Abstract
Genetic studies have demonstrated the involvement of the complement regulator factor H in nondiarrheal, nonverocytotoxin (i.e., atypical) cases of hemolytic uremic syndrome. Different factor H mutations have been identified in 10%–30% of patients with atypical hemolytic uremic syndrome (aHUS), and most of these mutations alter single amino acids in the C-terminal region of factor H. Although these mutations are considered to be responsible for the disease, the precise role that factor H plays in the pathogenesis of aHUS is unknown. We report here the structural and functional characterization of three different factor H proteins purified from the plasma of patients with aHUS who carry the factor H mutations W1183L, V1197A, or R1210C. Structural anomalies in factor H were found only in R1210C carriers; these individuals show, in their plasma, a characteristic high-molecular-weight factor H protein that results from the covalent interaction between factor H and human serum albumin. Most important, all three aHUS-associated factor H proteins have a normal cofactor activity in the proteolysis of fluid-phase C3b by factor I but show very low binding to surface-bound C3b. This functional impairment was also demonstrated in recombinant mutant factor H proteins expressed in COS7 cells. These data support the hypothesis that patients with aHUS carry a specific dysfunction in the protection of cellular surfaces from complement activation, offering new possibilities to improve diagnosis and develop appropriate therapies.
Introduction
Hemolytic uremic syndrome (HUS) is a microvasculature disorder leading to microangiopathic hemolytic anemia, thrombocytopenia, and acute renal failure. Most cases of HUS are associated with epidemics of diarrhea caused by verocytotoxin-producing bacteria. There are cases of HUS not associated with diarrhea, which are known as “atypical HUS” (aHUS [MIM 134370; MIM 235400]). Cases of aHUS are sometimes familial and generally have a poor outcome (for review, see Ruggenenti and Remuzzi 1998). Recently, it has been demonstrated that aHUS segregates with the factor H gene locus (HF1) (Warwicker et al. 1998) and that a significant number of patients with aHUS (10%–30%) carry mutations in HF1 (Warwicker et al. 1998; Buddles et al. 2000; Caprioli et al. 2001; Pérez-Caballero et al. 2001; Richards et al. 2001). Although early studies suggested the association between aHUS and deficiencies of factor H (Thompson and Winterborn 1981; Pichette et al. 1994; Ohali et al. 1998; Rougier et al. 1998), many patients with aHUS who have mutations in HF1 present normal complement profiles and have plasma levels of factor H that are either within the normal range or elevated (Warwicker et al. 1998; Caprioli et al. 2001; Pérez-Caballero et al. 2001; Richards et al. 2001).
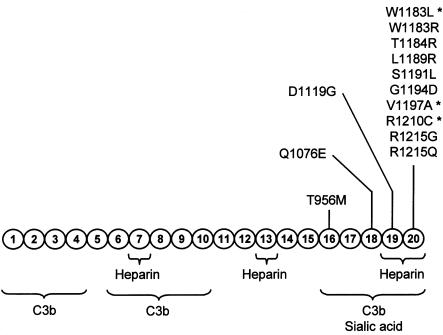
Factor H is a plasma protein (molecular weight 155,000 Da) that controls activation of the complement system in the fluid phase and on cellular surfaces. Factor H binds to C3b, accelerates the decay of the alternative-pathway C3 convertase, and acts as a cofactor in the factor I–mediated proteolytic inactivation of C3b (Weiler et al. 1976; Pangburn et al. 1977). It can also interact with polyanionic molecules (sialic acids or glycosaminoglycans) on certain cellular surfaces, conferring to them resistance to damage as a consequence of complement activation through the alternative pathway (Meri and Pangburn 1990; Pangburn et al. 1991, 2000). Factor H is composed of 20 repetitive units of 60 amino acids, and these units are named “short consensus repeats” (SCRs) (Ripoche et al. 1988). The functional domains in the factor H molecule have been delineated elsewhere (Sharma and Pangburn 1996). Factor H has three binding sites for C3b, in SCR1-4, SCR6-10, and SCR16-20. The C3b binding site in SCR1-4 is the only site essential for the cofactor activity, but deletion of any of the C3b binding sites significantly decreases factor H binding to C3b deposited on cellular surfaces (Sharma and Pangburn 1996). In addition, SCR7, SCR13, and SCR16-20 have been found to contain heparin and/or sialic acid binding sites (Pangburn et al. 1991; Blackmore et al. 1996, 1998; Ram et al. 1998).
The mutational screening of the HF1 gene in independent series of patients with aHUS has resulted in the identification of a remarkable clustering of missense mutations in the SCR16-20 region of factor H (Caprioli et al. 2001; Pérez-Caballero et al. 2001; Richards et al. 2001) (fig. 1). This region is involved in the binding of factor H to C3b deposited on cellular surfaces (Pangburn et al. 2000). Therefore, we and others have suggested a specific dysfunction in the protection of cellular surfaces by factor H in patients with aHUS (Caprioli et al. 2001; Pérez-Caballero et al. 2001; Richards et al. 2001; Zipfel 2001; Perkins and Goodship 2002).
Figure 1.
Functional domains and mutations in the factor H molecule, showing a diagram of the structure of human factor H with the 20 SCRs. Functional domains are indicated schematically. The location of the missense mutations thus far characterized in patients with aHUS (Warwicker et al. 1998; Caprioli et al. 2001; Pérez-Caballero et al. 2001; Richards et al. 2001; Perkins and Goodship 2002; Remuzzi et al. 2002b) is indicated, to illustrate that they are clustered in a specific factor H region that has been involved in the control of C3b deposited on surfaces. The mutations analyzed in the present article are indicated by asterisks.
