| - | + | ++ | +++ | ||||||
| Control | 12 | 0 (0.0) | 12 | 0 | 0 | 0 | |||
| Degeneration | 18 | 9 (50.0)1 | 0.004 | 9 | 4 | 4 | 1 | 13.7161 | < 0.050 |
| Precancerosis | 9 | 8 (88.9)1 | < 0.001 | 1 | 3 | 2 | 3 | 16.2291 | < 0.001 |
| HCC | 9 | 9 (100)1 | < 0.001 | 0 | 1 | 5 | 3 | 16.2991 | < 0.001 |
Compared with the control group by the Kruskal-Wallis test (HC = 25.5149, P < 0.001). IGF-IR: Insulin-like growth factor-I receptor; HCC: Hepatocellular carcinoma.
Quantitative analysis of circulating and liver IGF-IR
The levels of circulating or liver IGF-IR expression during hepatocyte malignant transformation are shown in Table 4. There was a rising tendency of serum or liver IGF-IR along with the morphological changes in hepatocarcinogenesis with HCC > precancerous > degeneration > control. The average serum IGF-IR level in the precancerous group was significantly higher than that in the control or degeneration group (P < 0.01); and the level in the HCC group was significantly higher than that in the control or degeneration group (F = 11.850, P < 0.001); The IGF-IR level in the livers in the HCC group was obviously higher than that in the control, degeneration or precancerous groups (F = 8.720, P < 0.001). Moreover, there was a positive correlation (r = 0.91, t = 14.222, P < 0.001) found between serum and liver IGF-IR, suggesting that the over-expression of liver IGF-IR release into blood and circulating IGF-IR monitor hepatocyte malignant transformation.
Table 4.
Insulin-like growth factor-I level in rat livers or sera during malignant transformation of hepatocytes
| Group | n |
Serum IGF-IR |
Liver IGF-IR |
||||
| nmol/mL | q | P value | nmol/mg wet liver | q | P value | ||
| Control | 12 | 1.03 ± 0.47 | 0.43 ± 0.17 | ||||
| Degeneration | 18 | 1.51 ± 0.461 | 1.486 | 0.318 | 0.55 ± 0.201 | 0.456 | 0.812 |
| Precancerosis | 9 | 1.92 ± 0.291 | 4.116 | 0.003 | 0.66 ± 0.141 | 0.578 | 0.748 |
| HCC | 9 | 2.43 ± 0.571 | 7.674 | < 0.001 | 0.96 ± 0.091 | 6.357 | < 0.001 |
Compared with the control group. IGF-IR: Insulin-like growth factor-I receptor; HCC: Hepatocellular carcinoma.
DISCUSSION
HCC is one of the most common cancers and causes of mortality[20,21]. A great deal of progress in understanding the mechanism(s) of hepatocarcinogenesis has been achieved in recent years[16,22]. IGFs imbalance affect the malignant transformation of hepatocytes, and the over-expressions of IGF-II and IGF-IR promote mitosis and cellular transformation and inhibiting apoptosis[10]. IGF-IR is a trans-membrane tyrosine kinase receptor located on chromosome 15q26.3 composed of two α and two β subunits linked by disulfide bonds. The extracellular α subunit is responsible for ligand binding, whereas the β subunit consists of a trans-membrane domain and a cytoplasmic tyrosine kinase domain. It is always expressed efficiently in rat nonparenchymal cells including Kupffer cells, stellate cells and increases the sensitivity of the hepatocytes to IGFs mitogenic effect. In this study, the dynamic models of HCC were applied to investigate the features of IGF-IR expression in hepatocarcinogenesis, the relationship between IGF-IR expression and hepatocytes malignant transformation, and explore the multistage pathogenesis of HCC.
