Abstract
The polymorphism of p53 codon 72, a transversion of G to C (Arg to Pro), has been demonstrated to be associated with the risk for lung cancer. However, individual studies conducted in Asians have provided conflicting and inconclusive findings. Thus, we performed a meta-analysis by pooling all currently available case–control studies to estimate the effect of p53 codon 72 Arg/Pro polymorphism on the development of lung cancer. The pooled odds ratios (ORs) with the corresponding 95 % confidence intervals (95 %CIs) were calculated to assess this effect. A total of 14 individual studies involving 7,929 cases and 5,924 controls were included into this meta-analysis according to the inclusion criteria. The overall OR for the dominant genetic model indicated that the p53 codon 72 Arg/Pro variant was positively correlated with lung cancer risk (ORArg/Pro + Pro/Pro vs. Arg/Arg = 1.14, 95 %CI 1.07–1.23, P OR < 0.001). Similar results were found in the stratified analysis of population-based studies. The histological types of lung cancer and smoking status seemed to exert no effect on the lung cancer risk. Sensitivity analysis confirmed the stability of the above findings. The updated meta-analysis suggests that the p53 codon 72 Arg/Pro polymorphism is a risk factor for lung cancer in the Asian population. However, the potential role of gene–environment interaction in lung cancer susceptibility needs further investigation in future studies with high quality.
Keywords: Lung cancer, P53 codon 72, Polymorphism, Meta-analysis
Introduction
Lung cancer is one of the leading causes of cancer-related death in the world and a major public health challenge during the past few decades [1]. Tobacco smoking and alcohol consumption have been well-established as risk factors for lung cancer [2]. Despite the obvious carcinogenic effects of smoking and alcohol consumption, not all exposed individuals develop lung cancer, suggesting that some other factors including genetic polymorphism may also contribute to the pathogenesis of lung cancer. A number of genetic polymorphisms have been demonstrated to alter the risk of this deadly disease independently or in combination with each other as well as the environmental exposures [2–4]. Examination of the genetic polymorphisms may help to interpret the variation in individual lung cancer risk.
The p53 tumor suppressor gene, being located on chromosome 17p13, is one of the most frequently mutated genes in human cancer with a predominance of missense mutations scattered over 200 codons [5]. The encoded p53 protein can be activated by a variety of cellular stresses, which plays pivotal roles in the maintenance of genomic stability, the regulation of apoptosis, and cell cycle by transactivating the downstream target genes [6]. Mutations of the p53 gene may result in loss of its tumor suppressor function and thus contribute to the carcinogenesis and/or tumor progression. The polymorphism of p53 codon 72, a transversion of G to C (Arg to Pro), has been shown to be related to the risk of some malignant tumors [7–9]. It has been confirmed that the two polymorphic variants, Arg and Pro, of p53 codon 72 differ biochemically and biologically [10, 11]. Therefore, the polymorphism of p53 codon 72 Arg/Pro may exert different effects on diverse types of cancer or even the same cancer in different populations. The homozygote genotype Arg/Arg was found to be significantly associated with an increased risk of cervical cancer in a previous meta-analysis [12]. Nevertheless, a recent meta-analysis by Liu et al. suggested that the p53 codon 72 Arg/Arg genotype played a protective role in gastric cancer risk among Asians [13]. Regarding the lung cancer risk, the effect of p53 codon 72 Arg/Pro polymorphism on the Asian patients remained unclear due to the conflicting and inconclusive findings across individual studies. The aim of this meta-analysis with a large amount of available data was to estimate the association of p53 codon 72 Arg/Pro polymorphism with lung cancer risk in the Asian population.
Materials and methods
Search strategy and identification of relevant studies
We performed a comprehensive search of the PubMed, Embase, Web of Science, and Wanfang databases to identify potentially relevant studies on the association between p53 codon 72 Arg/Pro polymorphism and lung cancer risk up to December 6, 2012. All eligible studies were retrieved. Reviews and the references of eligible studies were hand-searched for additional relevant publications. When more than one publications with overlapping data, the most recent or complete one was selected. The following terms were used in the literature search: p53, p53 codon 72, p53 codon 72 Arg/Pro, or rs1042522; and lung cancer, lung carcinoma, or lung; and polymorphism, polymorphisms, or mutation.
Inclusion criteria
Studies were included into the meta-analysis if they satisfied the following criteria: (1) assessing the association of p53 codon 72 Arg/Pro polymorphism with lung cancer risk; (2) applying a case–control design; (3) and providing the frequencies of both alleles and genotypes in cases and controls or available information to calculate them. Case-only studies and reviews were all excluded.
Data extraction
Data were carefully extracted independently by two investigators from all included studies according to the above inclusion criteria. Disagreements were resolved by discussion. The extracted data comprised the following items: surname of the first author, publication year, country of origin, summary characteristics of cases and controls, genotyping method, number of cases and controls, frequencies of the alleles and genotypes in cases and controls, matching factor, smoking status of subjects, source of controls, and the Hardy–Weinberg equilibrium (HWE) of genotype distribution among controls. Subjects were divided into smokers and nonsmokers based on their smoking history. According to the histological types of lung cancer, two subgroups of squamous cancer (SC) and adenocarcinoma (AC) were made. In addition, subgroups of hospital-based study and population-based study were assigned by the source of controls.
