Significance
Genomic structural variants (SVs) significantly contribute to human genetic variation and have been linked with numerous diseases. Compared with humans, the characterization of SVs occurring within and across nonhuman primates has lagged. We generated comprehensive massively parallel DNA sequencing-based SV maps in three nonhuman primate species and show that the rates of different SV formation mechanisms, such as nonallelic homologous recombination and Alu retrotransposition, vary significantly between the great apes and the rhesus macaque—leading to markedly different SV landscapes in these species. Linking gene expression data with species-specific gene duplications, we describe several instances where gene duplicates seem to lead to evolutionary innovation through the gain of gene expression in new tissues.
Keywords: genome evolution, retrotransposons, neofunctionalization, copy-number variation
Abstract
Although nucleotide resolution maps of genomic structural variants (SVs) have provided insights into the origin and impact of phenotypic diversity in humans, comparable maps in nonhuman primates have thus far been lacking. Using massively parallel DNA sequencing, we constructed fine-resolution genomic structural variation maps in five chimpanzees, five orang-utans, and five rhesus macaques. The SV maps, which are comprised of thousands of deletions, duplications, and mobile element insertions, revealed a high activity of retrotransposition in macaques compared with great apes. By comparison, nonallelic homologous recombination is specifically active in the great apes, which is correlated with architectural differences between the genomes of great apes and macaque. Transcriptome analyses across nonhuman primates and humans revealed effects of species-specific whole-gene duplication on gene expression. We identified 13 gene duplications coinciding with the species-specific gain of tissue-specific gene expression in keeping with a role of gene duplication in the promotion of diversification and the acquisition of unique functions. Differences in the present day activity of SV formation mechanisms that our study revealed may contribute to ongoing diversification and adaptation of great ape and Old World monkey lineages.
Genomic structural variants (SVs), including copy number variants and balanced SV forms (such as inversions), are a major source of human genetic variation (1, 2). The development of massively parallel sequencing (MPS) to characterize SVs (3–5) has enabled comprehensive analyses of origin and functional impact of SVs in humans (3, 6). Although SVs are presumed to play a major role in primate evolution and phenotypic variation (7) as well, empirical evidence showing such a role remains scarce (8). Comparative analyses of reference genome assemblies of the chimpanzee (9), orang-utan (10), and rhesus macaque (11) have provided some initial insights into large-scale structural changes in primate genome evolution (12). Microarray technology-based surveys have provided additional glimpses of the abundance of polymorphic unbalanced SVs (i.e., copy number variants) in different primate species, enabling the construction of SV maps at a resolution of tens to hundreds of kilobases (13–16).
Thus far, despite ongoing progress in assessing SNP variation in primates (10, 17–19), no study has leveraged MPS technology for ascertaining inter- and intraspecies SVs in different primates. We, therefore, performed MPS-based genome analyses in five individuals from each of these primate species, Pan troglodytes (chimpanzee), Pongo abelii (orang-utan), and Macaca mulatta (rhesus macaque), to construct comprehensive SV maps in these species. Our analyses have revealed marked differences in SV formation mechanism activities and further yielded a complex relationship between genomic copy number and gene expression patterns, with several gene duplications conferring tissue-specific gene expression changes.
Results
Nucleotide Resolution Genetic Variation Maps in Three Primate Species.
To construct high-resolution SV maps, we sequenced fibroblast-derived genomic DNA from five unrelated chimpanzee, orang-utan, and rhesus macaque individuals (Dataset S1) with 101-bp Illumina paired-end DNA reads. The average sequencing coverage ranged from 15× to 20× and was estimated to be sufficient for detecting 70–80% of deletion polymorphisms with >90% accuracy (3, 4). Algorithms developed for population-scale DNA variant analyses in humans (20) (SI Appendix) were applied to these nonhuman primates and yielded 6.6 million chimpanzee SNPs, 12.8 million orang-utan SNPs, and 13.8 million rhesus macaque SNPs (SI Appendix, Table S1) with a false discovery rate (FDR) of 1.2% and a false negative rate of 7% (Dataset S2). We further generated a map of short (<50 bases) insertions and deletions (indels), identifying 0.54 million, 0.95 million, and 1.21 million indels in these same species, respectively (SI Appendix, Table S1), with an overall FDR of 2.3% (Dataset S2).
