Abstract
We here report the molecular overlap of the linkage of three essential protein complexes that coordinate the formation of the mitotic spindle. These proteins are dynein, a large motor complex that moves machinery inside cells, and two of its regulators: a protein complex called dynactin, that is a dynein activator, and a protein called NudE whose depletion in mice produces a small brain and mental retardation. What is intriguing about the dynein/dynactin/NudE interplay is that dynactin and NudE bind to a common segment of dynein that is intrinsically disordered. Elucidating differences in their binding modes may explain how one regulator can be selected over the other even when both are present in the same cellular compartment. These results have far reaching impact not only in our understanding of processes essential for formation and orientation of the spindle but also offer a novel role for protein disorder in controlling cellular processes, and highlight the advantages of NMR spectroscopy in elucidating atomic level characterization of extremely complex dynamic cellular assemblies.
Dynein structure and regulation
Cytoplasmic dynein is a microtubule-based molecular motor that uses the energy derived from ATP hydrolysis to transport cellular cargo. Transport by dynein motors is essential in several aspects of cell behavior, including separation of chromosomes during mitosis, cell migration and transport of vesicles containing nutrients and other products [1]. Dynein is a massive 1.6 MDa protein complex composed of multiple subunits of different size. Figure 1 represents our current model for the configuration of its various subunits. There is no structure available for the entire complex, but new structural details of the dynein motor domain [2, 3], and of the light and intermediate chains [4-7] continue to emerge.
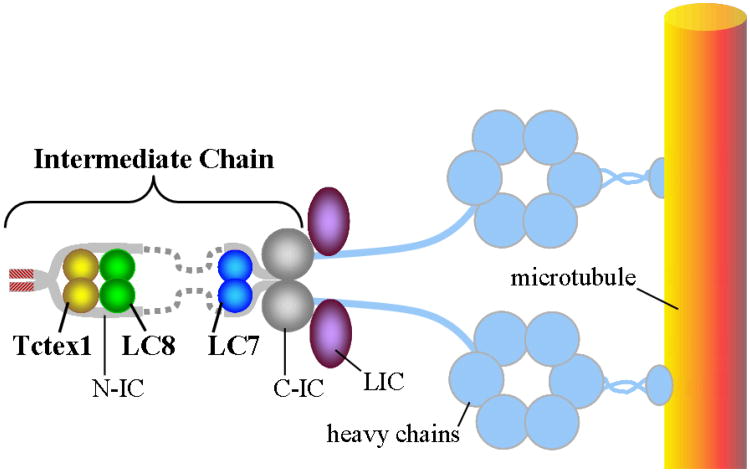
Figure 1.
Schematic representation of cytoplasmic dynein showing assembly of six proteins: the N-terminal domain of IC (gray bars) is intrinsically disordered; the C-terminal domain of IC (gray spheres) is predicted to be ordered; and the three homodimeric light chains are Tctex1 (yellow), LC8 (green), and LC7 (blue). Also shown are the light intermediate chains (LIC, purple), the heavy chain (light blue), and a microtubule (orange). The motor region of dynein consists of the heavy chain subunits that form a ring of AAA domains and a microtubule binding domain attached to the AAA ring by a flexible 15-nm coiled-coil stalk. The dotted gray bars are the disordered linkers connecting the IC segments that bind the light chains.
Commensurate with the numerous functions and activities of dynein within the cell, a multitude of adaptors and regulators have been identified [8]. The best characterized of the dynein regulatory proteins is dynactin; a multiprotein assembly that is essential for most, if not all cellular functions of the cytoplasmic dynein complex [9]. Dynactin is linked to dynein through its largest subunit, p150Glued [10]. Another dynein regulatory protein, NudE, first identified in A. nidulans as a protein required for even distribution of nuclei along the hyphae [11], is essential in processes including kinetochore and centrosome migration, organization of the Golgi complex, centrosome duplication and mitotic spindle positioning, and membrane transport [12-15].
NudE and p150Glued share a common binding motif on IC [16] which creates a paradox because dynactin, NudE and dynein co-localize in many cellular compartments [17], and raises the question as to how dynein regulation by either protein is coordinated. How does p150Glued binding to IC affect IC binding to NudE, and vice versa, and how does dynein select between different regulators?
Dynein intermediate chain is an intrinsically disordered protein
The dynein intermediate chain (IC) is central to the structure of the dynein motor. It is composed of two domains. The extended N-terminal domain (N-IC) is indicated by grey solid and dotted lines (Figure 1). The C-terminal domain (C-IC), which interacts with the heavy chain, is predicted to form a relatively ordered and compact β-propeller structure indicated by the grey globular shapes in Figure 1. Two copies of IC are present in every dynein motor, which are bridged by the three dimeric light chains Tctex1, LC8, and LC7. In addition to the light chains, N-IC contains interaction motifs for several other proteins known to be integral to the function of dynein. These include p150Glued, NudE, huntingtin and the ZW10 subunit of the Rod RZZ complex [8].