Statistical analysis
The pooled odds ratios (ORs) with the corresponding 95 % confidence intervals (95 %CIs) were used to evaluate the strength of the association between p53 codon 72 Arg/Pro polymorphism and lung cancer risk. Five genetic contrast models involving the allelic (Pro allele vs. Arg allele), homozygous (Pro/Pro vs. Arg/Arg), additive (Arg/Pro vs. Arg/Arg), recessive (Pro/Pro vs. Arg/Arg + Arg/Pro), and dominant (Arg/Pro + Pro/Pro vs. Arg/Arg) models were applied. The chi-square-based Q statistic test and I 2 test were adopted to estimate the between-study heterogeneity among all included studies, and the heterogeneity was statistically significant if the p value is less than 0.05 and the I 2 is more than 50 % [14, 15]. The fixed-effect model by the Mantel–Haenszel's method was used to calculate the pooled ORs when the between-study heterogeneity was significant [16]; otherwise, the random-effect model by DerSimonian and Laird's method was applied [17]. The pooled ORs were estimated by use of Z statistic test with the significance level set at p < 0.05. Subgroup analyses by source of controls, histological types of lung cancer, and smoking status were performed to further identify the association between p53 codon 72 Arg/Pro polymorphism and lung cancer susceptibility. Sensitivity analysis by sequential omission of any individual studies was also conducted to confirm the stability and reliability of the pooled results in the meta-analysis [18]. Both Begg's funnel plot and Egger's test were adopted to evaluate the publication bias in our meta-analysis [19, 20]. All statistical analyses were performed by STATA 12.0 software (StataCorp, College Station, TX, USA).
Results
Study characteristics
A total of 14 case–control studies on the association of p53 codon 72 Arg/Pro polymorphism and lung cancer risk in Asian population were retrieved based on the inclusion criteria [21–34]. The characteristics of all studies were summarized in Table 1. The included studies were mainly carried out in China, Japan, Korea, Singapore, and USA. Among the 14 publications, ten ones were published in English [21–30] and the others were in Chinese [31–34]. According to the source of controls, nine individual studies including 6,228 cases and 4,420 controls were categorized into population-based case–control studies, four involving 699 cases and 819 controls were hospital-based case–control studies, and still one with 1,687 subjects was conducted in a mixed population. The genotype distribution in controls of the included studies all agreed with HWE, except the two ones by Liu et al. and Shao et al., respectively. The genotype frequencies for Arg/Arg, Pro/Pro, and Arg/Pro of cases and controls were presented in Table 1 in detail.
Table 1.
Summary characteristic for all included studies in the meta-analysis
| First author | Year | Source of controls | Country | HWE | Genotype distribution in cases and controls | Matching factor | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pro/Pro | Arg/Pro | Arg/Arg | Pro/Pro | Arg/Pro | Arg/Arg | ||||||
| Murata M | 1996 | HCC | Japan | + | 22 | 89 | 80 | 37 | 131 | 99 | Age and gender |
| Wang YC | 1999 | HCC | China | + | 52 | 74 | 68 | 30 | 75 | 47 | Age |
| Pierce LM | 2000 | PCC | USA | + | 19 | 51 | 41 | 23 | 65 | 82 | Sex, ethnicity, and age |
| Hiraki A | 2003 | HCC | Japan | + | 24 | 99 | 68 | 43 | 106 | 90 | Age and gender |
| Zhang JH | 2003 | PCC | China | + | 32 | 45 | 21 | 27 | 69 | 40 | Age, gender, ethnicity, and residence |
| Shao GG | 2005 | PCC | China | NR | 48 | 16 | 24 | 37 | 42 | 33 | Ethnicity and residence |
| Sakiyama T | 2005 | Mixed | Japan | + | 144 | 460 | 398 | 73 | 310 | 302 | Age, race, and smoking history |
| Zhang X | 2006 | PCC | China | + | 279 | 506 | 321 | 264 | 731 | 425 | Age, gender, ethnicity, and residence |
| Jung HY | 2008 | PCC | Korea | + | 42 | 130 | 108 | 37 | 136 | 120 | NR |
| Li RN | 2009 | PCC | China | + | 17 | 58 | 50 | 22 | 42 | 37 | Ethnicity and residence |
| Chua HW | 2010 | HCC | Singapore | + | 26 | 69 | 28 | 31 | 88 | 42 | The hospital, age, and frequency |
| Wang W | 2010 | PCC | China | + | 35 | 45 | 44 | 20 | 58 | 50 | Ethnicity and residence |
| Piao JM | 2011 | PCC | Korea | + | 657 | 1,821 | 1,458 | 190 | 776 | 734 | Age and gender |
| Liu D | 2012 | PCC | China | NR | 79 | 137 | 144 | 119 | 115 | 126 | Age, gender, ethnicity, frequency, and residence |
HCC hospital-based case–control study, PCC population-based case–control study, HWE Hardy–Weinberg equilibrium, NR not reported
Meta-analysis results
Total studies
The pooled ORs of all included case–control studies revealed that there was a statistically significant association between the p53 codon 72 Arg/Pro polymorphism and lung cancer risk in the dominant model (ORArg/Pro + Pro/Pro vs. Arg/Arg = 1.14, 95 %CI 1.07–1.23, P OR < 0.001), while no significant association was found in other genetic models (ORPro allele vs. Arg allele = 1.11, 95 %CI 1.00–0.24, P OR = 0.062; ORPro/Pro vs. Arg/Arg = 1.23, 95 %CI 0.98–1.54, P OR = 0.077; ORArg/Pro vs. Arg/Arg = 1.07, 95 %CI 0.99–1.16, P OR = 0.072; ORPro/Pro vs. Arg/Arg + Arg/Pro = 1.22, 95 %CI 0.98–1.53, P OR = 0.073) (Table 2 and Fig. 1). However, individuals carrying the mutant Pro allele were more likely to develop lung cancer, although there is lack of statistical significance in the allelic, homozygous, additive, and recessive genetic models.