To gain further insight into the pathogenesis of aHUS, we have purified factor H from the plasma of patients with aHUS, and we have produced recombinant aHUS-associated factor H mutant proteins in COS7 cells. The functional activity of the purified factor H proteins was determined by utilization of two assays, analysis of the cofactor activity in the proteolysis of fluid-phase C3b by factor I, and analysis of the binding to surface-bound C3b. Our results demonstrate that the aHUS-associated factor H proteins have impaired binding to surface-bound C3b, providing the first experimental evidence that the missense mutations found in patients with aHUS have functional consequences.
Patients, Material, and Methods
Patients and Control Individuals
Three patients with aHUS who carry missense mutations in the HF1 gene and three unaffected control individuals were selected for the present studies. The clinical and genetic data on the patients are depicted in table 1. In brief, patient HUS2 is heterozygous for the W1183L mutation. Despite the HF1 mutation, patient HUS2 presented with normal complement profiles, and, interestingly, his plasma factor H levels were significantly elevated. Patient HUS29 is heterozygous for the R1210C mutation. Levels of factor H in patient HUS29 are at the lower end of the normal range. C3 levels were also slightly decreased. HUS2 and HUS29 plasmas are expected to contain a mixture of wild-type and mutant factor H proteins. Patient HUS3, who also has a factor H missense mutation (V1197A), is the only patient, in the series of patients with aHUS whom we studied, who had half-normal levels of factor H and a moderate hypocomplementemia with decreased AP50 and C3 levels. Genetic analyses demonstrated that the two copies of the factor H gene were mutated in patient HUS3 (Pérez-Caballero et al. 2001). One copy carries a deletion that is incompatible with expression of factor H, and the second copy carries the V1197A mutation in SCR20. As a consequence of these defects, patient HUS3 presents both—factor H deficiency and a V1197A mutant factor H that, in addition, should be the only factor H protein in the plasma of this patient. The V1197A mutation, like the R1210C mutation, seems to be a recurrent factor H mutation. It has also been found in heterozygosis in two other patients with aHUS in two independent studies (Caprioli et al. 2001; Richards et al. 2001).
Table 1.
Complement Profiles and Factor H Levels in Patients with aHUS
|
Levels of Complement Componentsand Functional Activitiesb |
|||||||
| Patient (Age) | Clinical Historya | CH50 | AP50 | C3(77–210 mg/dl) | C4(14–47 mg/dl) | H(17–50 mg/dl) | HF1 Mutations |
| HUS2 (25 years) | CRF, transplanted twice, hemodialysis | N | N | 71 | 30 | 116 | W1183L |
| HUS3 (55 years) | CRF, hemodialysis | N | L | 59 | 26 | 11c | V1197Ac |
| HUS29 (14 mo) | Recurrences, CRF, peritoneal dialysis | ND | ND | 58 | 28.5 | 19.8 | R1210C |
CRF = chronic renal failure.
Measured in serum samples. Complement activity was determined by hemolytic assays using antibody-sensitized sheep erythrocytes (CH50) or rabbit erythrocytes (AP50). C3 and C4 concentrations were determined by nephelometry (Immage, Beckmann). Factor H was quantitated by a sandwich ELISA, using goat polyclonal antibodies (Quidel) and a murine monoclonal antibody (Quidel), and by semiquantitative western blot analyses, as described elsewhere (Sánchez-Corral et al. 2000). N = normal value; L = low; ND = not done.
Heterozygous for an HF1 null allele.
Three control individuals, denoted as “N1,” “N2,” and “N3,” were included in these studies. Two of them were selected from a panel of unaffected individuals, for being homozygotes for the two major factor H polymorphic variants (Rodríguez de Córdoba and Rubinstein 1984). N1 is an HF1*1 (Tyr402) homozygote, and N2 is an HF1*2 (His402) homozygote. Serum and EDTA-blood samples were obtained from patients and control individuals after informed consent was obtained. EDTA plasma (10 ml) was prepared by centrifugation at 4°C and was kept frozen at −20°C until used.
Factor H Purification
Five to ten milliliters of EDTA plasma were dialysed against Tris-HCl (20 mM; pH 7.5) and NaCl (50 mM) and were applied to a heparin-Sepharose column. After extensive washes with dialysis buffer, the proteins bound to the column were eluted with an NaCl gradient (50–500 mM). Fractions containing factor H were identified by ELISA, were pooled, were dialysed against Tris-HCl (20 mM; pH 7.5) and NaCl (150 mM), and were incubated overnight at 4°C with Sepharose beads coated with a mouse monoclonal anti–human factor H antibody (35H9). Factor H was eluted from the Sepharose-35H9 column with NaSCN (3 M). NaSCN was removed from the eluate by using a Sephadex G-25 column, and the factor H sample was dialysed against Tris-HCl (20 mM; pH 7.5) and NaCl (50 mM) and was loaded into a Mono-Q column (HR 5/5; Amersham Biosciences). Factor H was eluted with an NaCl (50–500 mM) gradient and the fractions containing factor H were identified by SDS-PAGE. Factor H–albumin complexes were purified from factor H by using an anti-HSA (human serum albumin)–Sepharose column.
Expression and Purification of Recombinant Proteins
The full-length human factor H cDNA (3,926 bp; Entrez-Nucleotide accession number Y00716) was introduced in the eukaryote expression vector pCINeo (Stratagene). The aHUS-associated HF1 mutation W1183L was created in the HF1-pCINeo construct through site-directed mutagenesis, by using a commercial kit from Stratagene. The HF1 insert in the W1183L-pCINeo construct was completely sequenced in both strands, to confirm that no other mutations were present. Plasmid DNA was prepared using “concert maxipreps” (GibcoBRL) columns. COS7 cells grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum (GibcoBRL) were transfected with the HF1-pCINeo or W1183L-pCINeo constructs, by using lipofectin (GibcoBRL). Transfected cells were cultured in the presence of 1 mg/ml G-418 sulfate (Geneticin; GibcoBRL), and resistant cells were cloned by limiting dilution. Individual clones secreted factor H to the culture media at concentrations ranging from 0.2 to 2 μg/ml. Factor H recombinant proteins expressed in COS7 cells were purified from the cultured cells' supernatants by following the same protocol as that used to purify factor H from human plasma.