With its many interactions, N-IC appears to be the key modulator of dynein assembly and attachment to cargoes. N-IC is representative of intrinsically disordered proteins (IDPs), an emerging class of proteins lacking well-defined structure. IDPs include proteins that are completely disordered and those comprised of a mixture of ordered and disordered residues. Disordered structures are defined as flexible ensembles of conformations that are, on average, aperiodic, extended and not well packed by other protein atoms. IDPs play diverse roles in the promotion of supramolecular assembly and regulation of function in various binding partners, and are themselves highly amenable to regulation through post-translational modification. IDPs are often located at the center of biological complexes where they may act as scaffolds presenting multiple binding domains and promoting spatial orientations that facilitate protein-protein interactions (reviewed in [18]). Often these IDPs when present in complexes fold upon binding [19], or retain their disordered structure in the complex, a phenomenon, referred to as fuzziness [20].
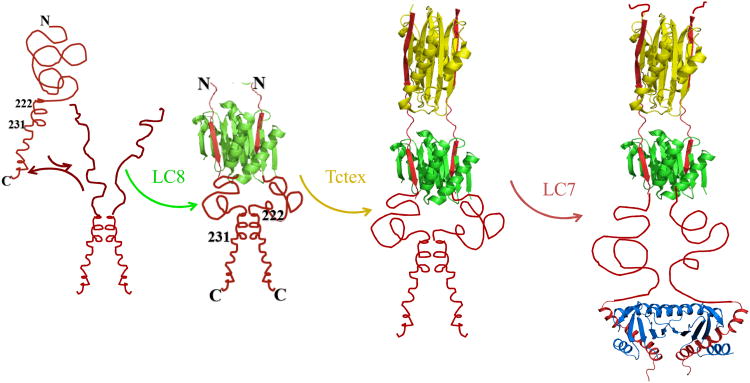
The assembly of the intrinsically disordered N-IC constitutes yet another class of disordered complexes. N-IC contains several disordered linear motifs that adopt unique structures when bound to dynein light chains. These induced structures complete the fold of the binding partners, while the linkers connecting the multiple linear motifs which are not involved in binding remain disordered [5, 6, 21, 22]. The linear motifs in dynein IC have the propensity to fold either as β-strands (recognitions motifs for Tctex1 and LC8) or α-helix (recognition motif for LC7), but only adopt this fold when bound to these partners. Figure 2 illustrates the different linear motifs in dynein IC in the apo and step-wise bound forms.
Figure 2.
Step-wise assembly model of N-IC with dimeric light chains. N-IC(red chain) is shown as disordered and primarily monomeric protein in equilibrium with a small percentage of dimer. This equilibrium can easily be shifted towards dimer as the local protein concentration is increased, or in the presence of self-association promoting residues. The minor population of dimer enhances the binding affinity of LC8 (green) which in turn enhances the binding affinity of Tctex1 (yellow). Upon LC7 binding (blue), the self association domain of IC that is populated in the presence of LC8 is shifted upstream and instead the helix-helix self-association is replaced by helix-LC7 association. Disordered IC adopts b-strands upon LC8 and tctex1 binding, and a helix-turn-helix upon LC7 binding. The linkers connecting the bound dimeric light chains remain disordered.
NMR spectroscopy and isothermal titration calorimetry as demonstrated below is a powerful combination for identifying recognition elements in disordered proteins and the energetics of multiple association steps.
Dynactin and NudE share common structural domains
The residues primarily involved in the interaction between dynein IC, dynactin p150Glued, and NudE include for all three proteins, segments whose secondary structure is predicted by standard sequence-based algorithms to be coiled-coil. NudE, has a predicted N-terminal coiled-coil domain and a largely unstructured C-terminal domain that associates with CENP-F, a nuclear matrix component required for kinetochore-microtubules interactions [23]. In solution, the N-terminal domain of NudE, nNudE, is a dimeric coiled-coil as predicted [24]. Solution studies of p150CC1 (residues 221-509) indicate a dimeric coiled-coil as predicted (data not shown). In contrast, NMR studies on apo IC1-143 which contains the binding sites for p150Glued, NudE, Tctex1 and LC8 show that while residues 3-36 are helical as predicted, there is no detectable population of stable coiled-coil, and the predominate conformations of unbound IC are disordered [21, 25]. In the apo form, IC1-143 is disordered but contains a short helical structure localized by NMR secondary chemical shifts to IC residues 1-40 [21].