Table 2.
Meta-analysis results for the p53 codon 72 polymorphism and lung cancer risk
| Group/subgroup | Cases/controls | Odds ratio | I 2 (%) | P H | |
|---|---|---|---|---|---|
| OR [95 %CI] | P OR | ||||
| Total studies | 7,929/5,924 | ||||
| Pro allele vs. Arg allele | 1.11 [1.00–1.24] | 0.062 | 70.6 | <0.001 | |
| Pro/Pro vs. Arg/Arg | 1.23 [0.98–1.54] | 0.077 | 70.9 | <0.001 | |
| Arg/Pro vs. Arg/Arg | 1.07 [0.99–1.16] | 0.072 | 21.9 | 0.216 | |
| Arg/Pro + Pro/Pro vs. Arg/Arg | 1.14 [1.07–1.23] | <0.001 | 32.6 | 0.115 | |
| Pro/Pro vs. Arg/Arg + Arg/Pro | 1.22 [0.98–1.53] | 0.073 | 76.2 | <0.001 | |
| Hospital-based studies | 699/819 | ||||
| Pro allele vs. Arg allele | 0.97 [0.84–1.12] | 0.687 | 0.0 | 0.606 | |
| Pro/Pro vs. Arg/Arg | 0.93 [0.69–1.27] | 0.655 | 0.0 | 0.460 | |
| Pro/Arg vs. Arg/Arg | 0.95 [0.76–1.19] | 0.666 | 28.1 | 0.244 | |
| Pro/Arg + Pro/Pro vs. Arg/Arg | 0.95 [0.77–1.18] | 0.631 | 0.0 | 0.550 | |
| Pro/Pro vs. Arg/Arg + Pro/Arg | 0.98 [0.75–1.28] | 0.887 | 44.2 | 0.146 | |
| Population-based studies | 6,228/4,420 | ||||
| Pro allele vs. Arg allele | 1.16 [0.99–1.35] | 0.059 | 77.7 | <0.001 | |
| Pro/Pro vs. Arg/Arg | 1.32 [0.97–1.78] | 0.073 | 77.6 | <0.001 | |
| Arg/Pro vs. Arg/Arg | 1.08 [0.99–1.18] | 0.077 | 28.2 | 0.193 | |
| Arg/Pro + Pro/Pro vs. Arg/Arg | 1.17 [1.07–1.27] | <0.001 | 42.1 | 0.087 | |
| Pro/Pro vs. Arg/Arg + Arg/Pro | 1.32 [0.98–1.77] | 0.069 | 81.8 | <0.001 | |
| Histological types | |||||
| SC | 1,062/3,199 | ||||
| Pro allele vs. Arg allele | 0.99 [0.65–1.50] | 0.951 | 92.0 | <0.001 | |
| Pro/Pro vs. Arg/Arg | 0.96 [0.40–2.28] | 0.919 | 91.6 | <0.001 | |
| Arg/Pro vs. Arg/Arg | 1.04 [0.88–1.23] | 0.648 | 44.4 | 0.145 | |
| Arg/Pro + Pro/Pro vs. Arg/Arg | 1.06 [0.75–1.49] | 0.762 | 74.1 | 0.009 | |
| Pro/Pro vs. Arg/Arg + Arg/Pro | 0.94 [0.52–1.69] | 0.827 | 86.8 | <0.001 | |
| AC | 1,602/3,199 | ||||
| Pro allele vs. Arg allele | 1.01 [0.84–1.21] | 0.935 | 65.7 | 0.033 | |
| Pro/Pro vs. Arg/Arg | 1.17 [0.96–1.44] | 0.125 | 59.7 | 0.059 | |
| Arg/Pro vs. Arg/Arg | 1.02 [0.88–1.19] | 0.805 | 11.3 | 0.336 | |
| Arg/Pro + Pro/Pro vs. Arg/Arg | 1.05 [0.91–1.21] | 0.491 | 49.2 | 0.116 | |
| Pro/Pro vs. Arg/Arg + Arg/Pro | 1.03 [0.73–1.47] | 0.853 | 71.2 | 0.002 | |
| Smoking status | |||||
| Smokers | 2,139/1,868 | ||||
| Pro allele vs. Arg allele | 0.92 [0.66–1.27] | 0.595 | 85.5 | <0.001 | |
| Pro/Pro vs. Arg/Arg | 0.86 [0.45–1.65] | 0.642 | 85.7 | <0.001 | |
| Arg/Pro vs. Arg/Arg | 0.95 [0.79–1.13] | 0.561 | 0.0 | 0.552 | |
| Arg/Pro + Pro/Pro vs. Arg/Arg | 1.06 [0.91–1.25] | 0.528 | 28.6 | 0.240 | |
| Pro/Pro vs. Arg/Arg + Arg/Pro | 1.08 [0.68–1.71] | 0.744 | 82.7 | <0.001 | |
| Nonsmokers | 1,247/2,139 | ||||
| Pro allele vs. Arg allele | 1.04 [0.92–1.18] | 0.521 | 48.3 | 0.102 | |
| Pro/Pro vs. Arg/Arg | 1.15 [0.90–1.46] | 0.272 | 0.0 | 0.521 | |
| Arg/Pro vs. Arg/Arg | 0.77 [0.52–1.15] | 0.207 | 64.7 | 0.023 | |
| Arg/Pro + Pro/Pro vs. Arg/Arg | 0.86 [0.61–1.20] | 0.371 | 59.3 | 0.043 | |
| Pro/Pro vs. Arg/Arg + Arg/Pro | 1.24 [0.89–1.74] | 0.195 | 42.6 | 0.083 | |
OR odds ratio, 95 %CI 95 % confidence interval, P H P value of heterogeneity analysis, SC squamous cancer, AC adenocarcinoma
Fig. 1.