Mimicking the detection of SVs in the 1000 Genomes Project (3), we integrated different approaches for the discovery of SVs ≥50 bases in size (SI Appendix, Figs. S1 and S2). We designed high-resolution custom array comparative genomic hybridization (aCGH) platforms (>9 million probes; effective SV calling resolution of ∼500 bp) for each species and hybridized two samples from each species to guide and verify our SV discovery framework (SI Appendix). Based on these independent aCGH experiments, we devised species-specific protocols for variant filtering to account for the differences in quality of each primate reference genome assembly (SI Appendix, Fig. S3). Overall, we identified 2,680, 4,983, and 3,905 polymorphic deletions in the chimpanzee, orang-utan, and macaque, respectively (Fig. 1 and SI Appendix, Table S1). Random resampling and deletion calling in sets of five humans drawn from the 1000 Genomes Project sample set indicated a relatively low margin of error originating from sampling individuals (SD of ∼19%) with respect to the total number of SVs discovered (SI Appendix). To assess the quality of our deletion callset, we verified 42 of 50 randomly sampled variant sites using PCR (SI Appendix, Fig. S1C and Dataset S2). We further evaluated the allelic state of deletions and were able to assign high-confidence genotypes for 35%, 19%, and 50% of the deletions in chimpanzees, orang-utans, and macaques, respectively. Our ability to genotype was, notably, influenced by the amount and size of gaps in each of the reference genome assemblies. The orang-utan reference genome, for instance, harbors a marked excess of small gaps (SI Appendix, Fig. S3B), leading to a notable reduction in deletions genotyped at high confidence, although PCR assays showed a high accuracy for SVs discovered in this species (Dataset S2). Based on the concordance of these genotypes with our high-density tiling aCGH experiments, we estimate a FDR of <15% for deletion genotyping (SI Appendix). We also verified the expected loss of single nucleotide variant heterozygosity in unique (one copy) deleted regions, further substantiating the quality of our calls (SI Appendix, Fig. S4A). SV regions showed a slightly reduced depth of coverage (SI Appendix, Fig. S4B) of uniquely mappable reads (at a similar level as in human data) (3) as expected given the general preponderance of SVs occurring in repeat-rich regions that display reduced read mappability. In addition to inferring deletions, we also inferred 1,499, 1,095, and 807 polymorphic and 1,910, 540, and 625 fixed duplications in these primate species (SI Appendix, Table S1), showing high concordance (>80%) with our aCGH data (SI Appendix).