PAGE and Immunoblotting
PAGE in the presence of SDS was performed using the buffer system described by Laemmli (1970). Reduction of samples was achieved in the presence of 700 μM β-mercaptoethanol (Sigma), and incubation was achieved at 100°C for 5 min. Gels were stained with 1% Coomassie brilliant blue R-250 (Sigma) or with 12 mM silver nitrate reagent. Western blot analysis was performed on nitrocellulose membranes, by using anti–factor H monoclonal antibody 35H9, as described elsewhere (Sánchez-Corral et al. 2000).
Fluid-Phase C3b Cofactor-Activity Assay
The fluid-phase cofactor activity of factor H was determined in a C3b proteolytic assay (Fujita and Nussenzweig 1979), using purified proteins. In brief, different amounts (10, 50, and 250 ng) of purified factor H were added to 2 μg of C3b and 0.4 μg of factor I and were incubated at 37°C for 1 h in Tris-HCl (20 mM; pH 7.5) and NaCl (150 mM). Proteolysis of C3b was determined by analyzing the cleavage of the α′ chain and the generation of the α65, α45, and α43 fragments using 8% SDS-PAGE under reducing conditions. Human C3 was purified from plasma, and C3b was generated by limited trypsin digestion (Sánchez-Corral et al. 1989). Factor I was a generous gift of Dr. R. Sim (MRC Immunochemistry Unit, Oxford University, Oxford, United Kingdom).
Surface-Bound C3b–Binding Assay
The binding of factor H to surface-bound C3b was determined using an ELISA method. In brief, 96-well polystyrene microtiter plates (Costar) were coated overnight at 4°C with 0.4 μg of purified C3b in 100 μl of coupling buffer (0.1 M NaHCO3 [pH 9.5]). Plates were washed once in TNT buffer (50 mM Tris/HCl [pH 7.4], 150 mM NaCl, and 0.2% Tween 20), and the wells were blocked with 1% BSA in TNT buffer for 1 h at room temperature. After the washing in TNT buffer, purified factor H at different concentrations in 100 μl of 1% BSA in TNT buffer was added to each well in duplicate and was allowed to interact with the surface-bound C3b for 1 h at room temperature. After four washes in TNT buffer, 100 μl of a rabbit anti–human factor H polyclonal antibody generated in the laboratory was added to each well and was incubated for 1 h at room temperature. This antibody lacks reactivity against human C3b. After three washes in TNT buffer, 100 μl of a 1/1,000 dilution of goat anti–rabbit immunoglobulin G antibody coupled with horseradish peroxidase (DAKO) was added and kept for 30 min at room temperature. The plates were washed five times, the enzymatic reaction was developed with OPD (DAKO) and was stopped with 0.1 M H2SO4, and absorbance was determined by reading at 492 nm. The concentration of the factor H preparations used in these experiments was determined in duplicates of the samples added to the C3b-coated plates in a parallel ELISA using the rabbit anti–factor H polyclonal antibody and the monoclonal antibody 35H9.
Results and Discussion
Although genetic studies have clearly demonstrated the involvement of the complement regulator factor H in aHUS and many different HF1 mutations have already been identified in a significant number of patients, the precise role that factor H plays in the pathogenesis of aHUS is still unknown. To get further insight into the molecular basis of aHUS and to characterize the functional consequences of the aHUS-associated factor H mutations, we selected three patients with aHUS (HUS2, HUS3, and HUS29) and three control individuals (N1, N2, and N3) (see the “Patients, Material, and Methods” section) and purified their factor H to homogeneity. Isolation of factor H from EDTA-plasma samples was achieved in three chromatographic steps: heparin-Sepharose column, affinity chromatography using the anti–factor H monoclonal antibody 35H9, and fast protein liquid chromatography on a Mono-Q column. All of the factor H proteins were obtained with similar yields and presented with a comparable degree of purification. During the purification procedures, we observed that the factor H present in the aHUS plasmas was fully retained in the heparin-Sepharose column and that it was eluted as a single peak with the NaCl gradient. Moreover, the elution profiles in the three patients with aHUS and the three control plasmas were identical.
