Abstract
Myoclonus-dystonia (M-D) is a movement disorder characterized by rapid muscle contractions and sustained twisting and repetitive movements and has recently been associated with mutations in the ε-sarcoglycan gene (SGCE). The mode of inheritance is autosomal dominant with reduced penetrance upon maternal transmission, suggesting a putative maternal imprinting mechanism. We present an apparently sporadic M-D case and two patients from an M-D family with seemingly autosomal recessive inheritance. In both families, we detected an SGCE mutation that was inherited from the patients' clinically unaffected fathers in an autosomal dominant fashion. Whereas, in the first family, RNA expression studies revealed expression of only the mutated allele in affected individuals and expression of the normal allele exclusively in unaffected mutation carriers, the affected individual of the second family expressed both alleles. In addition, we identified differentially methylated regions in the promoter region of the SGCE gene as a characteristic feature of imprinted genes. Using a rare polymorphism in the promoter region in a family unaffected with M-D as a marker, we demonstrated methylation of the maternal allele, in keeping with maternal imprinting of the SGCE gene. Loss of imprinting in the patient with M-D who had biallelic expression of the SGCE gene was associated with partial loss of methylation at several CpG dinucleotides.
Introduction
Myoclonus-dystonia (M-D [MIM 159900]) is a movement disorder characterized by a combination of rapid, brief muscle contractions (myoclonus) and sustained twisting and repetitive movements, resulting in abnormal postures (dystonia). M-D usually starts in the 1st or 2nd decade of life and is usually inherited in an autosomal dominant fashion. Males and females are equally affected (Gasser 1998; Klein 2002). A major M-D gene was located on chromosome 7q21 in the majority of tested families (Nygaard et al. 1999; Klein et al. 2000; Asmus et al. 2001; Vidailhet et al. 2001) and was recently identified as the ε-sarcoglycan gene (SGCE [GenBank accession number NM_003919; MIM 604149]), with five different heterozygous loss-of-function mutations in six German families with M-D (Zimprich et al. 2001).
The function of the SGCE protein, a transmembrane glycoprotein, is still elusive in this context of a nondegenerative movement disorder of the CNS. The other members of the sarcoglycan family—α-, β-, γ-, and δ- sarcoglycan—compose a complex that is an important structure of dystrophin-associated glycoprotein assembly in striated muscles.
The murine Sgce gene is primarily transcribed from the paternal allele and is therefore maternally imprinted (Piras et al. 2000). Interestingly, pedigree analysis of several families with M-D showed a marked difference in penetrance, depending on the parental origin of the disease allele. This indicated a putative maternal imprinting mechanism in humans, as well, that may be responsible for the reduced penetrance of the condition in individuals who inherited the mutated allele from their mother (Zimprich et al. 2001).
Genomic imprinting is defined as the epigenetic marking of certain subregions of the parental genome in mammals, resulting in the expression of imprinted genes of one allele only, according to their parent of origin (Reik and Walter 2001). Genomic imprinting requires an epigenetic control mechanism, such as differential methylation of CpG dinucleotides depending on their parental origin (Razin and Cedar 1994). Here we present the clinical and genetic analysis of two families with M-D with novel SGCE mutations and provide molecular evidence for paternal expression of the SGCE gene, by means of cDNA analysis and bisulfite sequencing.
Subjects and Methods
Subjects
All patients and unaffected family members underwent a standardized neurological examination performed by a movement-disorders specialist. The diagnosis of M-D was established according to the recently modified clinical criteria (Gasser 1998; Klein 2002). In family V, the mode of inheritance appeared to be recessive, with two affected sisters from unaffected parents, and the patient in family S seemed to have a sporadic case of the disorder.