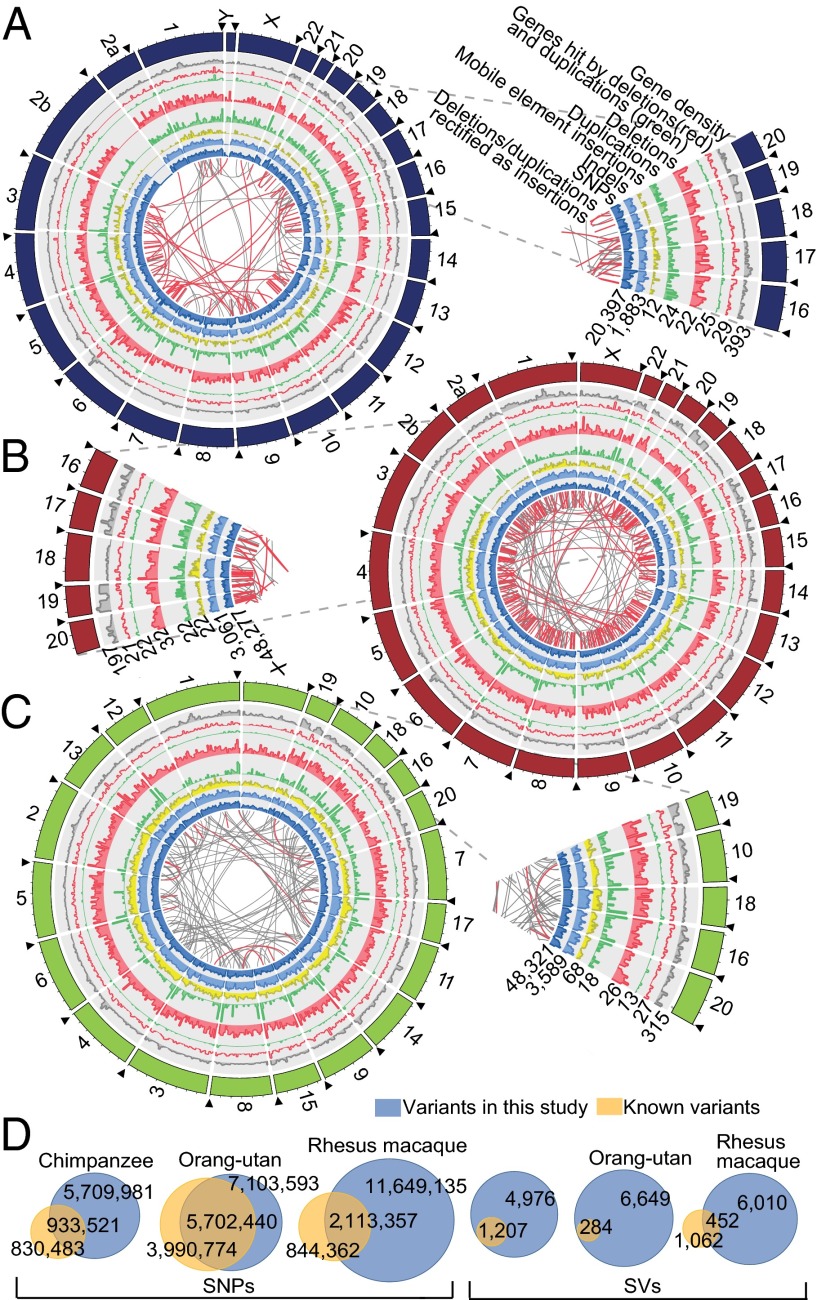
Fig. 1.
Overview of genomic sequence variants in three nonhuman primate species. Circos plots illustrate the genome-wide distribution of genomic sequence variants in (A) chimpanzee, (B) orang-utan, and (C) macaque. Black arrowheads mark the start of the chromosomes. Macaque chromosomes are sorted according to orthology with respect to human. The missing part of chromosome 2b in chimpanzee is caused by a large telomeric reference genome gap. Connecting lines at the inside of each plot depict the movement of duplicative insertions (i.e., deletions and duplications rectified as insertions based on assessment of the ancestral state of the respective loci) (22). Red connecting lines indicate NAHR events, and gray connecting lines indicate non-NAHR events (MEIs were excluded in this graph). Pie slices zoom into the respective circos plots. Heights for different variant types in the circos plots are relative to the abundance of the respective variant type along the genome (numbers at the lower edge of the pie slices indicate the maximum value in a bin for each variant type in the whole subcircle). The bin size is 5 Mb. (D) Venn diagrams depict the proportion of variants that were previously reported. For this analysis, we made use of the Single Nucleotide Polymorphism database (dbSNP) and previously published aCGH-based surveys. Nonreference MEIs were not considered in the depicted Venn diagram.
We further identified polymorphic mobile element insertions (MEIs) (21) in these species. We mapped 764, 2,548, and 15,566 unique MEIs (nonreference MEIs) not annotated in the reference genomes of chimpanzee, orang-utan, and macaque, respectively (SI Appendix, Fig. S4C and Table S1). We validated 42 of 49 (86%) randomly selected unique MEIs by PCR (SI Appendix, Fig. S4D). Together with transposable elements previously annotated in the respective reference genomes, which we inferred to be polymorphically absent/present in some individuals (reference MEIs) (SI Appendix), we ascertained 858, 2,863, and 16,690 polymorphic MEIs in chimpanzees, orang-utans, and macaques, respectively (SI Appendix, Table S1).
Relative Genomic and Functional Impact of Structural Variation in Primates.