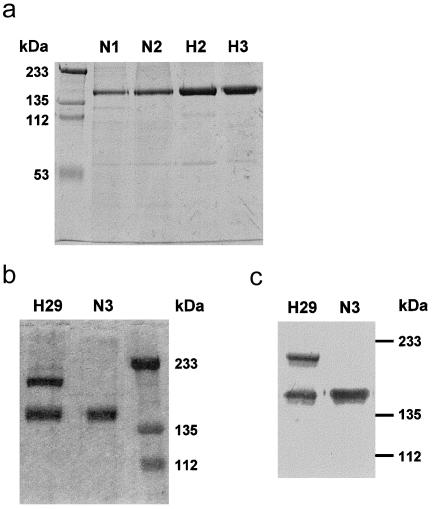
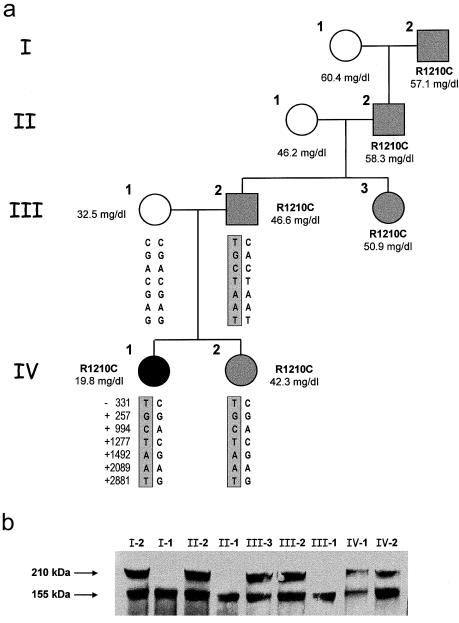
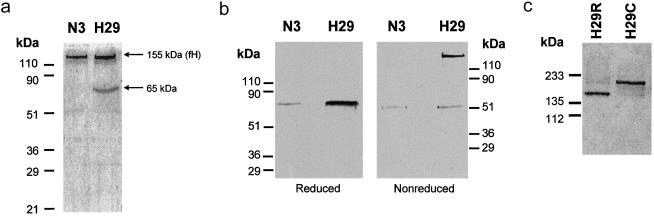
Analysis of the purified proteins by SDS-PAGE did not show any differences between the molecular sizes of the W1183L and V1197A aHUS-associated and control factor H proteins (fig. 2a). However, the SDS-PAGE analysis of the factor H proteins purified from the R1210C heterozygotes showed two bands, of 155 kDa and 210 kDa, respectively. Both bands present comparable intensities when stained with 1% Coomassie (fig. 2b), and both are equally well recognized by monoclonal and polyclonal anti–human factor H antibodies in western blot experiments (fig. 2c). The R1210C mutation has previously been reported in two unrelated Italian patients with aHUS—one familial and one sporadic (Caprioli et al. 2001). In the three pedigrees, incomplete penetrance of the disease and a characteristic high-molecular-weight factor H band in R1210C carriers are observed (fig. 3) (Caprioli et al. 2001). The 210-kDa factor H band segregates with the R1210C mutation in four generations in the HUS29 family, suggesting that this high-molecular-weight band corresponds to the product of the R1210C mutant factor H allele (fig. 3). To further characterize the 210-kDa factor H band, we analyzed the factor H protein purified from R1210C heterozygotes in SDS-PAGE under reducing conditions (fig. 4a). These analyses revealed the presence of an additional protein band with an electrophoretic mobility comparable to that of HSA (65 kDa), suggesting that the 210-kDa band results from the covalent interaction between factor H and HSA. Western blot analyses of the factor H proteins purified from patient HUS29 demonstrated that, in fact, the 210-kDa band is recognized by a polyclonal antibody against HSA and that, on reduction with β-mercaptoethanol, the 65-kDa protein band that originates from the 210-kDa band is recognized by these antibodies as HSA (fig. 4b).
Figure 2.
Electrophoretic analysis of control and aHUS-associated factor H proteins. All samples included in the present analyses were run nonreduced in 8% polyacrylamide gels in the presence of SDS. a, 8% polyacrylamide gel stained with 1% Coomassie, to illustrate that no differences were observed between the factor H proteins purified from control individuals N1 and N2 (1 μg) and patients HUS2 and HUS3 (2 μg). b, 8% polyacrylamide gel stained with 1% Coomassie, showing that two protein bands, of 210 kDa and 155 kDa (factor H), were copurified from patient HUS29. c, Western blot using the 35H9 monoclonal anti–factor H antibody, to illustrate that the two protein bands purified from patient HUS29 are recognized as factor H. Factor H purified from control individual N3 was used as a reference of wild-type factor H in this experiment.
Figure 3.
Segregation of the R1210C mutation in family HUS29. a, Pedigree of family HUS29, illustrating the segregation of the R1210C mutation. Levels of factor H in plasma are indicated for each member of the family. Nonaffected R1210C carriers are shown in gray. Seven SNPs in the factor H gene, including one in the promoter region (−331T/C), were analyzed to determine the segregation of the maternal and paternal factor H alleles in the patient and her sister, but the mother was homozygous for the seven SNPs. b, Western blot analysis of 1 μl of plasma from each HUS29 family member by using the anti–human factor H monoclonal antibody 35H9. Plasma samples are not reduced. The positions of the two factor H bands are indicated with arrows. Notice the decreased levels of both factor H alleles in the patient (IV-1).
Figure 4.
Characterization of the high-molecular-weight factor H associated with the R1210C mutation. a, 10% SDS-PAGE of the factor H (fH) proteins purified from patient HUS29 and control individual N3. Samples were reduced with 10% β-mercaptoethanol. A gel stained with 1% Coomassie is shown. b, Western blot analyses of reduced and nonreduced factor H proteins purified from patient HUS29 and control individual N3, using a rabbit anti-HSA polyclonal antibody. c, SDS-PAGE analysis of the wild-type (H29R; flow through) and mutated (H29C; retained) factor H proteins purified from patient HUS29, using anti-HSA affinity chromatography. Samples were not reduced. A silver-stained 10% polyacrylamide gel is shown.
Interestingly, the factor H R1210C mutation described here is very similar to that responsible for the anomalous C1 inhibitor (C1-INH) protein reported by Laurell and Mårtensson (1971) in a patient with angioneurotic edema. In that report, an abnormal C1-INH protein disulfide bonded to serum albumin—known as “Da mutation”—also resulted from a missense mutation that changes an arginine residue at position 444 to cysteine (Laurell and Mårtensson 1971; Skriver et al. 1989). The C1-INH–albumin heterodimers lack the capacity that normal C1-INH has in the inhibition of the activated C1 complexes (Laurell and Mårtensson 1971).