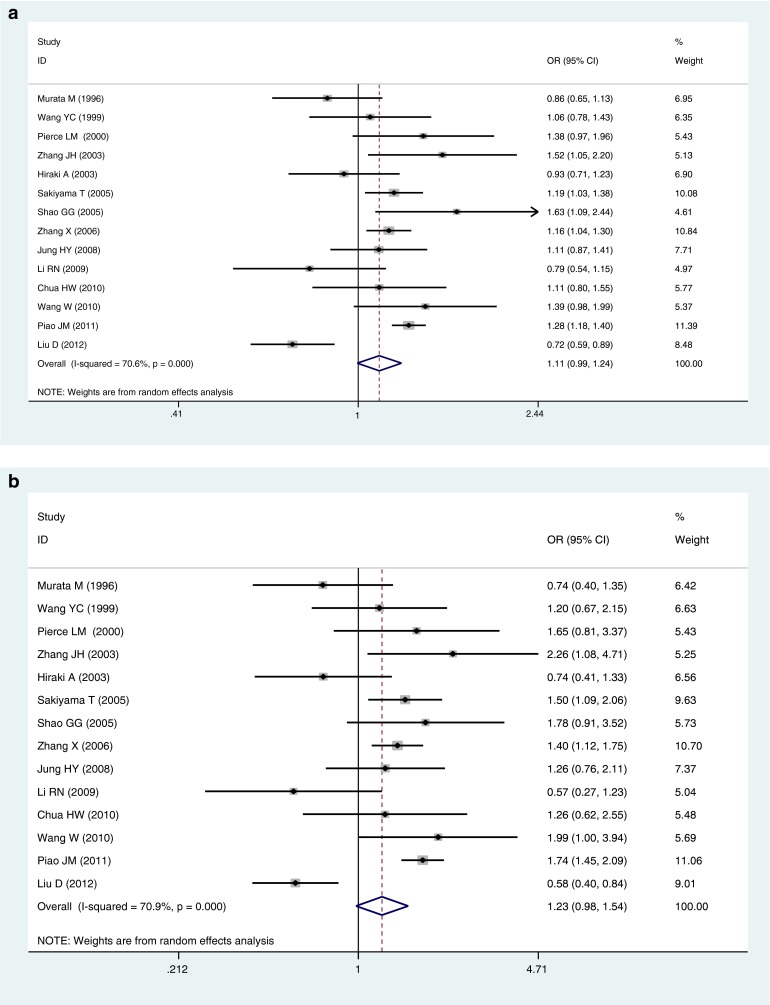
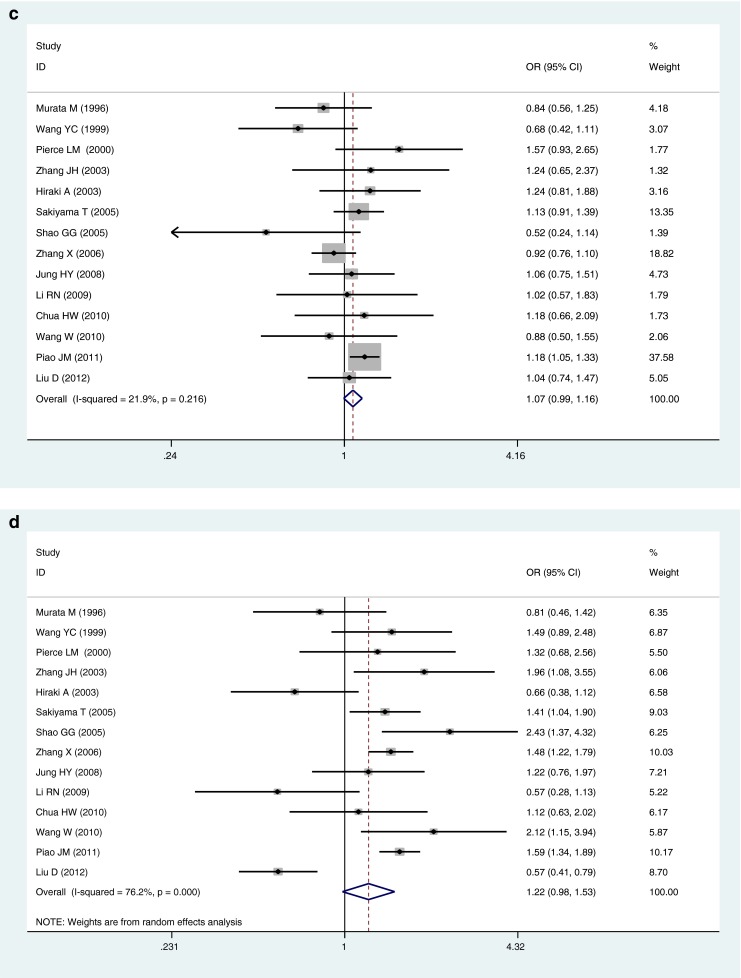
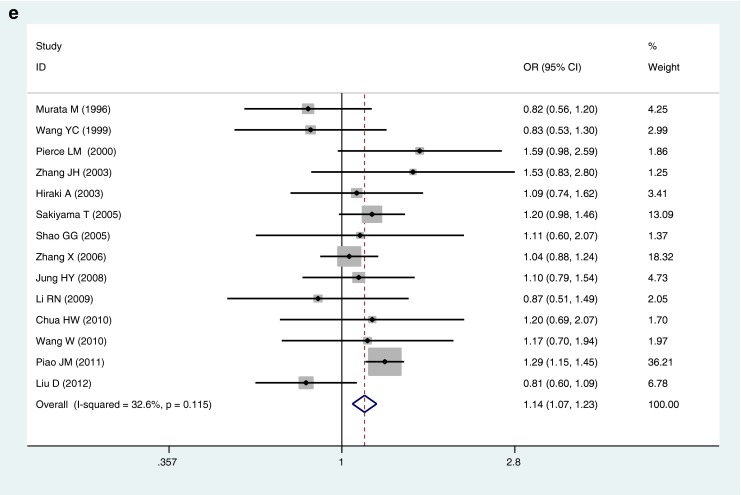
Forest plots for the association of P53 codon 72 polymorphism and lung cancer risk in total studies. a Pro allele vs. Arg allele, the allelic model. b Pro/Pro vs. Arg/Arg, the homozygous model. c Arg/Pro vs. Arg/Arg, the additive model. d Pro/Pro vs. Arg/Arg + Arg/Pro, the recessive model. e Arg/Pro + Pro/Pro vs. Arg/Arg, the dominant model
Subgroup analysis by source of controls
In stratifying analysis by source of controls, significantly increased risk was found when comparing the combined genotype Arg/Pro + Pro/Pro with Arg/Arg in population-based studies (ORArg/Pro + Pro/Pro vs. Arg/Arg = 1.17, 95 %CI 1.07–1.27, P OR < 0.001). Similarly, a positive association was found in the allelic, homozygous, additive, and recessive models, but there was no statistical significance (Table 2). Interestingly, the variant of p53 codon 72 Arg/Pro was negatively associated with lung cancer risk in the subgroup analysis of hospital-based studies (Table 2). Nevertheless, the finding may be a chance, in that there were only four individual hospital-based studies with a total of 699 cases and 819 controls included into our meta-analysis.
Subgroup analysis by histological types of lung cancer
Among individuals with SC and AC of lung cancer, no statistically significant association of the p53 codon 72 Arg/Pro polymorphism with lung cancer risk was observed under all genetic contrast models (Table 2). Additionally, patients with SC had lower frequencies of Pro/Pro genotype and Pro allele in comparison with the AC patients, although there was no statistical significance (Table 2).