When combining our deletion, duplication, and MEI sets, we inferred a total of 6,947, 9,481, and 22,027 SVs in these species. We used previously published aCGH data to assess which portion of our deletion and duplication calls had been previously reported, an analysis that revealed 90% of these calls were not previously reported (Fig. 1D and Dataset S3). Assessment of the relative genomic impact of DNA variants revealed marked species-specific differences. Comparing the number of SNPs between two individuals of a given primate species, 3.2, 6.6, and 7.3 Mb genomic sequence differed, on average, among chimpanzees, orang-utans and macaques, respectively, whereas 18.8, 19.4, and 11.8 Mb genomic sequence of these species differed at the level of SVs (SI Appendix, Fig. S7C). Hence, great apes have fewer but larger SVs, causing this variant class to have the largest impact on genomic variation, whereas macaques show an abundance of small SVs.
We first assessed the functional impact of SVs by intersecting our SV lists with annotated genes, promoters, and noncoding RNAs. Overall, we identified 933 SVs (326 deletions, 603 duplications, and 4 MEIs) intersecting protein-coding sequences and fewer intersecting noncoding RNAs (SI Appendix, Fig. S5). Deletions and MEIs, but not duplications, were significantly depleted from coding loci based on simulations [P < 0.001 (deletions), P < 0.05 (MEIs), and P > 0.50 (duplications); permutation-based empirical P value]. We further identified 92,901 nonsynonymous SNPs and 12,804 indels intersecting with protein-coding loci, significantly less than would be expected if these variants were distributed uniformly along the genome (P < 0.001; permutation-based empirical P value). This finding suggests that coding SNPs, indels, deletions, and MEIs are under strong purifying selection. Although significantly more genes were affected by nonsynonymous coding SNPs and indels in macaques (P = 2.3 × 10−10 for SNPs, P = 1.4 × 10−6 for indels, two-sided Fisher's exact test), we observed a significantly reduced number of genes affected by SVs compared with the great apes (P = 0.012; two-sided Fisher's exact test) (SI Appendix, Fig. S5 and Dataset S3). We examined the effects of purifying selection on the distribution of SVs across primate genomes. Site frequency spectrum analysis revealed no major genome-wide differences in purifying selection (SI Appendix, Fig. S6) (i.e., there was no indication that selection acts in profoundly different ways in macaques as opposed to great apes). Thus, purifying selection alone is unlikely to explain the differences in the relative DNA sequence impact of SVs in different primates.
Differences in Genome Architecture Are Linked with Species-Specific SV Landscapes.
Because distinct SV formation mechanisms tend to be associated with specific variant size spectra (3, 22, 23), we hypothesized that differences in the activity of SV formation mechanisms may account for the size differences and if true, have shaped the species-specific SV landscapes. We assessed SVs mapped at nucleotide resolution for SV formation based on sequence analysis (SI Appendix) to distinguish MEIs, nonallelic homologous recombination (NAHR), variable number of tandem repeat expansion or contraction, and nonhomology-associated rearrangements (such as nonhomologous end joining or microhomology-mediated break-induced replication) (22, 24). Indeed, analysis of MEIs showed a markedly higher Alu activity in macaques as opposed to great apes (P < 2.2 × 10−16; two-sided Fisher's exact test) (Fig. 2 A and B) consistent with earlier reports (9), leading to a pronounced increase of small SVs [i.e., ∼300 bases in size (the size of Alu elements)] in macaques (SI Appendix, Fig. S7A). According to previous reports, ∼15% of human SVs comprise MEIs, similar to the great apes (3). In the great apes that we studied, the relative abundance of polymorphic LINE/L1 elements surpassed Alu elements, with the L1Pt family in chimpanzees and the L1PA3 family in orang-utans dominating the respective MEI landscapes, whereas the AluMacYa3 was the most dominant MEI subfamily in macaques (subfamily assignments based on reference MEIs) (SI Appendix, Fig. S7B). Polymorphic Alu insertions were found at a proportionally lower rate in orang-utans compared with chimpanzees (from 43% of all MEIs in chimpanzees to 6% in orang-utans; P < 2.6 × 10−100, two-sided Fisher's exact test) (Fig. 2B), in keeping with earlier reports based on whole-genome alignment and PCR (10, 25).