Similar to the case of the C1-INH Da mutation, on the basis of the analyses of the factor H proteins purified from the R1210C carriers, we conclude that the high-molecular-weight factor H band present in the plasma of the R1210C heterozygotes results from the covalent interaction between factor H and HSA, most likely through the additional cysteine residue at position 1210. These findings allowed us to purify the products of the mutated factor H allele (210-kDa band; denoted as “H29C”) and the normal factor H allele (155-kDa band; denoted as “H29R”) in the factor H preparations from patient HUS29, by affinity chromatography using an anti-HSA antibody (fig. 4c).
The capacity that the factor H proteins purified from the patients with aHUS and unaffected individuals have to control activation of the complement system was estimated using two different assays—analysis of the cofactor-activity of factor H in the proteolysis of fluid-phase C3b by factor I and analysis of the binding of factor H to surface-bound C3b.
The factor H proteins purified from patients HUS2 and HUS3 presented normal cofactor activity in the proteolysis of fluid-phase C3b by factor I. These activities were indistinguishable from the activity of the factor H proteins purified from the control individuals (fig. 5). Similarly, the cofactor activities of the two factor H alleles of patient HUS29 (H29R and H29C) were normal and comparable to the control proteins (fig. 5).
Figure 5.
Analysis of the cofactor activity of factor H in the proteolysis of fluid-phase C3b by factor I. a, Analysis by 8% SDS-PAGE under reducing conditions of the proteolysis of C3b by factor I in the presence of factor H. Factor H samples include purified factor H from two control individuals (N1 and N2), purified factor H from patients with aHUS (HUS2 [H2] and HUS3 [H3]), purified factor H corresponding to the wild-type (H29R) and mutated (H29C) alleles of patient HUS29, and purified recombinant wild-type factor H (r-factor H) and W1183L mutant factor H (r-W1183L) expressed in COS7 cells. Each panel includes three different concentrations of factor H. Identification of the C3b fragments is depicted in the panel corresponding to N2. The cofactor activity of factor H results in cleavage of the α′ chain of C3b in the fragments α65 and α45/α43. The top left panel corresponds to the control lanes of C3b alone and C3b plus factor I in the absence of factor H. b, Densitometric analysis of the proteolysis of C3b by factor I in the presence of factor H. Stained gels were scanned in a GS-800 Calibrated Densitometer (Bio-Rad), and the bands were quantified with ImageQuant, version 3.3 (Molecular Dynamics). The percentage of the C3b cleavage was determined as the ratio of the values corresponding to the α65 and the α′ bands. The code for the samples is indicated at bottom.
The recombinant factor H proteins purified from the COS7 cells' culture supernatants—wild-type and W1183L mutant factor H—also presented cofactor activity that was comparable to the activity of the factor H proteins purified from plasma (fig. 5). These analyses indicate that the recombinant factor H proteins produced in COS7 cells are fully active and that the W1183L mutation has no effect on the cofactor activity.
As a whole, these data suggest that patients with aHUS who carry the W1183L, V1197A, and R1210C mutations in factor H have normal capacity to control activation of the complement in plasma. This is in agreement with the clinical laboratory findings in most patients with aHUS who show complement profiles in plasma that would be incompatible with a classical factor H deficiency (Pérez-Caballero et al. 2001). Since V1197A mutant factor H is the only factor H in the plasma of patient HUS3, these data also indicate that the moderate hypocomplementemia in HUS3 is probably unrelated to the V1197A mutation and is most likely due to this patient's carrying a null HF1 allele and presenting half-normal levels of factor H.
In contrast with the apparently normal cofactor activity, the ability of the factor H proteins to interact with surface-bound C3b in the three aHUS-associated factor H proteins is highly diminished as compared with the three control factor H proteins (fig. 6a). The C3b-binding assay is a simple test that measures direct binding of purified factor H to purified C3b immobilized on microtiter plates (see the “Patients, Material, and Methods” section). In this assay, a reduced binding of factor H to the microtiter wells indicates that the factor H has reduced capacity to bind surface-bound C3b or, alternatively, that not all of the factor H present in the factor H preparation has capacity to bind to surface-bound C3b.
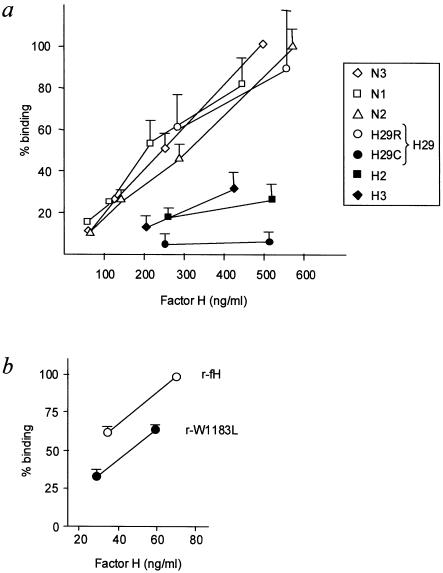
Figure 6.
Binding of factor H to surface-bound C3b. a, Analysis of factor H purified from plasma. Included were purified factor H from three control individuals (N1, N2, and N3), purified factor H from patients with aHUS (HUS2 [H2] and HUS3 [H3]), and purified factor H corresponding to the wild-type (H29R) and mutated (H29C) factor H alleles of patient HUS29. In the present analyses, the concentration of factor H ranged between 50 and 500 ng/ml. The results are expressed as the percentage of the binding observed for factor H from the control individual N3 at 500 ng/ml. Values are the mean ± SD of at least three independent experiments. b, Analysis of recombinant factor H proteins, showing the binding of purified recombinant wild-type factor H (r-fH) and W1183L mutant factor H (r-W1183L). Two different concentrations of recombinant factor H were used. The results are expressed as the percentage of the binding observed for recombinant wild-type factor H (r-fH) at the highest concentration. Values are the mean ± SD of at least three independent experiments.