Mutation Analysis
After obtaining informed consent, we collected a blood sample from each available member of families V and S, and both DNA and RNA were extracted directly from the blood. All 12 exons and flanking intron regions of the SGCE gene were tested for mutations by means of SSCP analysis, followed by cycle sequencing of PCR products in cases of observed band shifts, through use of an automated sequencing machine (LI-COR). In addition, 100 chromosomes from both unrelated patients with M-D and unaffected control individuals were screened for the presence of exonic polymorphisms in the coding region of the SGCE gene, by SSCP. Furthermore, we screened 44 individuals from 20 families by sequencing, as well as 242 control chromosomes of unrelated individuals by SSCP analysis, for the presence of three known polymorphisms in the promoter region (−911G→C, −1062G→A, and −1067A→G [Grabowski et al. 2002]). Finally, 260 control chromosomes from individuals of central European origin were investigated for the presence of sequence changes in the 3′ UTR, by SSCP analysis. Primers and PCR conditions are summarized in table 1.
Table 1.
Primer Sequences and PCR Conditions for Exon Amplification of the SCGE Gene
| Region andPrimer Name | Sequence(5′→3′) | AnnealingTemperature(°C) | No. ofCycles | Lengthof PCRProduct(bp) |
| Promotor: | ||||
| PromUF | GTGCCATGCTTTACAAACAGATAAG | |||
| PromUR | CAGCTCATGTACCTCTGCAGTTC | 66 | 35 | 376 |
| Exon 1: | ||||
| Ex 1 F | CTGATGCTGAACTGGCCAAG | |||
| Ex 1 R | AGAGAGGCTGGTGCCCAAAG | 60 | 35 | 304 |
| Exon 2: | ||||
| Ex 2 Fs | CTGAATTATCAAGGGCGTATC | |||
| Ex 2 Rs | CCATTTGAAATAATGTTAATG | 56 | 35 | 295 |
| Exon 3: | ||||
| Ex 3 Fs | AGACAGAATGTTTTGATTGAAAC | |||
| Ex 3 Rs | ACCACCATCAGGTAAGTTTAG | 60 | 35 | 355 |
| Exon 4: | ||||
| Ex 4 F | TTCTCATTGCCCAGAGAAGG | |||
| Ex 4 R | TCAGTTATATTAGGTATGTGGC | 58 | 35 | 339 |
| Exon 5: | ||||
| Ex 5 Fs | CTTCATTAAAGATATGCATGC | |||
| Ex 5 Rs | ATAAGTTTGATAAGATCACCG | 58 | 35 | 306 |
| Exon 6: | ||||
| Ex 6 Fs | TAAATCCTGCTTTTAAGGTGG | |||
| Ex 6 Rs | TTATTCCTAAAAGCAGTTCAG | 58 | 35 | 335 |
| Exon 7: | ||||
| Ex 7 F | AAGAATGCTTTAGTGTATCCAG | |||
| Ex 7 Rs | TTGTTATCTTAGCAGGATCTC | 58 | 35 | 348 |
| Exon 8: | ||||
| Ex 8 F | GACAATGTCAGCATTTCCAC | |||
| Ex 8 R | GTTTTAGTTTCTACCCCTCCT | 58 | 35 | 309 |
| Exon 9: | ||||
| Ex 9 F | CAAATTGATGACCCATCAGGC | |||
| Ex 9 Rs | CATGCATATTAATAATTATGGCTC | 60 | 35 | 295 |
| Exon 10: | ||||
| Ex10 Fs | TAATGTAGCCTAGTGGCCAC | 55 | 40 | 448 |
| Ex10 Rs | AGCCAACTTCATGACTTCTAG | |||
| Exon 11: | ||||
| Ex 11 F | CTGGGGTCATAGTTTACCCG | |||
| Ex11 Rs | ATTTGGTGAAGATAAAGCTTC | 60 | 35 | 249 |
| Exon 12: | ||||
| Ex 12 F | GATGGAAACTTTCTCCTTGCC | |||
| Ex 12 R | CAACATGCATAACATATGCCAG | 60 | 35 | 230 |
| 3′ UTR: | ||||
| 3UTR F | TGCTCAAAATCTAAAAGTATCCAT | |||
| 3UTR R | ATTTCACCAGTCATTAGGCTTCA | 55 | 35 | 372 |
cDNA Analysis
For cDNA analysis, we reverse transcribed RNA from all available family members, through use of random primers (SuperScript; Invitrogen). To amplify mutation-containing regions, we employed a nested PCR assay, using primers shown in table 2, followed by direct cycle sequencing of the amplicons. In addition, a melting-curve analysis of PCR products was performed with specific mutation-sensitive hybridization probes on the LightCycler (Roche Diagnostics) (table 3). In exon 5, the hybridization probes were complementary to the wild-type sequence, and, thus, the melting temperature of the probe-target hybrid was lower for the mutant allele (mismatch). For exon 7, the hybridization probes were designed to be complementary to the mutant sequence, resulting in a higher melting temperature of the probe-target hybrid of the mutant allele (perfect match). For visualization of the melting temperatures as a peak, the software took the first negative derivative (−dF/dT) of the melting curve.