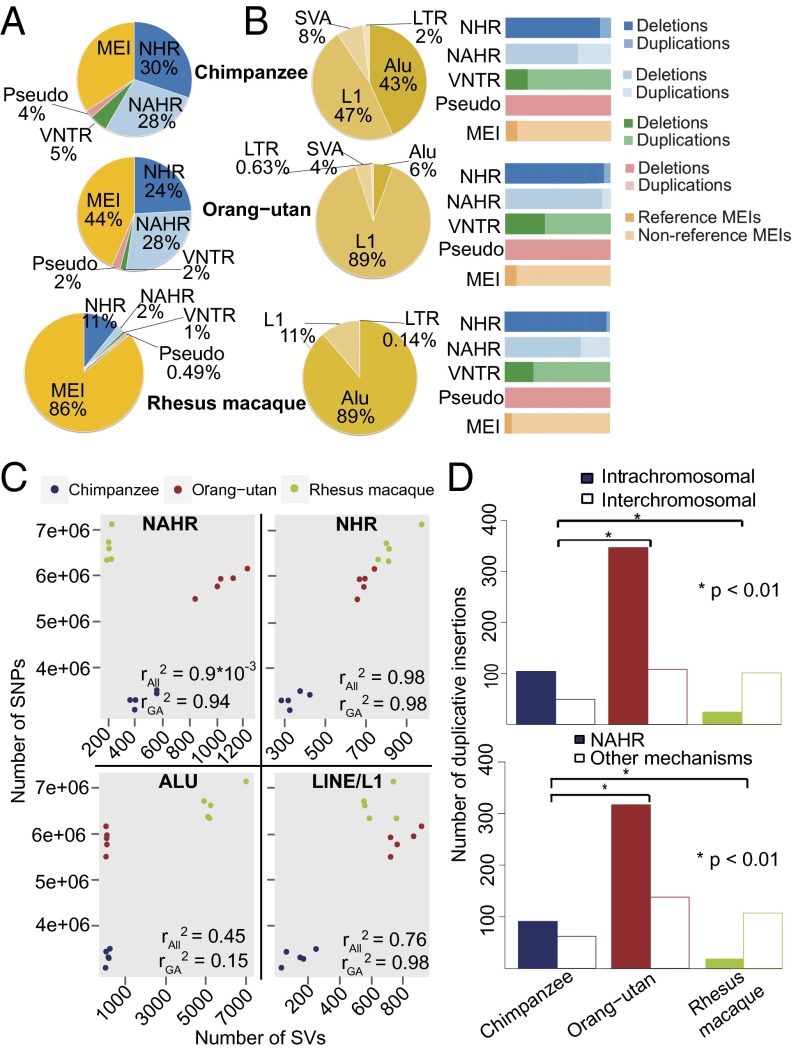
Fig. 2.
Differences between SV formation mechanisms among primate species. (A) Relative proportion of SV formation mechanisms observed in each species. (B, Left) Breakdown of MEIs identified as reference or nonreference transposable element insertions. LTR, endogenous retrovirus-associated long terminal repeats; SVA, SINE–variable number of tandem repeat–Alu composite mobile elements. (B, Right) Breakdown of SV formation mechanisms. Pseudo stands for inferred processed pseudogenes. (C) Correlation in the abundance of SNPs and SVs formed by different mechanisms. Dots represent different samples. rAll2, Pearson correlation coefficient for all three studied primate species; rGA2, Pearson correlation coefficient for studied great ape species. (D, Upper) Breakdown of intrachromosomal and interchromosomal duplicative insertions (P value computed using a two-sided Fisher's exact test). (D, Lower) Breakdown of duplicative insertions mediated by NAHR and other mechanisms.
Furthermore, we noted striking differences in the activity of NAHR events between the great apes and macaques (Fig. 2A). Specifically, 28% of the chimpanzee and orang-utan SVs were inferred to be formed by NAHR compared with only 2% of the macaque SVs (P < 2.2 × 10−16; two-sided Fisher's exact test) (Fig. 2A). In humans, NAHR has been reported to contribute to 22–28% of SVs (3, 22, 23), suggesting an overall similar rate of NAHR-based SV formation throughout great ape species, including humans. We reasoned that, if SNP and SV mutation rates are approximately similar across primate species, numbers of observed SNPs and SVs should correlate. Indeed, we observed a strong correlation between the number of SNPs detected in each of 15 primate samples and the number of nonhomology-associated rearrangement and LINE/L1 events (r2 values of 0.98 and 0.76, respectively) (Fig. 2C). Weaker correlation or no correlation was observed between SNPs and Alu element insertions (r2 = 0.45) as well as between SNPs and NAHR events (r2 = ∼0), further supporting the notion that Alu and NAHR formation rates have changed considerably in recent primate evolution (SI Appendix, Table S2).