Recognition sites identified by NMR and ITC
A combination of NMR spectroscopy and titration calorimetry show that the binding site of p150Glued on IC corresponds to two segments: region 1 composed of residues 1-41, a sequence predicted to have coiled-coil secondary structure, and region 2, composed of residues 46-75, a predominantly unstructured segment with nascent helical propensity [21]. These binding regions were identified by peak disappearance in NMR spectra of 15N labeled IC upon titration with unlabeled p150Glued. The differentiation between region 1 and 2 is due to their disappearance at different ratios during the titration. No new peaks appear for the bound form, either due to the large 100 kDa complex or to exchange broadening associated with the dynamic nature of the complex, or to combination of both. The peaks that are retained in the spectrum belong to a segment that is completely disordered and thus has different relaxation behavior than the bound complex. Similar experiments with NudE show a different pattern of peak disappearance indicating that only region 1 is involved in binding.
Since the characterization of the bound complex is hampered by disappearance of the peaks at the binding site, an additional complementary technique is used to verify the binding boundaries. Smaller constructs that correspond to region 1 alone, and those that contain regions 1 and 2 were made and their binding to both p150 and NudE was characterized by ITC and compared to a larger domain of IC. With NudE, region 1 retains the full binding affinity, while with p150Glued, both regions 1 and 2 are required to match the binding affinity of NudE observed with region 1 alone [21, 24].
Features of this shared binding segment are: First, there are two noncontiguous IC recognition sequences for p150, only one for NudE. Second, these regions are helical in nature; region 1 is likely coiled-coil in the p150/IC and NudE/IC complexes as inferred from patterns of spectral exchange broadening and sequence-based structure prediction of the apo protein, while region 2 becomes helical with p150Glued but more disordered with NudE. Third, the intervening residues between IC regions interacting with dynactin remain disordered in the complex. Fourth, the affinity of one protein to IC is different due to pre-binding of the other. Spectra of IC when p150CC1 is titrated in a pre-formed nNudE/IC binary complex show that p150CC1 can displace nNudE and result in even more pronounced peak disappearance than with p150CC1 alone, suggesting that the binding affinity to p150Glued increases in the presence of NudE. In contrast, in the reciprocal experiment, NudE in excess does not appear to compete with p150CC1. The observation that the binding affinity to p150 increases in the presence of NudE even though a stable ternary complex is not formed, is quite puzzling, and so is the ability of p150Glued to out-compete NudE and not vice versa, especially when both bind with similar affinity.
Structure of assembled IC
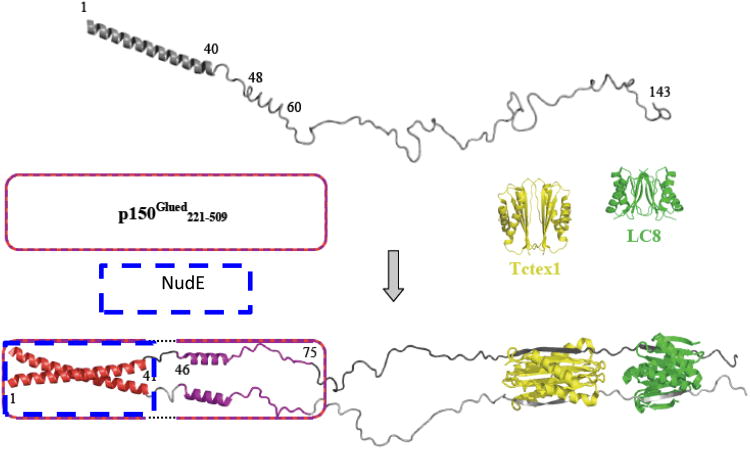
Our interpretive model for IC in its assembled state with p150Glued or NudE and the light chains (Figure 3) is based upon a combination of structural data from X-ray crystallography (structure of Tctex1 and LC8 bound to a short segment of IC), dynamics and chemical shift mapping information from NMR spectroscopy (sites where p150 and NudE bind and the retained disorder in the linkers) as well as sequence-based prediction of structural propensity (coiled-coil structure of the p150/NudE binding region). Both helices of unbound IC (Figure 3, top) are included in the multi-region binding footprint of p150Glued, with the first comprising the entirety of region 1. The proposed coiled coil conformation for region 1 in the bound state derives from the complex exchange processes and sequence-based prediction of a coiled-coil in both IC and the p150Glued, or NudE; the coiled-coil assemblage depicted in the model could represent an IC/IC coiled-coil packed on p150Glued/p150Glued coiled-coil or NudE/NudE coiled-coil. The second, nascent helix in apo IC is contained within the p150Glued recognition motif in region 2, and shows less attenuation of peak intensity than those of region 1, presumably due to less complex exchange broadening processes, likely chemical exchange between the free and p150Glued–bound states of IC as well as structural fluctuation between nascent and fully-formed helix within IC. Thus the nascent helical structure depicted in the model for apo IC is proposed to persist and perhaps stabilize in the bound state, while residues ∼67-75 of region 2, which show less attenuation of peak intensity suggests retained disorder in this part of region 2 in bound p150Glued-IC. With NudE, there is no significant change in intensity in region 2 suggesting that this region remains flexible and does not interact with NudE.