Table 2.
Primer Sequences and PCR Conditions Used for Nested PCR Assay of cDNA
| Primer Name (Exon) | Sequence(5′→3′) | AnnealingTemperature(°C) | No. ofCycles | Lengthof PCRProduct(bp) |
| 3Fi (3 forward) | GCGAGATTAGTAATGATCCC | |||
| 7Ri (7 reverse) | ACGCCTTCCCGTCGGCAGCAC | 58 | 30 | 805 |
| 4Fi (4 forward) | AACTGCCTACAACAGGCGCAC | |||
| 6Ri (6 reverse) | CAATGAAATTTTGCACCAGTC | 58 | 30 | 433 |
| 5Fi (5 forward) | CGTTGCCATATCAAGCAGAATTC | |||
| 9RiE (9 reverse) | TGCATCAATGGCATGTTTGTG | 58 | 30 | 776 |
| 6Fi (6 forward) | GATGTAGTCAAGAAATGGAGC | |||
| 9RiA (9 reverse) | GATTTCTGAATAGCACTGTGATG | 58 | 30 | 361 |
Table 3.
Primers and Specific Hybridization Probes for Melting-Curve Analysis
|
Melting Temperature(°C) |
||||||||
| Wild Type |
Mutant |
|||||||
| Primer Name (Exon) | Sequencea(5′→3′) | gDNA | cDNA | gDNA | cDNA | AnnealingTemperature(°C) | No.ofCycles | LengthofPCRProduct(bp) |
| SGCE 5F (4, 5 forward) | TTAATATAATGTCTGCAGAAGACT | |||||||
| SGCE 5R (6 reverse) | TGTGGATTTTCAACTTCTCGTA | 57 | 30 | 285 | ||||
| SGCE 5 Sensor [A] (5) | CCCTGCCACCCCTGTCTAG-F | |||||||
| SGCE 5 Anchor (5) | L-CCGATGTGATGTTTATGGCGTTCAG-P | 63.8 | 63.5 | 60.2 | 58.4 | |||
| SGCE 7F (6, 7 forward) | GGTGCAAAATTTCATTGGTTG | |||||||
| SGCE 7R (7 reverse) | GACCAGTGCCACTGCCGA | 57 | 30 | 191 | ||||
| SGCE sensor delT (7) | L-TCCTAATTACACTGGC_GTGCC-P | |||||||
| SGCE 7 anchor (7) | TTCTTTGAAAAGCAGAGACTATTACACGGA-F | 58.3 | 59.3 | 63.1 | 64.8 | |||
Sequences of primers and FRET hybridization probes used for the melting analysis of gDNA and mRNA (cDNA). The sensor probe for exon 5 is in the sense orientation and is wild-type–specific [A], and the sensor probe for exon 7 is in the antisense orientation and is mutant-specific [delT]. The sites of the mutations are underlined. “F” indicates a 3′-terminal fluorescein-label (donor dye), “L” is the 5′-terminal acceptor dye (LightCycler Red 640; Roche Diagnostics), and “P” is a 3′-terminal phosphate group preventing polymerase probe extension.