Hepatoma models were successfully induced in male SD rats for investigating the dynamic expression and alteration features of IGF-IR and IGF-IR-mRNA during hepatocytes malignant transformation (Table 1). At an early stage, the morphological changes of hepatocytes showed the granule-like degeneration appeared in the cytoplasm and a large heterogeneous nucleus occasionally. At the intermediate stage, the nuclear chromatin was denser and the ratio of nucleus to cytoplasm increased. And at the later stage, the structure of hepatic tissue disappeared, the medium large and the nuclear chromatin was denser, and the ratio of nucleus to cytoplasm increased (Figure 1). Hepatocytes were transformed into malignant cells with a large synthesis of nucleic acid. Moreover, at the stage of precancerous to hepatoma formation with the progressing increasing of liver total RNA and IGF-IR mRNA were significantly higher than those in the control or degeneration group (Table 2 and Figure 2), indicated that the activation and transcription of hepatoma related genes be closely connected with nucleic acid synthesis and IGF-IR participate in hepatocarcinogenesis[23,24].
Significant abnormalities of IGF-IR gene transcription and expression could be detected during the process of hepatocytes malignant transformation. Recent researches showed that HCC include the over-expression of IGF-II[13], which have close relationship with the degree of differentiation[25]. It exerts its actions mainly via IGF-IR. The combination then leads to activation of the phosphatidylinositol3-kinase and the Ras/mitogen activated protein kinase pathways and contributes to mitosis, proliferation, transformation and anti-apoptosis. IGF-II mRNA is over-expressed at early stage of HCC formation[13]. Immunohistochemistry of dynamic models revealed that the expression levels of IGF-IR in rat hepatic tissues and sera assumed a rising tendency during the development of HCC (Table 3, Figure 3). The average level of IGF-IR from the precancerous groups was higher than that in the normal and degeneration groups. The over-expression of liver IGF-IR release into the blood and circulating IGF-IR could be detected (Table 4), suggesting that HCC is a high-IGF-IR expressing tumor along with the malignant transformation of hepatocytes and its abnormality should be a potential molecular marker for HCC development[26,27].
HCC has exhibit numerous genetic abnormalities as well as epigenetic alterations including modulation of DNA methylation. Molecular factors are involved in the process of HCC development and metastasis. Several laboratories have implicated constitutive activation of miRNA as one of the early key events involving in neoplastic progression of the liver. Recent studies in vivo revealed that interfering with signaling via IGF-IR has an antitumor effect by inducing apoptosis and inhibiting cell growth[13,28]. Further studies will permit us to analyze mechanism of human hepatocarcinogenesis and pay attention to these areas. However, the combination of pathological features and some biomarkers with high sensitivity and specificity for early diagnosis of HCC seems to be more practical up to present.
COMMENTS
Background
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death, especially in the inshore area of the Yangtze River. Insulin-like growth factor (IGF) signaling is specifically required for HCC progression, especially in IGF-II and IGF-I receptor (IGF-IR) which are the very important signaling contributors.
Research frontiers
IGF-IR is an important regulatory factor necessary for cell growth and differentiation, provides mitogenic signal for tumor growth, exhibits a very potent anti-apoptotic activity, induces vascular endothelial growth factor expression, and able to maintain the cell phenotype transformation in carcinogenesis. However, the features of IGF-IR expression during hepatocytes malignant transformation have not yet been reported.
Innovations and breakthroughs
The dynamic models of rat hepatoma were applied to investigate the features of IGF-IR expression in hepatocarcinogenesis for the first time. The IGF-IR expression was progressively increased during hepatocyte malignant transformation at mRNA or protein level, and there was a positive correlation between sera and liver IGF-IR, indicating that the IGF-IR over-expression in liver tissues release into blood and circulating IGF-IR monitor occurrence of rat hepatoma.
Applications
Overexpression of IGF-IR might participate in rat hepatocarcinogenesis and its abnormality should be an early molecular marker for hepatocyte malignant transformation.
Peer review
This paper was aimed at investigating the expression of IGF-IR during hepatocarcinogenesis induced by 2-fluorenylacetamide in Sprague-Dawley rats. Increase in IGF-IR mRNA and protein incidence and expression are reported in preneoplastic and neoplastic tissue. It is an interesting animal model study for the pathway of the IGF-IR in the HCC.