Figure 3.
A model for assembly of dynein intermediate chain with p150Glued, NudE, and light chains. The model depicts apo IC:1-143 (top) as a primarily disordered and monomeric ensemble of conformations with one defined helical region encompassing the N-terminal 40 residues and another region of nascent helicity encompassing residues ∼48-60, as determined from NMR measurements. Segments of IC for which NMR spectral characteristics are altered by p150Glued binding are indicated in red (Region 1, residues 1-41) and purple (Region 2, residues 46-75). The p150Glued construct is represented as a box with a red striped border, while the NudE construct has a blue border. In the IC-bound state, dotted lines in the box indicate that p150Glued does not interact with IC residues 42-45 (Linker 1). The IC•Tctex1•LC8 subcomplex portion of the model is from a crystal structure (24) (PDB entry 3FM7), with Tctex1 (yellow) and LC8 dimers (green), and the corresponding IC segments in grey (one subunit light grey, the other dark grey). All structures, including apo Tctex1 (PDB entry 1YGT) and apo LC8 (PDB entry 3BRI) were generated using PyMOL (Delano). The figure was adapted from [21].
Thus the bound IC structure differs depending on the partners with which it binds, and the difference is localized to what we term region 2, a stretch of a sequence that is highly disordered and susceptible to posttranslational modification and to alternative splicing.
Disordered linkers for versatility in regulation
Biological evidence suggests that NudE is required for localization of p150Glued at the nuclear envelope in prophase [26] implying that binding of NudE to IC enhances p150 binding. Our in vitro experiments support this observation; in the presence of both p150Glued and NudE, binding of p150Glued is tighter than when NudE is absent. This observation can be explained by evoking the ensemble properties common for intrinsically disordered proteins. Since both complexes associate with moderate IC-partner affinity, IC-NudE and IC-p150Glued complexes are rapidly interconverting ensembles of intrinsically disordered apo IC conformations and IC-partner complexes. Some population of the IC-NudE ensemble will have region 2 more exposed and more readily accessible to p150Glued binding. With region 2 bound to a protein – much larger than a residue-level modification – there may well be a shift in region 1 to conformations more favorable to p150Glued binding and/or less favorable to NudE binding. Then by mass action, in a mixture of NudE and p150Glued, an IC complex with the latter would be more highly populated.
Equally puzzling observation is the localization of both dynactin and NudE in the same cellular compartments. How is regulation of IC by dynactin versus NudE coordinated when both are present, and what determines which protein binds? Our in vitro results suggest that events that modify region 2 but do not significantly affect region 1 could interfere with p150Glued binding but have limited effect on NudE binding. Thus, residue-specific structural changes in and near region 2 likely either diminish or enhance dynactin binding, with limited effect on NudE binding. Disorder in region 2 and in the long linker following region 2 promotes local modifications like phosphorylation and splicing. The disorder in the 4-resiude linker separating region 1 and 2 minimizes effects on region 1 from residue-level modification in and near region 2 while its short length makes it likely that protein binding to region 2 affects the average structure and NudE affinity of region 1. The consequence of disorder in this small bi-segmental IC domain is that a modification localized to a short stretch in IC can modulate selection among multiple complex regulators of dynein function.
Acknowledgments
Funding: This work was supported, in whole or in part, by National Institutes of Health Grant GM 084276.