Bisulfite Sequencing
By bisulfite treatment of genomic DNA (gDNA), all unmethylated cytosine residues are converted to uracil, while methylated residues remain unaltered in this process. After subsequent methylation-specific PCR (MSP) and sequencing, all unmethylated cytosine residues appear as thymidines in the sequence. Bisulfite treatment of gDNA was performed using a protocol based on that of Frommer et al. (1992). In brief, 2 μg of DNA were alkaline denatured and treated overnight (for 16 h) with hydrochinone (500 μM) and sodium bisulfite (2.6 M). For desalting and the final desulfonation reaction, we used the CpGenome DNA Modification Kit (Intergen). Bisulfite-treated DNA was dissolved in 35 μl of 10-mM Tris-HCl (pH 8.0).
MSP was performed using Taq polymerase and the primers listed in table 4. PCR products were visualized on ethidium bromide–stained agarose gels and were extracted using Perfectprep Gel Cleanup (Eppendorf). PCR products were cloned in the pCR2.1Topo vector, through use of Topoisomerase (Topo TA-Cloning Kit; Invitrogen) to separate the parental alleles. PCR product–containing plasmids were isolated using the QIAprep Spin Miniprep Kit (Qiagen) and were sequenced using vector-specific primers (table 4).
Table 4.
MSP Primers and Primers for Plasmid Sequencing
| Primer Name (Region) | Sequence 5′→3′ | AnnealingTemperature(°C) | No. ofCycles | Length ofPCR Product(bp) |
| PromMF (promoter SGCE) | GTGTTATGTTTTATAAATAGATAAG | |||
| PromMR (promoter SGCE) | CAACTCATATACCTCTACAATTC | 59 | 35 | 376 + insert |
| Imp3F (promoter SGCE) | TTTGTTTTGGGGTATAGTTAGTGAAG | |||
| Imp3R (promoter SGCE) | ATACAACCTATATAAAACTCTCAATC | 57 | 35 | 392 + insert |
| M13 F (plasmid) | CACGACGTTGTAAAACGAC | |||
| M13 R (plasmid) | GGATAACAATTTCACACAGG | 60 | 35 | 227 + insert |
Genotyping
Haplotype analysis of the SGCE region on chromosome 7q was performed with the following polymorphic DNA markers (marker positions are given in parentheses): D7S2212 (95.43 cM), D7S2410 (100.81 cM), D7S1820 (105.92 cM), Col1A2mfd (106.46 cM), Col1A2 (106.46 cM), D7S2482 (108.59 cM), and S7S821 (109.12 cM) (Center for Medical Genetics Web site).
Results
Patients
In both families, the affected individuals showed characteristic clinical signs of M-D. Ages at onset for the three affected females were 4 and 9.5 years (family V) and 2 years (family S). All three patients showed typical myoclonus and dystonia.
Mutations in the SGCE Gene
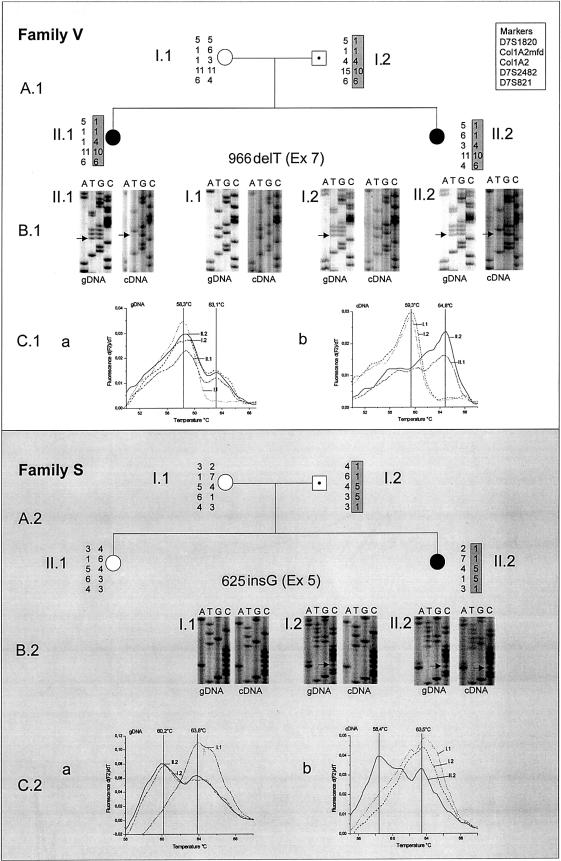
In family V, we detected a 1-bp deletion in exon 7 (966delT) (fig. 1B.1), resulting in frameshift at amino acid 322 and protein truncation at position 333. In family S, we found a 1-bp insertion in exon 5 (625insG), resulting in a missense protein from amino acid 210 onward, with protein truncation after position 216 (fig. 1B.2).