The V1197A mutant factor H should be the only factor H protein in the plasma of patient HUS3, and, thus, the decreased binding of the factor H preparation from this patient to the C3b-coated plates (fig. 6a) is interpreted as a decreased binding of the mutant V1197A factor H protein to the surface-bound C3b. Similarly, figure 6a shows that the purified factor H–albumin heterodimers (H29C) do not bind to surface-bound C3b, whereas the product of the wild-type allele (H29R) in the R1210C heterozygotes binds normally to the C3b-coated plates. Binding of serum albumin to the regions of factor H that are needed to interact with surface-bound C3b (SCR20) most likely represents a serious steric hindrance that makes these surfaces inaccessible and prevents binding of the factor H–albumin heterodimers to the C3b-coated plates.
In contrast, the reduced factor H binding to the C3b-coated plates that was observed in the case of the factor H preparations from patient HUS2 (fig. 6a) most likely indicates that not all of the factor H present in these samples has capacity to bind to surface-bound C3b. These data were interpreted as suggesting that the mutant W1183L factor H protein has impaired interaction with surface-bound C3b. In agreement with this conclusion, the recombinant W1183L mutant factor H protein shows a significantly lower capacity to bind to surface-bound C3b than the recombinant wild-type factor H protein does (fig. 6b), providing direct evidence that this mutation interferes with the interaction between factor H and surface-bound C3b.
As a whole, these results represent the first evidence of a functional defect in the factor H associated with aHUS and illustrate a potential limitation, in the three patients with aHUS, to control complement activation on cellular surfaces. It is remarkable that, despite showing an impaired interaction with surface-bound C3b, the recombinant W1183L mutant factor H produced in COS7 cells shows more capacity to bind to surface-bound C3b than the factor H preparations from patient HUS2 do (fig. 6). This is most likely related to the fact that all of the recombinant factor H proteins produced in COS7 cells show much more affinity for surface-bound C3b than the equivalent proteins purified from plasma do. Whether this increased affinity is a consequence of different factor H glycosylation in COS7 cells is presently unknown.
On the basis of structural predictions, it has recently been proposed that aHUS-associated factor H mutations may result in the failure of the mutant factor H to interact with polyanionic cell surfaces, therefore preventing activation of the alternative pathway of complement activation on these surfaces (Perkins and Goodship 2002). The data reported here clearly demonstrate that the factor H preparations from patients with aHUS who carry different mutations show decreased binding to surface-bound C3b as compared with unaffected control individuals. Whether impaired recognition of surface-bound C3b by factor H is a common feature among aHUS-associated mutations is presently unknown. The findings reported here may offer a simple test, to distinguish between normal and functionally impaired factor H, that will improve the molecular diagnosis of aHUS, complementing the mutational analysis of the HF1 gene.
The patients with aHUS who are included in the present studies carry factor H mutations—two of which are in heterozygosis—that cause loss of binding of factor H to surface-bound C3b. This suggests that development of aHUS requires activation of the complement system and implies that factor H functional haploinsufficiency results in an inefficient protection of the cellular surfaces. According to these ideas, a suboptimal concentration of wild-type factor H in patients with aHUS, which is perhaps sufficient to control basal complement activation, upon a situation that triggers complement activation (i.e., infection, immune complexes, cytotoxic drugs, etc.), cannot maintain the integrity of the microvasculature cell membranes and results in tissue damage and destruction. It has recently been shown that mice with factor H deficiency and uncontrolled C3 activation develop membranoproliferative glomerulonephritis (Pickering et al. 2002). Interestingly, these animals, which show severe hypocomplementemia, do not develop aHUS, supporting the idea that aHUS requires an active complement system.
Family HUS29 also illustrates that the aHUS phenotype cannot be fully explained only on the basis of the presence of a single factor H genetic mutation. Figure 3 shows that, in this pedigree, there are five members, in addition to the patient, who carry the R1210C mutation in heterozygosis but that none of them has been affected by the disease. Similar findings have been observed in other pedigrees with aHUS that carry this and other mutations in the factor H gene (Warwicker et al. 1998; Caprioli et al. 2001; Pérez-Caballero et al. 2001).
The incomplete penetrance of the disease in carriers of the factor H mutation suggests that aHUS is a complex disorder that involves environmental influences, genetic modifier loci, or a combination of both of these factors. Infection, pregnancy, toxins, immune complexes, and cytotoxic or immunosuppressive drugs are probably among the environmental factors triggering aHUS.
The von Willebrand factor cleaving protease (ADAMTS13) deficiency could be one of the genetic factors contributing to the aHUS phenotype (Remuzzi et al. 2002a). In patient HUS29, however, tests to assess for von Willebrand factor have been normal, including testing for multimers (data not shown). Genetic factors determining variations in the plasma levels of factor H could also explain the incomplete penetrance that the disease has in R1210C carriers in the HUS29 pedigree studied here. In this respect, whereas factor H levels are at the lower part of the normal range (19.8 mg/dl) in patient HUS29, factor H levels are at the higher part of or above the normal range (42.3–58.3 mg/dl) in the nonaffected R1210C carriers in the HUS29 pedigree. Interestingly, the mother of patient HUS29 shows intermediate plasma levels of factor H (32.5 mg/dl). The possibility that the intermediate plasma levels of factor H in the mother and the decreased factor H levels in the patient could be due to the presence of a low-expression factor H maternal allele segregating to the patient seems unlikely, because expression of both factor H alleles in the patient is decreased as compared with other R1210C carriers (fig. 3b). However, it is possible that genetic factors influencing the plasma levels of factor H contribute to the pathogenesis of aHUS. In an unaffected individual, the threefold range of variation in the factor H plasma concentration (17–50 mg/dl) is probably irrelevant, but in an individual carrying an aHUS-associated mutant factor H allele in heterozygosis, a two- to threefold reduction in the total levels of plasma factor H could be critical.