Footnotes
Supported by The Society Development of Nantong, HS2012039; Jiangsu Health Projects, BL2012053, K201102; the Priority Academic Program Development of Jiangsu, and the International S and T Cooperation Program, 2013DFA32150 of China
P- Reviewers Corrales FJ, Chiu KW, Feo F, Ishikawa T, Li YY S- Editor Gou SX L- Editor A E- Editor Ma S
References
- 1.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273.e1. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao D, Jiang D, Huang Z, Lu J, Tao Q, Yu Z, Meng X. Abnormal expression of hepatoma specific gamma-glutamyl transferase and alteration of gamma-glutamyl transferase gene methylation status in patients with hepatocellular carcinoma. Cancer. 2000;88:761–769. [PubMed] [Google Scholar]
- 4.Abdalla MA, Haj-Ahmad Y. Promising Candidate Urinary MicroRNA Biomarkers for the Early Detection of Hepatocellular Carcinoma among High-Risk Hepatitis C Virus Egyptian Patients. J Cancer. 2012;3:19–31. doi: 10.7150/jca.3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Augello C, Vaira V, Caruso L, Destro A, Maggioni M, Park YN, Montorsi M, Santambrogio R, Roncalli M, Bosari S. MicroRNA profiling of hepatocarcinogenesis identifies C19MC cluster as a novel prognostic biomarker in hepatocellular carcinoma. Liver Int. 2012;32:772–782. doi: 10.1111/j.1478-3231.2012.02795.x. [DOI] [PubMed] [Google Scholar]
- 6.Daniel M, Peek GW, Tollefsbol TO. Regulation of the human catalytic subunit of telomerase (hTERT) Gene. 2012;498:135–146. doi: 10.1016/j.gene.2012.01.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047–2063. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 8.Freise C, Ruehl M, Erben U, Neumann U, Seehofer D, Kim KY, Trowitzsch-Kienast W, Stroh T, Zeitz M, Somasundaram R. A hepatoprotective Lindera obtusiloba extract suppresses growth and attenuates insulin like growth factor-1 receptor signaling and NF-kappaB activity in human liver cancer cell lines. BMC Complement Altern Med. 2011;11:39. doi: 10.1186/1472-6882-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ubagai T, Kikuchi T, Fukusato T, Ono Y. Aflatoxin B1 modulates the insulin-like growth factor-2 dependent signaling axis. Toxicol In Vitro. 2010;24:783–789. doi: 10.1016/j.tiv.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 10.Scharf JG, Dombrowski F, Ramadori G. The IGF axis and hepatocarcinogenesis. Mol Pathol. 2001;54:138–144. doi: 10.1136/mp.54.3.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian J, Yao D, Dong Z, Wu W, Qiu L, Yao N, Li S, Bian Y, Wang Z, Shi G. Characteristics of hepatic igf-ii expression and monitored levels of circulating igf-ii mRNA in metastasis of hepatocellular carcinoma. Am J Clin Pathol. 2010;134:799–806. doi: 10.1309/AJCPTFDSE2V3LCZP. [DOI] [PubMed] [Google Scholar]
- 12.Qiu LW, Yao DF, Zong L, Lu YY, Huang H, Wu W, Wu XH. Abnormal expression of insulin-like growth factor-II and its dynamic quantitative analysis at different stages of hepatocellular carcinoma development. Hepatobiliary Pancreat Dis Int. 2008;7:406–411. [PubMed] [Google Scholar]
- 13.Tovar V, Alsinet C, Villanueva A, Hoshida Y, Chiang DY, Solé M, Thung S, Moyano S, Toffanin S, Mínguez B, et al. IGF activation in a molecular subclass of hepatocellular carcinoma and pre-clinical efficacy of IGF-1R blockage. J Hepatol. 2010;52:550–559. doi: 10.1016/j.jhep.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexia C, Fallot G, Lasfer M, Schweizer-Groyer G, Groyer A. An evaluation of the role of insulin-like growth factors (IGF) and of type-I IGF receptor signalling in hepatocarcinogenesis and in the resistance of hepatocarcinoma cells against drug-induced apoptosis. Biochem Pharmacol. 2004;68:1003–1015. doi: 10.1016/j.bcp.2004.05.029. [DOI] [PubMed] [Google Scholar]