Figure 1.
Pedigrees of families V and S. A.1 and A.2, Pedigrees with haplotypes in the SGCE region on chromosome 7q21. Affected individuals are shaded in black; unaffected mutation carriers are indicated with a black dot. The mutated allele inherited from each of the fathers is highlighted in gray. B.1 and B.2, Sequences at the gDNA and cDNA levels, with the 966delT in exon 7 and 625insG in exon 5 (reverse strand is shown) indicated with arrows. C.1 and C.2, Melting curves generated with gDNA (a) and cDNA (b).
mRNA Expression Studies
At the cDNA level, sequence analysis revealed only one expressed allele in the mutation carriers in family V. The affected individuals expressed only the mutated allele, whereas the unaffected father expressed only the wild-type allele (fig. 1B.1). These results were confirmed by melting-curve analysis: the unaffected mutation-negative mother showed one melting-temperature peak at the gDNA level, whereas all three mutation carriers had two melting-temperature peaks, indicative of both the mutated and the wild-type alleles (fig. 1C.1a). At the cDNA level (fig. 1C.1b), both affected children had only the high melting-temperature peak (i.e., the mutated allele), whereas their parents had the low melting-temperature peak, corresponding to the wild-type allele.
In family S, the mutation-carrying father also expressed only the normal allele, whereas the affected daughter showed expression of both parental alleles, as shown by two different approaches (cDNA sequencing [fig. 1B.2] and melting-curve analysis [fig. 1C.2]). Contamination with gDNA was excluded through use of an exon-spanning nested PCR assay, and all experiments were performed in triplicate, with the same results. No other mutations were found in the coding region of the SGCE gene of the affected daughter in family S.
Search for Polymorphisms
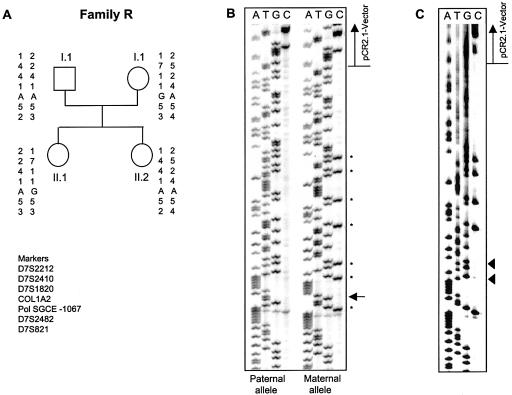
One of 20 families unaffected with M-D (family R) carried the −1067A→G polymorphism in the promoter region. In this family, the mother and the elder daughter were heterozygous for the polymorphism (A/G), whereas the father and the younger daughter were homozygous for the wild-type allele (A/A) (fig. 2). The same polymorphism was detected by SSCP analysis in 7 of 121 unrelated individuals, for whom no relatives were available. Neither of the two other known polymorphisms in the promoter region was detected in any individual studied. Also, we did not identify any polymorphisms in the transcribed regions of SGCE, including the exons and the 3′ UTR.
Figure 2.
Methylation studies. A, Pedigree of family R, with haplotypes in the SGCE chromosomal region. B, Sequences of two plasmids containing either the paternal (left) or maternal (right) allele of individual II.1 from family R, after bisulfite treatment. The polymorphism is indicated with an arrow. Sequences demonstrate that the maternal allele (with G) was methylated, since several Cs (indicated by asterisks) escaped conversion to Ts. In contrast, the paternal allele was unmethylated. C, Sequence of plasmid 5 of the PromM region derived from the index patient in family S, demonstrating the loss of two methylated Cs (indicated by triangles).