References
- 1.Vallee RB, Williams JC, Varma D, Barnhart LE. Dynein: An ancient motor protein involved in multiple modes of transport. J Neurobiol. 2004;58(2):189–200. doi: 10.1002/neu.10314. [DOI] [PubMed] [Google Scholar]
- 2.Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331(6021):1159–65. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8 A crystal structure of the dynein motor domain. Nature. 2012;484(7394):345–50. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 4.Benison G, Karplus PA, Barbar E. Structure and dynamics of LC8 complexes with KXTQT-motif peptides: swallow and dynein intermediate chain compete for a common site. J Mol Biol. 2007;371(2):457–68. doi: 10.1016/j.jmb.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 5.Hall J, Karplus PA, Barbar E. Multivalency in the assembly of intrinsically disordered Dynein intermediate chain. J Biol Chem. 2009;284(48):33115–21. doi: 10.1074/jbc.M109.048587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall J, Song Y, Karplus PA, Barbar E. The crystal structure of dynein intermediate chain-light chain roadblock complex gives new insights into dynein assembly. J Biol Chem. 2010;285(29):22566–75. doi: 10.1074/jbc.M110.103861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams JC, Roulhac PL, Roy AG, Vallee RB, Fitzgerald MC, Hendrickson WA. Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc Natl Acad Sci U S A. 2007;104(24):10028–33. doi: 10.1073/pnas.0703614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10(12):854–65. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill SR, Schroer TA, Szilak I, Steuer ER, Sheetz MP, Cleveland DW. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1991;115(6):1639–50. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holzbaur EL, Hammarback JA, Paschal BM, Kravit NG, Pfister KK, Vallee RB. Homology of a 150K cytoplasmic dynein-associated polypeptide with the Drosophila gene Glued. Nature. 1991;351(6327):579–83. doi: 10.1038/351579a0. [DOI] [PubMed] [Google Scholar]
- 11.Efimov VP, Morris NR. The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J Cell Biol. 2000;150(3):681–8. doi: 10.1083/jcb.150.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44(2):279–93. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 13.Lam WK, Felmlee MA, Morris ME. Monocarboxylate transporter-mediated transport of gamma-hydroxybutyric acid in human intestinal Caco-2 cells. Drug Metab Dispos. 2010;38(3):441–7. doi: 10.1124/dmd.109.030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang H, Zhan HJ, Wang BG, Pan Y, Hao XS. Change in expression of apoptosis genes after hyperthermia, chemotherapy and radiotherapy in human colon cancer transplanted into nude mice. World J Gastroenterol. 2007;13(32):4365–71. doi: 10.3748/wjg.v13.i32.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wainman A, Creque J, Williams B, Williams EV, Bonaccorsi S, Gatti M, Goldberg ML. Roles of the Drosophila NudE protein in kinetochore function and centrosome migration. J Cell Sci. 2009;122(Pt 11):1747–58. doi: 10.1242/jcs.041798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenney RJ, Weil SJ, Scherer J, Vallee RB. Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J Biol Chem. 2011;286(45):39615–22. doi: 10.1074/jbc.M111.289017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallee RB, McKenney RJ, Ori-McKenney KM. Multiple modes of cytoplasmic dynein regulation. Nat Cell Biol. 2012;14(3):224–30. doi: 10.1038/ncb2420. [DOI] [PubMed] [Google Scholar]
- 18.Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in scaffold proteins: getting more from less. Prog Biophys Mol Biol. 2008;98(1):85–106. doi: 10.1016/j.pbiomolbio.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19(1):31–8. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuxreiter M, Tompa P. Fuzzy complexes: a more stochastic view of protein function. Adv Exp Med Biol. 2012;725:1–14. doi: 10.1007/978-1-4614-0659-4_1. [DOI] [PubMed] [Google Scholar]
- 21.Morgan JL, Song Y, Barbar E. Structural dynamics and multiregion interactions in dynein-dynactin recognition. J Biol Chem. 2011;286(45):39349–59. doi: 10.1074/jbc.M111.296277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyarko A, Barbar E. Light chain-dependent self-association of dynein intermediate chain. J Biol Chem. 2011;286(2):1556–66. doi: 10.1074/jbc.M110.171686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vergnolle MA, Taylor SS. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr Biol. 2007;17(13):1173–9. doi: 10.1016/j.cub.2007.05.077. [DOI] [PubMed] [Google Scholar]
- 24.Nyarko A, Song Y, Barbar E. Intrinsic disorder in dynein intermediate chain modulates its interactions with NudE and dynactin. J Biol Chem. 2012 doi: 10.1074/jbc.M112.376038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nyarko A, Hare M, Hays TS, Barbar E. The intermediate chain of cytoplasmic dynein is partially disordered and gains structure upon binding to light-chain LC8. Biochemistry. 2004;43(49):15595–603. doi: 10.1021/bi048451+. [DOI] [PubMed] [Google Scholar]
- 26.Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo T, Gatti X, Vallee R, Ellenberg J, Doye V. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol. 2011;192(5):855–71. doi: 10.1083/jcb.201007118. [DOI] [PMC free article] [PubMed] [Google Scholar]