Subgroup analysis by smoking status
When stratifying by smoking status, the p53 codon 72 Arg/Pro variant was not related to increased or decreased risk of lung cancer in mutant Pro carriers no matter smoking or not (Table 2). Besides, the nonsmokers had higher frequencies of Pro/Pro genotype and Pro allele compared with the smokers, but lack statistical significance (Table 2).
Heterogeneity and sensitivity analyses
The between-study heterogeneity was significant in allelic, homozygous, and recessive models among total studies (Pro allele vs. Arg allele, I 2 = 70.6, P H < 0.001; Pro/Pro vs. Arg/Arg, I 2 = 70.9, P H < 0.001; Pro/Pro vs. Arg/Arg + Arg/Pro, I 2 = 76.2, P H < 0.001) (Table 2); consequently, the random-effect model was used to estimate the pooled ORs. In reverse, the pooled ORs for Arg/Pro vs. Arg/Arg and Arg/Pro + Pro/Pro vs. Arg/Arg were evaluated by fixed-effect model (Table 2). We made subgroup analysis to identify the between-study heterogeneity among the included studies. No significant heterogeneity was observed among the hospital-based studies, but the population-based ones (Table 2). Sensitivity analysis by sequential omission of individual studies one at a time confirmed the stability and reliability of the results in our meta-analysis (data not shown).
Publication bias
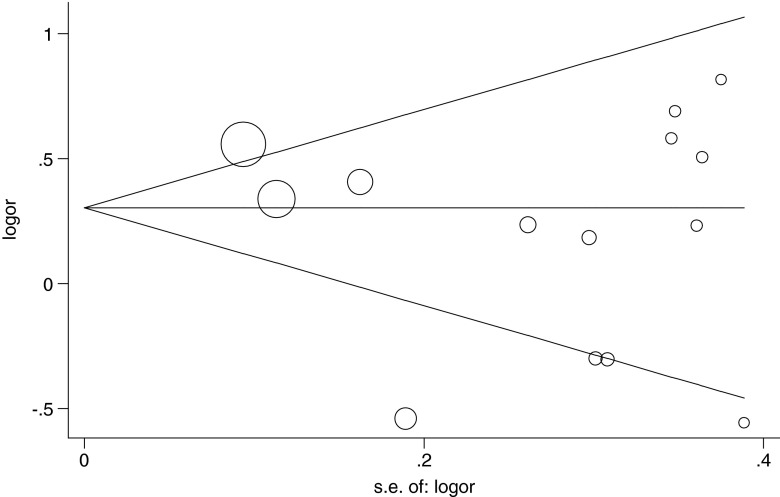
The publication bias was evaluated by Begg's funnel plot and Egger's test. No visual asymmetry was found in the funnel plot analysis (Fig. 2), suggesting no publication bias among the included studies. In addition, the results of Egger's tests for all genetic models also did not indicate publication bias in the present meta-analysis (data not shown).