Imprinting Studies
Through bisulfite sequencing of the PromM region (nucleotide positions −1148 to −773 relative to the start ATG), we identified a total of 25 methylated cytosines at all CpG dinucleotides in a 376-bp part of the promoter region of the SGCE gene. These methylated cytosines were present on one parental allele only (fig. 2B). Using a polymorphism in the same region, we demonstrated in family R (which was unaffected with M-D) that the maternal allele, marked by the polymorphism (G), was methylated in 5/5 clones. In contrast, the paternal allele, as indicated by the wild-type A, was unmethylated (all cytosines converted to thymidine) in another 5/5 clones (fig. 2B). Thus, in this family, all of the maternal CpG dinucleotides were methylated in the region studied, whereas none of the paternal CpG dinucleotides was methylated (table 5).
Table 5.
Results of Sequencing of Cloned PCR Products after Bisulfite Conversion, Showing the Methylation Status in the PromM Region
|
No. of |
||||
| FamilyandIndividual | CompletelyUnmethylatedInserts | CompletelyMethylatedInserts | HypomethylatedInserts | Total No.of ClonesStudied |
| Family S: | ||||
| II.2 | 5 | 14 | 5 | 24 |
| I.1 | 8 | 6 | 10 | 24 |
| I.2 | 5 | 6 | 0 | 11 |
| Family R: | ||||
| II.1 | 5 | 5 | 0 | 10 |
Since we observed biallelic expression of the SGCE gene in the affected members of family S, we performed imprinting studies on this family, as well. The index patient also showed completely methylated (14/24 clones) and completely unmethylated (5/24 clones) alleles in the studied PromM region. In addition, we found hypomethylated alleles in 5/24 clones. The PromM PCR-product inserts contained 1 (twice), 3, 7, or 21 converted Cs, indicating that these CpG dinucleotides were unmethylated (table 5; example shown in fig. 2C). Similarly, another region of the promoter (Imp3) that is even closer to the translational start site (nucleotide positions −625 to −234 relative to the start ATG) also showed completely methylated (2/8 clones), completely unmethylated (2/8 clones), and hypomethylated (4/8 clones) alleles. The Imp3 region contained 28 differentially methylated CpG dinucleotides. Hypomethylated clones had 1, 2, 24, or 26 converted Cs (data not shown). The converted Cs appeared to be randomly spread over both regions studied. Unfortunately, parental origin of the alleles could not be determined, because of the lack of polymorphisms in family S.
Analysis of the methylation pattern in the father of this family, who showed expression of the wild-type allele only, revealed completely methylated (6/11) and completely unmethylated (5/11) alleles for the PromM region (table 5). In contrast, the mother had completely methylated (6/24), completely unmethylated (8/24), and hypomethylated (10/24) alleles in the same region (table 5). As in the daughter, hypomethylation occurred at random positions and variable frequencies (1 [three times], 2 [twice], 22, and 24 [four times] converted Cs in the 10 hypomethylated clones).
In addition, we found a few single-base-pair substitutions, at random positions in the inserts of all plasmids sequenced, that were generated from three different individuals with and without M-D. These sequence changes are most likely due to errors introduced by the Taq polymerase. We calculated the error rate for the Taq and the conditions used as 1:800 (0.13%).
Haplotype Analysis
In both families with M-D, affected individuals inherited the mutation-bearing allele from their unaffected fathers in an autosomal dominant fashion, as demonstrated by haplotype analysis (fig. 1A.1 and 1B.1). In family R (not affected with M-D), paternity was confirmed by haplotype analysis (fig. 2A).