Reduced plasma levels of factor H are not a general explanation of the incomplete penetrance of the disease in carriers of factor H mutations and are in apparent contradiction with cases such as that in patient HUS2, whose plasma factor H levels are significantly elevated over the normal range (table 1). We believe that an extended half-life of the mutant factor H protein in plasma could result in a reduction in the proportion of the wild-type factor H; this reduction would account for both the unexpectedly low binding, to surface-bound C3b, of the HUS2 factor H preparations and the increased plasma levels of factor H in this patient. We are currently designing experiments to determine the proportion of wild-type factor H in the plasma of patient HUS2. Additionally, it would be interesting to investigate whether an impaired interaction between factor H and surface-bound C3b alters the clearance of factor H from plasma.
Although there is currently no definitive explanation for the incomplete penetrance of aHUS in carriers of the factor H mutations, the analysis of the HUS29 pedigree and the functional characterization of the factor H aHUS-associated mutations reported here reinforce the current view (Caprioli et al. 2001; Pérez-Caballero et al. 2001; Richards et al. 2001), which is that exogenous administration of wild-type factor H or other complement inhibitors should be considered as a potential treatment for aHUS. In fact, at age 14 mo, patient HUS29, who had three recurrences of HUS since the initial presentation at age 6 mo and progressed to end-stage kidney disease, has had no relapses (hematologic) in 4 mo of treatment with weekly fresh-frozen plasma infusions. Moreover, a recent work by Remuzzi et al. (2002b) reports the success of a combined liver and kidney transplantation in a patient with aHUS who carries the W1183R factor H mutation, further supporting the factor H replacement therapy.
In conclusion, our results demonstrate that the complement regulatory protein factor H, purified from three patients with aHUS, has impaired recognition of surface-bound C3b. These findings are in accordance with the hypothesis that patients with aHUS have a specific dysfunction in the protection of cellular surfaces from complement activation. Although these studies justify the exogenous administration of wild-type factor H to the patients with aHUS, further analysis will be needed to determine whether impaired recognition of surface-bound C3b by factor H is a common feature among patients with aHUS and to explain the incomplete penetrance of the disease in carriers of aHUS-associated factor H mutations.
Acknowledgments
We thank the families with aHUS and all their clinicians—in particular, Drs. Ángel Alonso and María Auxiliadora Bajo for their collaboration and donation of blood samples. We would also like to thank S. Vara de Rey, L. Gulliksen, and the DNA Sequencing Laboratory at the Centro de Investigaciones Biológicas for their contribution to this work. We wish to acknowledge Drs. S. Lamas, K. Heath, T. Goodship, and J. Goodship for helpful discussions related to this work. This research was supported by the Spanish Comision Interministerial de Ciencia y Tecnologia (SAF2002-01083) and by the Comunidad de Madrid (08.6/0028.1/2000). P.S.-C., D.P.-C., O.H., and E.G. were awarded grants from the Fondo de Investigaciones Sanitarias, Glaxo Wellcome, the i3p program of the Consejo Superior de Investigaciones Científicas, and the Ministerio de Ciencia y Tecnologia, respectively.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Entrez-Nucleotide, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide (for factor H cDNA sequence [accession number Y00716])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for autosomal dominant [MIM 134370] and recessive [MIM 235400] HUS)
References
- Blackmore TK, Hellwage J, Sadlon TA, Higgs N, Zipfel PF, Ward HM, Gordon DL (1998) Identification of the second heparin-binding domain in human complement factor H. J Immunol 160:3342–3348 [PubMed] [Google Scholar]
- Blackmore TK, Sadlon TA, Ward HM, Lublin DM, Gordon DL (1996) Identification of a heparin binding domain in the seventh short consensus repeat of complement factor H. J Immunol 157:5422–5427 [PubMed] [Google Scholar]
- Buddles MR, Donne RL, Richards A, Goodship J, Goodship TH (2000) Complement factor H gene mutation associated with autosomal recessive atypical hemolytic uremic syndrome. Am J Hum Genet 66:1721–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli J, Bettinaglio P, Zipfel PF, Amadei B, Daina E, Gamba S, Skerka C, Marziliano N, Remuzzi G, Noris M (2001) The molecular basis of familial hemolytic uremic syndrome: mutation analysis of factor H gene reveals a hot spot in short consensus repeat 20. J Am Soc Nephrol 12:297–307 [DOI] [PubMed] [Google Scholar]
- Fujita T, Nussenzweig V (1979) The role of C4-binding protein and β1H in proteolysis of C4b and C3b. J Exp Med 150:267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–684 [DOI] [PubMed] [Google Scholar]
- Laurell AB, Mårtensson U (1971) C1 inactivator protein complexed with albumin in plasma from a patient with angioneurotic edema. Eur J Immunol 1:146–149 [DOI] [PubMed] [Google Scholar]
- Meri S, Pangburn MK (1990) Discrimination between activators and nonactivators of the alternative pathway of complement: regulation via a sialic acid/polyanion binding site on factor H. Proc Natl Acad Sci USA 87:3982–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohali M, Shalev H, Schlesinger M, Katz Y, Kachko L, Carmi R, Sofer S, Landau D (1998) Hypocomplementemic autosomal recessive factor H deficiency with decreased factor H. Pediatr Nephrol 12:619–624 [DOI] [PubMed] [Google Scholar]