Fig. 2.
Begg's funnel plot with pseudo-95 % confidence limits for estimating the publication bias
Discussion
The p53 codon 72 Arg/Pro variant, the most informative polymorphism of p53 gene, has been found to be significantly associated with an increased risk of lung cancer in a previous meta-analysis [35]. However, not all publications included into the meta-analysis agreed with HWE, and only eight individual studies were conducted in the Asian population. Moreover, they did not assess the role of p53 codon 72 Arg/Pro polymorphism in lung cancer risk concerning the histological types of lung cancer and smoking status among Asians. In the present meta-analysis, a significant relationship of the p53 codon 72 Arg/Pro variant with lung cancer risk was identified under the dominant genetic model in overall (ORArg/Pro + Pro/Pro vs. Arg/Arg = 1.14) and the subgroup analyses of population-based studies (ORArg/Pro + Pro/Pro vs. Arg/Arg = 1.17) in Asians. A slightly but not statistically significant association was found in the allelic, homozygous, additive, and recessive contrast models. Additionally, no statistically significant correlation was observed under all genetic contrast models in subgroup analyses according to the histological types of lung cancer and smoking status.
Lung cancer risk increases with cigarette smoking and other environmental exposures. Zhou et al. found that smoking could modify the effects of X-ray cross-complementing group 1 and excision repair cross-complementing group 2 polymorphisms on the risk for lung cancer, indicating a gene–environmental interaction in the lung carcinogenesis [36]. It has been well-established that there is a range of genetic susceptibility to lung cancer risk, such as microsomal epoxide hydrolase 1, matrix metalloproteinase-1, and glutathione S-transferase P1 [37–39]. In addition, the polymorphism of p53 codon 72 Arg/Pro also has been demonstrated to modify the risk for lung cancer among Asians in many previous case–control studies [21–34]. Murata et al. initially reported that the Arg/Arg homozygote seemed to be susceptible to smoking-unrelated lung cancer, while the Pro/Pro homozygote did not exert effects on the risk of this deadly disease [21]. A recent study by Liu et al. revealed that the frequencies of Pro/Pro genotype and Pro allele were lower in lung cancer patients and might modify the risk for smoking-related lung cancer in a Chinese population [30]. Reversely, our meta-analysis of all included studies showed that the Pro/Pro genotype and Pro allele were predominant in Asians with lung cancer, although there is lack of statistical significance. Interestingly, the mutant Pro/Pro genotype and Pro allele of p53 codon 72 played a protective but not statistically significant role in lung cancer risk in subgroup analysis of hospital-based studies. Nevertheless, there were only four individual hospital-based studies involving 699 cases and 819 controls included in this meta-analysis. Studies with small sample size were insufficient enough in statistical power to determine a true association. When stratifying by the histological types of lung cancer, patients with SC had lower frequencies of Pro/Pro genotype and Pro allele compared with patients with AC, although there was no statistical significance. When considering the stratified analysis based on the smoking status, the frequencies of Pro/Pro genotype and Pro allele were lower but not statistically significant in smoking-related lung cancer patients, similar to the finding by Liu et al. [30].
The frequencies of p53 codon 72 alleles and haplotypes differ across ethnicities [40], which may be the leading cause for different effects of the p53 codon 72 Arg/Pro polymorphism on lung cancer risk in different ethnicities. Significantly increased risks for lung cancer were found among Asians, but not Africans for both the homozygote Pro/Pro and the Pro allele carriers in a previous meta-analysis by Li et al. [41]. Similarly, the present meta-analysis with more available data suggests that the p53 codon 72 Arg/Pro variant is a risk factor for lung cancer in Asians. The inconsistent findings among ethnicities may be attributed to different genetic backgrounds and environments. Furthermore, the effect of p53 codon 72 Arg/Pro polymorphism on lung cancer risk differs in Asians. Jung et al. demonstrated that the p53 codon 72 Arg/Pro polymorphism was not significantly associated with lung cancer susceptibility in a Korean population [27]. On the contrary, the p53 codon 72 polymorphism was confirmed to be related to an elevated risk of lung cancer in another Korean population [29]. Different study design, sample size, genotyping method, and source of controls may be responsible for the conflicting findings among individual studies.
To the best of our knowledge, tobacco smoking is a major risk factor for lung cancer. It can affect carcinogenesis by interacting with the genetic polymorphisms. Tobacco carcinogens have been shown to exert a direct mutagenic action on DNA, particularly on p53 [42]. The study by Liu et al. suggested that the p53 codon 72 polymorphism modified the smoking-related lung cancer risk, indicating the smoking–gene interplay in lung carcinogenesis [30]. Nevertheless, we found that the p53 codon 72 Arg/Pro polymorphism was not significantly associated with the risk of lung cancer no matter among smokers or nonsmokers. Future studies with high quality and large sample size may further explore the potential role of gene–environment interactions in lung cancer risk.
Some limitations should be acknowledged when elucidating the results of our meta-analysis. Firstly, the genotype distribution of p53 codon 72 among controls was not all in agreement with HWE among total included studies. Secondly, as above mentioned, the appreciable association of smoking–gene interplay with lung cancer risk needs further confirmation in more future studies with high quality. Last but not the least, the p53 codon 72 Arg/Pro polymorphism may modify the risk for lung cancer among Asians in combination with other genetic polymorphisms, such as cytochrome P-450 1A1 [43] and p21 Ser31Arg [44]. Thus, the possible effect of gene–gene interactions on lung cancer risk is recommended to be further investigated.