In all species analyzed in our study, NAHR-mediated SVs were, on average, larger than other SV classes (defining variant classes based on formation mechanism; P < 0.05, permutation-based empirical P value) (SI Appendix, Fig. S7A). Accordingly, an increase in the rate of NAHR, leading to a higher number of NAHR-mediated SVs, contributed to the high nucleotide-level impact of SVs in the great apes. Similarly, the increased functional impact of SVs in great apes, with a large number of genes being affected by SVs in these species, is in part attributable to the high rate of NAHR (SI Appendix, Table S2). A plausible explanation for the high rate of NAHR in great apes is the burst of recent segmental duplications that is thought to have occurred during great ape evolution (12), with segmental duplications representing mediators of NAHR (24). Indeed, our assessment of comparable segmental duplication maps in the species that we studied (SI Appendix) showed that segmental duplications comprise 4.7–5.4% of the genomes of great apes compared with only 1.6% of the macaque genome (i.e., 2.6- to 3.4-fold relative increase; P < 0.0008, two-sided Fisher's exact test). We additionally delineated the ancestral allelic state (i.e., ancestral vs. derived allele) of SVs mapped at nucleotide resolution based on sequence analysis (3, 22). These analyses showed an excess of intrachromosomal over interchromosomal duplicative insertions (i.e., SVs arising from the insertion of duplicated sequence) in great apes and a marked depletion of intrachromosomal duplicative insertions in macaques (Fig. 2D and SI Appendix, Fig. S7D) (P < 0.01, two-sided Fisher's exact test). Because the formation of NAHR-mediated SVs frequently involves intrachromosomal segmental duplications (26), we conclude that intrinsic genomic differences between the macaque genome and the genomes of great apes (i.e., segmental duplication content and architecture) may be linked with the relative reduction of NAHR in macaque. Accordingly, we also observed a positive correlation between the amount of NAHR-formed SVs and the amount of intrachromosomal segmental duplications, with the most NAHR events inferred to be mediated by intrachromosomal segmental duplications in orang-utans (32%) and the fewest NAHR events inferred to be mediated by intrachromosomal segmental duplications in macaques (8%).
Interspecies Gene Duplications Can Impact Gene Expression and Coincide with Expression Acquisition in Unique Tissues.
With gene duplications being presumed to have a major impact on primate evolution (12), we looked more closely at fixed duplications affecting protein-coding genes (8, 27) by analyzing the sequence depth of coverage for 18,571 orthologous genes [available in the evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) database] (28) in chimpanzees, orang-utans, macaques, and humans (SI Appendix). We identified 1,963 fixed gene duplications affecting 1,078 orthologous genes, including whole (i.e., gene encompassing; 226 events) and partial (gene intersecting; 852 events) gene duplications (Dataset S4). Reanalysis of previously published cross-species aCGH data designed to assess highly conserved (mostly exonic) loci in the genome (29) enabled us to verify 52 of 68 (76%) fixed whole-gene duplications. Additionally, we verified two fixed gene duplications (DIP2C and SH3TC1) by FISH and four of five previously unreported duplications by quantitative real-time PCR (qPCR) (Fig. 3A, SI Appendix, Fig. S8A, and Dataset S2).