- Pangburn MK, Atkinson MAL, Meri S (1991) Localization of the heparin-binding site on complement factor H. J Biol Chem 266:16847–16853 [PubMed] [Google Scholar]
- Pangburn MK, Pangburn KLW, Koistinen V, Meri S, Sharma AK (2000) Molecular mechanisms of target recognition in an innate immune system: interactions among factor H, C3b, and target in the alternative pathway of human complement. J Immunol 164:4742–4751 [DOI] [PubMed] [Google Scholar]
- Pangburn MK, Schreiber RD, Müller-Eberhard HJ (1977) Human complement C3b inactivator: isolation, characterization, and demonstration of an absolute requirement for the serum protein β1H for cleavage of C3b and C4b in solution. J Exp Med 146:257–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Caballero D, González-Rubio C, Gallardo ME, Vera M, López-Trascasa M, Rodríguez de Córdoba S, Sánchez-Corral P (2001) Clustering of missense mutations in the C-terminal region of factor H in atypical hemolytic uremic syndrome. Am J Hum Genet 68:478–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins SJ, Goodship THJ (2002) Molecular modelling of the C-terminal domains of factor H of human complement: a correlation between haemolytic uraemic syndrome and a predicted heparin binding site. J Mol Biol 316:217–224 [DOI] [PubMed] [Google Scholar]
- Pichette V, Querin S, Schurch W, Brun G, Lehner-Netsch G, Delage JM (1994) Familial hemolytic-uremic syndrome and homozygous factor H deficiency. Am J Kidney Dis 24:936–941 [DOI] [PubMed] [Google Scholar]
- Pickering MC, Cook HT, Warren J, Bygrave AE, Moss J, Walport MJ, Botto M (2002) Uncontrolled C3 activation causes membranoproliferative glomerulonephritis in mice deficient in complement factor H. Nat Genet 31:424–428 [DOI] [PubMed] [Google Scholar]
- Ram S, Sharma AK, Simpson SD, Gulati S, McQuillen DP, Pangburn MK, Rice PA (1998) A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J Exp Med 187:743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remuzzi G, Galbusera M, Noris M, Canciani MT, Daina E, Bresin E, Contaretti S, Caprioli J, Gamba S, Ruggenenti P, Perico N, Mannucci PM (2002a) von Willebrand factor cleaving protease (ADAMTS13) is deficient in recurrent and familial thrombotic thrombocytopenic purpura and hemolytic uremic syndrome as well. Blood 100:778–785 [DOI] [PubMed] [Google Scholar]
- Remuzzi G, Ruggenenti P, Codazzi D, Noris M, Caprioli J, Locatelli G, Gridelli B (2002b) Combined kidney and liver transplantation for familial haemolytic uraemic syndrome. Lancet 359:1671–1672 [DOI] [PubMed] [Google Scholar]
- Richards A, Buddles MR, Donne RL, Kaplan BS, Kirk E, Venning MC, Tielemans CL, Goodship JA, Goodship THJ (2001) Factor H mutations in hemolytic uremic syndrome cluster in exons 18–20, a domain important for host cell recognition. Am J Hum Genet 68:485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoche J, Day AJ, Harris TJR, Sim RB (1988) The complete amino acid sequence of human complement factor H. Biochem J 249:593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez de Córdoba S, Rubinstein P (1984) Genetic polymorphism of human factor H (β1H). J Immunol 132:1906–1908 [PubMed] [Google Scholar]
- Rougier N, Kazatchkine MD, Rougier JP, Fremeaux-Bacchi V, Blouin J, Deschenes G, Soto B, Baudouin V, Pautard B, Proesmans W, Weiss E, Weiss L (1998) Human complement factor H deficiency associated with hemolytic uremic syndrome. J Am Soc Nephrol 9:2318–2326 [DOI] [PubMed] [Google Scholar]
- Ruggenenti P, Remuzzi G (1998) The hemolytic uremic syndrome. Kidney Int Suppl 66:S54–S57 [PubMed] [Google Scholar]
- Sánchez-Corral P, Antón LC, Alcolea JM, Marqués G, Sánchez A, Vivanco F (1989) Separation of active and inactive forms of the third component of human complement, C3, by fast protein liquid chromatography (FPLC). J Immunol Methods 122:105–113 [DOI] [PubMed] [Google Scholar]
- Sánchez-Corral P, Bellavia D, Amico L, Brai M, Rodríguez de Córdoba S (2000) Molecular basis for factor H and FHL-1 deficiency in an Italian family. Immunogenetics 51:366–369 [DOI] [PubMed] [Google Scholar]
- Sharma AK, Pangburn MK (1996) Identification of three physically and functionally distinct binding sites for C3b in human complement factor H by deletion mutagenesis. Proc Natl Acad Sci USA 93:10996–11001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriver K, Radziejewska E, Silbermann JA, Donaldson VH, Clarck-Bock S (1989) CpG mutations in the reactive site of human C1 inhibitor. J Biol Chem 264:3066–3071 [PubMed] [Google Scholar]
- Thompson R, Winterborn M (1981) Hypocomplementemia due to a genetic deficiency of β1H globulin. Clin Exp Immunol 46:110–119 [PMC free article] [PubMed] [Google Scholar]
- Warwicker P, Goodship THJ, Donne RL, Pirson Y, Nicholls A, Ward RM, Turnpenny P, Goodship JA (1998) Genetic studies into inherited and sporadic hemolytic uremic syndrome. Kidney Int 53:836–844 [DOI] [PubMed] [Google Scholar]
- Weiler JM, Daha MR, Austen KF, Fearon DT (1976) Control of the amplification convertase of complement by the plasma protein β1H. Proc Natl Acad Sci USA 73:3268–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel PF (2001) Hemolytic uremic syndrome: how do factor H mutants mediate endothelial damage? Trends Immunol 22:345–348 [DOI] [PubMed] [Google Scholar]