Discussion
In both families with M-D, we identified novel mutations in the recently identified SGCE gene (Zimprich et al. 2001) and demonstrated autosomal dominant inheritance of these mutations in a putative sporadic case and in a family with a proposed recessive mode of inheritance. Pseudosporadic or pseudorecessive inheritance could be explained by reduced penetrance. In both families, the father carried the mutated allele but was clinically unaffected because he expressed only the wild-type copy while the mutated allele was silent. Interestingly, reduced penetrance of the disorder is found particularly in the offspring of affected women, as observed in clinical-genetic pedigree analyses (Zimprich et al. 2001). To explain this phenomenon, a possible maternal imprinting mechanism was proposed that leads to paternal expression of the mutated allele only (Zimprich et al. 2001). As shown in other imprinted disorders, imprinted mutations can be silently transmitted for generations until they are passed through the “expressing” parental sex to the affected offspring (Viljoen and Ramesar 1992). It is likely that the fathers from families V and S inherited the mutated alleles from their mothers, who were not available for molecular genetic studies.
Paternal Expression of the Human SGCE Gene in Family V
This is the first report, to our knowledge, of molecular evidence of paternal expression of the human SGCE gene. As demonstrated by cDNA sequencing and melting-curve analysis, both daughters from family V expressed only the paternal, mutated allele and were therefore clinically affected.
Maternal Imprinting of the Human SGCE Gene
Methylation of CpG dinucleotides correlates with transcriptional repression of genes, including imprinted genes. Mechanisms for imprinting include differentially methylated regions (DMRs) of promoters and of imprinting-control regions (Reik and Murrell 2000). By investigating a part of the promoter region of the human SGCE gene, we identified such DMRs. A polymorphism in family R served to distinguish the parental alleles after cloning and demonstrated that the maternal allele is methylated. This is indicative of silencing of this allele, as shown by cDNA analysis in family V, and explains reduced penetrance of the disorder when the condition is inherited from the mother.
Loss of Imprinting in Family S
Interestingly, the affected daughter of family S showed biallelic expression of the SGCE gene, probably because of loss of imprinting (LOI). Potential reasons for LOI are removal of DMRs or mutations in regions critical for regulation of imprinting (Ohta et al. 1999). Although differential methylation was observed in the promoter region in the affected daughter of family S, we identified several clones (9/32) with an incomplete methylation pattern, which is highly unlikely to represent an artifact. Interestingly, hypomethylated alleles were also identified in the mother but not in the father. Therefore, we assumed that all completely unmethylated alleles were inherited from the father and that maternal alleles were hypomethylated or completely methylated, in the sense of a methylation mosaic. It remains to be investigated whether the partial loss of methylation in the regions studied leads to the biallelic expression in the index patient of family S. It is conceivable that biallelic expression also occurs in the mother; however, this could not be investigated, because of the lack of sequence alterations in the transcribed region. The cause for the occurrence of hypomethylation in family S remains to be elucidated and will be further studied. It is possible that there is a mutation in a cis-acting regulating element necessary for imprinting of the SGCE chromosomal region in family S.
Interestingly, alterations in the imprinting pattern that coincide with expression of the maternal allele seem to occur in a considerable percentage of individuals. Analysis of published pedigrees identified at least 4 of 62 patients (6%) who inherited the disease from their mother and at least three asymptomatic carriers who inherited the mutated allele from their father (Zimprich et al. 2001). Remarkably, a weak expression of the maternal allele was also observed in mice (Piras et al. 2000). It remains to be investigated how frequently biallelic expression of the gene occurs in humans.
On the basis of our clinical data, there was no obvious phenotypic difference between the affected individuals of families S and V, in spite of their different SGCE expression patterns. Coexpression of the mutated and wild-type allele in the affected daughter of family S suggests that the former confers a dominant-negative effect or a dominant gain of function.
In conclusion, we provide molecular evidence for paternal expression of the human SGCE gene caused by a maternal imprinting mechanism, explaining reduced penetrance. We show biallelic expression of the SGCE gene in one affected individual, suggestive of LOI and possibly related to loss of methylation at several CpG dinucleotides.