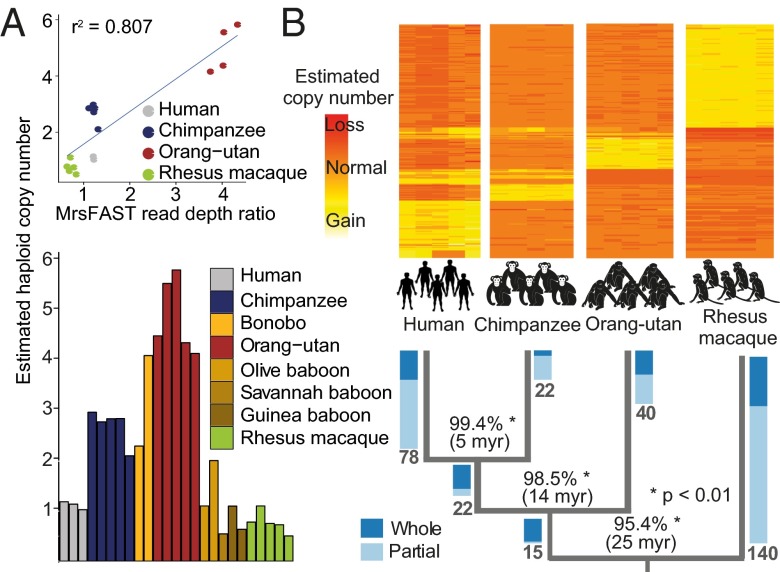
Fig. 3.
Investigation of fixed gene duplications. (A) qPCR verification of SUZ12 gene duplication. (Upper) Correlation of micro-read substitution-only Fast Alignment Search Tool (mrsFast) read-depth ratios (x axis) and estimated haploid copy numbers by qPCR (y axis). (Lower) qPCR results in an extended panel with eight different primate species. (B) Timing of the occurrence of fixed gene duplications in primate evolution. The heat map depicts mrsFAST read-depth ratios of fixed gene duplications that were timed. Rows represent timed orthologous genes, and columns represent individual samples. Yellow colors indicate higher read-depth ratios (>1) corresponding to a gain. Orange colors indicate read-depth ratios of ~1 corresponding to two diploid copies. Red colors indicate read-depth ratios < 1 corresponding to an inferred loss. In Lower, numbers in bold at tree edges represent the numbers of genes timed for a specific tree branch. Blue-colored bars represent ratios of timed whole- and partial gene duplications on a specific branch. Percentages at tree branches depict the mean sequence identity between duplicated paralogs on each of the branches (computed based on segmental duplication overlap).
We comprehensively investigated 317 gene duplications with complete orthology information along the primate phylogenetic tree and inferred the time of duplication emergence in primate evolution (Fig. 3B). The ratio between whole/partial gene duplications increased with the inferred age of the duplication event, with more ancient fixed events corresponding to whole-gene duplications (Fig. 3B). This observation suggests that whole-gene duplications more often have no selective consequence or evolve under positive selection, whereas partial gene duplications may more often display negative fitness effects and hence, show a more rapid decay. We performed gene category enrichment analysis using GeneCodis (30) on fixed gene duplications and observed significant enrichments of genes involved in immunity-, development-, and metabolism-associated processes (SI Appendix, Fig. S8B), functional categories that may play a prominent role in adaptive evolution (7, 31). Duplications affecting similar processes were previously reported to contribute to the evolution of nonprimate species, including the mouse and the fruit fly (32).
We also assessed whether duplicated genes are linked with changes in gene expression (6, 33) by sequencing the transcriptomes of the fibroblast-derived cell lines used for genomic DNA sequencing (SI Appendix). Analysis of these data showed an overall increase in expression levels for whole-gene duplications (P = 6.059 × 10\x{2212}5, two-sided Kolmogorov–Smirnov test) (Fig. 4A and SI Appendix, Fig. S9 A and B), whereas no significant increase was observed for partial gene duplications. However, even among the whole-gene duplications, a notable positive correlation (adjusted r > 0.5) between DNA- and RNA-based read depth was observed for only a minority (14 of 64 genes with annotation in at least three species and expression in fibroblasts; 21.9%) (SI Appendix, Fig. S9B). Hence, a proportional relationship between fixed gene duplicate copy number and expression (34) seems to represent an exception rather than a rule. Instead, dosage compensatory mechanisms (6, 33) or a lack of cis regulatory sequence context to enable gene duplicate expression in the tissue, where the parental gene is expressed (35, 36), may explain the observed relationship between gene expression and gene copy number for whole-gene duplications.