In summary, our meta-analysis shows a significant association between the p53 codon 72 Arg/Pro polymorphism and lung cancer risk in the Asian population. However, we failed to identify this association regarding the smoking status and histological types of lung cancer patients. In addition, the potential effect of gene–gene and gene–environment interactions on lung cancer development needs further investigation in future studies.
Acknowledgments
Conflicts of interest
None
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Steliga MA, Dresler CM. Epidemiology of lung cancer: smoking, secondhand smoke, and genetics. Surg Oncol Clin N Am. 2011;20:605–18. doi: 10.1016/j.soc.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Kiyohara C, Yoshimasu K, Takayama K, Nakanishi Y. EPHX1 polymorphisms and the risk of lung cancer: a huge review. Epidemiology. 2006;17:89–99. doi: 10.1097/01.ede.0000187627.70026.23. [DOI] [PubMed] [Google Scholar]
- 4.Torok S, Hegedus B, Laszlo V, Hoda MA, Ghanim B, Berger W, et al. Lung cancer in never smokers. Future Oncol. 2011;7:1195–211. doi: 10.2217/fon.11.100. [DOI] [PubMed] [Google Scholar]
- 5.Szymanska K, Hainaut P. TP53 and mutations in human cancer. Acta Biochim Pol. 2003;50:231–8. [PubMed] [Google Scholar]
- 6.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–31. doi: 10.1016/S0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 7.Yu H, Huang YJ, Liu Z, Wang LE, Li G, Sturgis EM, et al. Effects of MDM2 promoter polymorphisms and p53 codon 72 polymorphism on risk and age at onset of squamous cell carcinoma of the head and neck. Mol Carcinog. 2011;50:697–706. doi: 10.1002/mc.20806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Qasem A, Toulimat M, Tulbah A, Elkum N, Al-Tweigeri T, Aboussekhra A. The p53 codon 72 polymorphism is associated with risk and early onset of breast cancer among Saudi women. Oncol Lett. 2012;3:875–8. doi: 10.3892/ol.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding C, Yu H, Qin H. TP53 codon 72 polymorphism with hepatocellular carcinoma: a metaanalysis. J Int Med Res. 2012;40:446–54. doi: 10.1177/147323001204000206. [DOI] [PubMed] [Google Scholar]
- 10.Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19:1092–100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreau F, Matlashewski G. Molecular analysis of different allelic variants of wild-type human p53. Biochem Cell Biol. 1992;70:1014–9. doi: 10.1139/o92-145. [DOI] [PubMed] [Google Scholar]
- 12.Koushik A, Platt RW, Franco EL. p53 codon 72 polymorphism and cervical neoplasia: a meta-analysis review. Cancer Epidemiol Biomark Prev. 2004;13:11–22. doi: 10.1158/1055-9965.EPI-083-3. [DOI] [PubMed] [Google Scholar]
- 13.Liu KJ, Qi HZ, Yao HL, Lei SL, Lei ZD, Li TG, et al. An updated meta-analysis of the p53 codon 72 polymorphism and gastric cancer risk. Mol Biol Rep. 2012;39:8265–75. doi: 10.1007/s11033-012-1674-0. [DOI] [PubMed] [Google Scholar]
- 14.Cochran WG. The comparison of percentages in matched samples. Biometrika. 1950;37:256–66. doi: 10.1093/biomet/37.3-4.256. [DOI] [PubMed] [Google Scholar]
- 15.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 17.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 18.Attia J, Thakkinstian A, D’Este C. Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol. 2003;56:297–303. doi: 10.1016/S0895-4356(03)00011-8. [DOI] [PubMed] [Google Scholar]
- 19.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuck AE, Rubenstein LZ, Wieland D. Bias in meta-analysis detected by a simple, graphical test. Asymmetry detected in funnel plot was probably due to true heterogeneity. BMJ. 1998;316:469. doi: 10.1136/bmj.316.7129.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murata M, Tagawa M, Kimura M, Kimura H, Watanabe S, Saisho H. Analysis of a germ line polymorphism of the p53 gene in lung cancer patients; discrete results with smoking history. Carcinogenesis. 1996;17:261–4. doi: 10.1093/carcin/17.2.261. [DOI] [PubMed] [Google Scholar]
- 22.Wang YC, Chen CY, Chen SK, Chang YY, Lin P. p53 codon 72 polymorphism in Taiwanese lung cancer patients: association with lung cancer susceptibility and prognosis. Clin Cancer Res. 1999;5:129–34. [PubMed] [Google Scholar]
- 23.Pierce LM, Sivaraman L, Chang W, Lum A, Donlon T, Seifried A, et al. Relationships of TP53 codon 72 and HRAS1 polymorphisms with lung cancer risk in an ethnically diverse population. Cancer Epidemiol Biomark Prev. 2000;9:1199–204. [PubMed] [Google Scholar]
- 24.Hiraki A, Matsuo K, Hamajima N, Ito H, Hatooka S, Suyama M, et al. Different risk relations with smoking for non-small-cell lung cancer: comparison of TP53 and TP73 genotypes. Asian Pac J Cancer Prev. 2003;4:107–12. [PubMed] [Google Scholar]