- 15.Weng CJ, Hsieh YH, Tsai CM, Chu YH, Ueng KC, Liu YF, Yeh YH, Su SC, Chen YC, Chen MK, et al. Relationship of insulin-like growth factors system gene polymorphisms with the susceptibility and pathological development of hepatocellular carcinoma. Ann Surg Oncol. 2010;17:1808–1815. doi: 10.1245/s10434-009-0904-8. [DOI] [PubMed] [Google Scholar]
- 16.Lee JY, Han CY, Yang JW, Smith C, Kim SK, Lee EY, Kim SG, Kang KW. Induction of glutathione transferase in insulin-like growth factor type I receptor-overexpressed hepatoma cells. Mol Pharmacol. 2007;72:1082–1093. doi: 10.1124/mol.107.038174. [DOI] [PubMed] [Google Scholar]
- 17.Breuhahn K, Schirmacher P. Reactivation of the insulin-like growth factor-II signaling pathway in human hepatocellular carcinoma. World J Gastroenterol. 2008;14:1690–1698. doi: 10.3748/wjg.14.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aleem E, Nehrbass D, Klimek F, Mayer D, Bannasch P. Upregulation of the insulin receptor and type I insulin-like growth factor receptor are early events in hepatocarcinogenesis. Toxicol Pathol. 2011;39:524–543. doi: 10.1177/0192623310396905. [DOI] [PubMed] [Google Scholar]
- 19.Yao DF, Dong ZZ, Yao DB, Wu XH, Wu W, Qiu LW, Wang HM, Meng XY. Abnormal expression of hepatoma-derived gamma-glutamyltransferase subtyping and its early alteration for carcinogenesis of hepatocytes. Hepatobiliary Pancreat Dis Int. 2004;3:564–570. [PubMed] [Google Scholar]
- 20.El-Serag HB, Marrero JA, Rudolph L, Reddy KR. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134:1752–1763. doi: 10.1053/j.gastro.2008.02.090. [DOI] [PubMed] [Google Scholar]
- 21.Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26 Suppl 1:144–152. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- 22.Nussbaum T, Samarin J, Ehemann V, Bissinger M, Ryschich E, Khamidjanov A, Yu X, Gretz N, Schirmacher P, Breuhahn K. Autocrine insulin-like growth factor-II stimulation of tumor cell migration is a progression step in human hepatocarcinogenesis. Hepatology. 2008;48:146–156. doi: 10.1002/hep.22297. [DOI] [PubMed] [Google Scholar]
- 23.Lin RX, Wang ZY, Zhang N, Tuo CW, Liang QD, Sun YN, Wang SQ. Inhibition of hepatocellular carcinoma growth by antisense oligonucleotides to type I insulin-like growth factor receptor in vitro and in an orthotopic model. Hepatol Res. 2007;37:366–375. doi: 10.1111/j.1872-034X.2007.00055.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhao H, Desai V, Wang J, Epstein DM, Miglarese M, Buck E. Epithelial-mesenchymal transition predicts sensitivity to the dual IGF-1R/IR inhibitor OSI-906 in hepatocellular carcinoma cell lines. Mol Cancer Ther. 2012;11:503–513. doi: 10.1158/1535-7163.MCT-11-0327. [DOI] [PubMed] [Google Scholar]
- 25.Rehem RN, El-Shikh WM. Serum IGF-1, IGF-2 and IGFBP-3 as parameters in the assessment of liver dysfunction in patients with hepatic cirrhosis and in the diagnosis of hepatocellular carcinoma. Hepatogastroenterology. 2011;58:949–954. [PubMed] [Google Scholar]
- 26.Yao DF, Dong ZZ, Yao M. Specific molecular markers in hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2007;6:241–247. [PubMed] [Google Scholar]
- 27.Lu YY, Yao DF, Wu XH. Expression of insulin-like growth factor-I receptor and its clinical pathological characteristics in human hepatocellular carcinoma and its para-carcinoma. Nantong Daxue Xuebao. 2008;28:169–171. [Google Scholar]
- 28.Yao WF, Liu JW, Sheng GL, Huang DS. Blockade of IGF-IR exerts anticancer effects in hepatocellular carcinoma. Mol Med Rep. 2011;4:719–722. doi: 10.3892/mmr.2011.486. [DOI] [PubMed] [Google Scholar]