Acknowledgments
We would like to thank the patients and family members who participated in this study. This work was supported by a research grant from the Fritz Thyssen Foundation (to N.K., J.G., and C.K.). We gratefully acknowledge Drs. T. Strom and T. Gasser, for sharing unpublished primer information.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SGCE [accession number NM_003919])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SGCE [MIM 604149] and M-D [MIM 159900])
References
- Asmus F, Zimprich A, Naumann M, Berg D, Bertram M, Ceballos-Baumann A, Pruszak-Seel R, Kabus C, Dichgans M, Fuchs S, Müller-Myhsok B, Gasser T (2001) Inherited myoclonus-dystonia syndrome: narrowing the 7q21-q31 locus in German families. Ann Neurol 49:121–124 [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA 89:1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser T (1998) Inherited myoclonus-dystonia syndrome. Adv Neurol 78:325–334 [PubMed] [Google Scholar]
- Grabowski M, Zimprich A, Lorenz-Depiereux B, Asmus F, Bauer M, Kalscheuer VM, Gasser T, Meitinger T, Strom TM (2002) Epsilon-sarcoglycan (SGCE), the gene mutated in myoclonus-dystonia syndrome, is imprinted. Paper presented at the 34th European Society of Human Genetics Conference, Strassbourg, France, May 25–28 [Google Scholar]
- Klein C (2002) Myoclonus and myoclonus-dystonias. In: Pulst S (ed) Genetics of movement disorders. Academic Press, San Diego, pp 449–469 [Google Scholar]
- Klein C, Schilling K, Saunders-Pullman RJ, Garrels J, Breakefield XO, Brin MF, deLeon D, et al (2000) A major locus for myoclonus-dystonia maps to chromosome 7q in eight families. Am J Hum Genet 67:1314–1319 [PMC free article] [PubMed] [Google Scholar]
- Nygaard TG, Raymond D, Chen C, Nishino I, Greene PE, Jennings D, Heiman GA, Klein C, Saunders-Pullman RJ, Kramer P, Ozelius LJ, Bressman SB (1999) Localization of a gene for myoclonus-dystonia to chromosome 7q21-q31. Ann Neurol 46:794–798 [DOI] [PubMed] [Google Scholar]
- Ohta T, Gray TA, Rogan PK, Buiting K, Gabriel JM, Saitoh S, Muralidhar B, Bilienska B, Krajewska-Walasek M, Driscoll DJ, Horsthemke B, Butler MG, Nicholls RD (1999) Imprinting-mutation mechanisms in Prader-Willi syndrome. Am J Hum Genet 64:397–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras G, El Kharroubi A, Kozlov S, Escalante-Alcalde D, Hernandez L, Copeland NG, Gilbert DJ, Jenkins NA, Stewart CL (2000) Zac1 (Lot1), a potential tumor suppressor gene, and the gene for ε-sarcoglycan are maternally imprinted genes: identification by a subtractive screen of novel uniparental fibroblast lines. Mol Cell Biol 20:3308–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A, Cedar H (1994) DNA methylation and genomic imprinting. Cell 77:473–476 [DOI] [PubMed] [Google Scholar]
- Reik W, Murrell A (2000) Genomic imprinting: silence across the border. Nature 405:408–409 [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J (2001) Evolution of imprinting mechanisms: the battle of the sexes begins in the zygote. Nat Genet 27:255–256 [DOI] [PubMed] [Google Scholar]
- Vidailhet M, Tassin J, Durif F, Nivelon-Chevallier A, Agid Y, Brice A, Dürr A (2001) A major locus for several phenotypes of myoclonus-dystonia on chromosome 7q. Neurology 56:1213–1216 [DOI] [PubMed] [Google Scholar]
- Viljoen D, Ramesar R (1992) Evidence for paternal imprinting in familial Beckwith-Wiedemann syndrome. J Med Genet 29:221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Grabowski M, Asmus F, Naumann M, Berg D, Bertram M, Scheidtmann K, Kern P, Winkelmann J, Müller-Myhsok B, Riedel L, Bauer M, Müller T, Castro M, Meitinger T, Strom TM, Gasser T (2001) Mutations in the gene encoding ε-sarcoglycan cause myoclonus-dystonia syndrome. Nat Genet 29:66–69 [DOI] [PubMed] [Google Scholar]