- 25.Sakiyama T, Kohno T, Mimaki S, Ohta T, Yanagitani N, Sobue T, et al. Association of amino acid substitution polymorphisms in DNA repair genes TP53, POLI, REV1 and LIG4 with lung cancer risk. Int J Cancer. 2005;114:730–7. doi: 10.1002/ijc.20790. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Miao X, Guo Y, Tan W, Zhou Y, Sun T, et al. Genetic polymorphisms in cell cycle regulatory genes MDM2 and TP53 are associated with susceptibility to lung cancer. Hum Mutat. 2006;27:110–7. doi: 10.1002/humu.20277. [DOI] [PubMed] [Google Scholar]
- 27.Jung HY, Whang YM, Sung JS, Shin HD, Park BL, Kim JS, et al. Association study of TP53 polymorphisms with lung cancer in a Korean population. J Hum Genet. 2008;53:508–14. doi: 10.1007/s10038-008-0278-y. [DOI] [PubMed] [Google Scholar]
- 28.Chua HW, Ng D, Choo S, Lum SS, Li H, Soh LY, et al. Effect of MDM2 SNP309 and p53 codon 72 polymorphisms on lung cancer risk and survival among non-smoking Chinese women in Singapore. BMC Cancer. 2010;10:88. doi: 10.1186/1471-2407-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piao JM, Kim HN, Song HR, Kweon SS, Choi JS, Yun WJ, et al. p53 codon 72 polymorphism and the risk of lung cancer in a Korean population. Lung Cancer. 2011;73:264–7. doi: 10.1016/j.lungcan.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 30.Liu D, Wang F, Guo X, Wang Q, Wang W, Xu H, et al. Association between p53 codon 72 genetic polymorphisms and tobacco use and lung cancer risk in a Chinese population. Mol Biol Rep. 2013;40(1):645–9. doi: 10.1007/s11033-012-2103-0. [DOI] [PubMed] [Google Scholar]
- 31.Zhang JH, Li Y, Wang R, Wen DG, Wu ML, He M. P53 gene polymorphism with susceptibility to esophageal cancer and lung cancer in Chinese population. Zhonghua Zhong Liu Za Zhi. 2003;25:365–7. [PubMed] [Google Scholar]
- 32.Shao GG, Liu LL, Xu CJ. Association of polymorphism in TP53 gene with susceptibility and radiation sensitivity of non-small-cell-lung cancer in Chinese population. J Jilin Univ. 2005;31:97–9. [Google Scholar]
- 33.Li RN. Study of the relationship between repair gene single nucleotide polymorphisms and genetic susceptibility of lung cancer [D]. Wanfang Doctoral Dissertation Thesis, Zhongshan University. 2009. [Article in Chinese]. http://d.wanfangdata.com.cn/Thesis_Y1476782.aspx. Accessed 12 Jan 2013.
- 34.Wang W, Hao CF, Wu YJ, Wu YM. Effects of p53 Arg/Pro gene polymorphism and smoking on the occurrence of lung cancer in Henan Han population. J Zhengzhou Univ. 2010;45:746–8. [Google Scholar]
- 35.Dai S, Mao C, Jiang L, Wang G, Cheng H. P53 polymorphism and lung cancer susceptibility: a pooled analysis of 32 case–control studies. Hum Genet. 2009;125:633–8. doi: 10.1007/s00439-009-0664-3. [DOI] [PubMed] [Google Scholar]
- 36.Zhou W, Liu G, Miller DP, Thurston SW, Xu LL, Wain JC, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2, smoking, and lung cancer risk. Cancer Epidemiol Biomark Prev. 2003;12:359–65. [PubMed] [Google Scholar]
- 37.Li X, Hu Z, Qu X, Zhu J, Li L, Ring BZ, et al. Putative EPHX1 enzyme activity is related with risk of lung and upper aerodigestive tract cancers: a comprehensive meta-analysis. PLoS One. 2011;6:e14749. doi: 10.1371/journal.pone.0014749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao XY, Wang XD, Zang DY. MMP1-1607 1 G/2G polymorphism and lung cancer risk: a meta-analysis. Tumour Biol. 2012;33:2385–92. doi: 10.1007/s13277-012-0502-4. [DOI] [PubMed] [Google Scholar]
- 39.Feng X, Zheng BS, Shi JJ, Qian J, He W, Zhou HF. Association of glutathione S-transferase P1 gene polymorphism with the susceptibility of lung cancer. Mol Biol Rep. 2012;39:10313–23. doi: 10.1007/s11033-012-1908-1. [DOI] [PubMed] [Google Scholar]
- 40.Sjalander A, Birgander R, Kivela A, Beckman G. p53 polymorphisms and haplotypes in different ethnic groups. Hum Hered. 1995;45:144–9. doi: 10.1159/000154275. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Qiu LX, Shen XK, Lv XJ, Qian XP, Song Y. A meta-analysis of TP53 codon 72 polymorphism and lung cancer risk: evidence from 15,857 subjects. Lung Cancer. 2009;66:15–21. doi: 10.1016/j.lungcan.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 42.Rodin SN, Rodin AS. Human lung cancer and p53: the interplay between mutagenesis and selection. Proc Natl Acad Sci U S A. 2000;97:12244–9. doi: 10.1073/pnas.180320897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan C, Xu HY, Zhang CY, Zhang H, Chen CM, Zhang WM, et al. Effect of CYP1A1 MSPI polymorphism on the relationship between TP53 mutation and CDKN2A hypermethylation in non-small cell lung cancer. Arch Med Res. 2011;42:669–76. doi: 10.1016/j.arcmed.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Lin G, Fang F, Yu XJ, Yu L. Meta-analysis of the relationship between p21 Ser31Arg polymorphism and lung cancer susceptibility. Genet Mol Res. 2011;10:2449–56. doi: 10.4238/2011.October.13.2. [DOI] [PubMed] [Google Scholar]