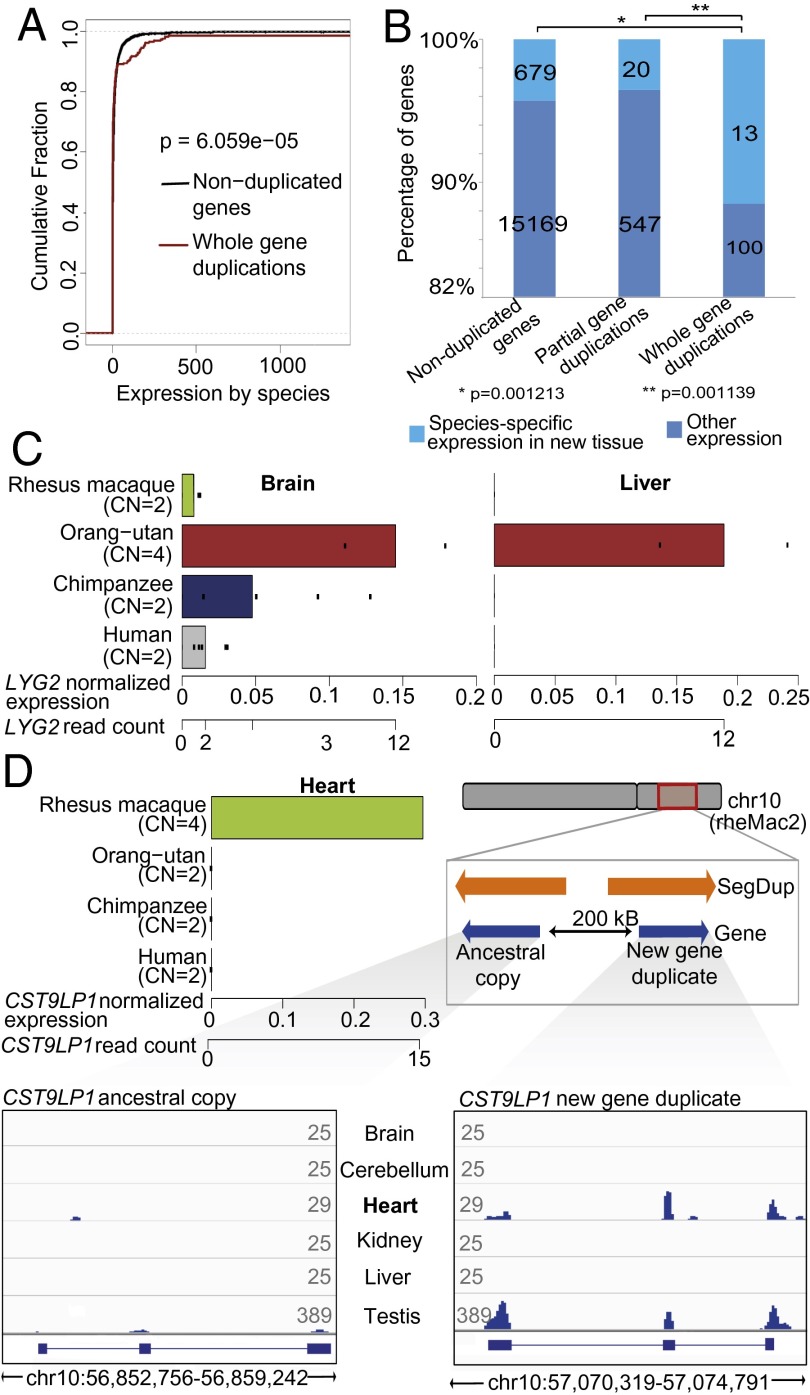
Fig. 4.
Fixed gene duplications coincide with gains of gene expression in tissues. (A) Enrichment of overexpressed genes in fixed whole-gene duplications. Red line, fibroblast cell line-based gene expression values in species with fixed whole-gene duplicates; black line, gene expression values from species harboring nonduplicated orthologous genes (P value based on two-sided Kolmogorov–Smirnov test). (B) Whole-gene duplications more often coincide with detected expression in unique tissues than partial gene duplications or nonduplicated genes (P value based on two-sided Fisher's exact test). (C) Inferred LYG2 expression gain in orang-utan liver coinciding with LYG2 duplication. Bars depict mean expression values of genes based on reads mapping without mismatches. Dots represent individual expression measurements. (D) Inferred CST9LP1 expression gain in macaque heart. Locus-specific analysis on macaque chromosome 10 (Lower Right) revealed that expression in the heart is originating from the derived CST9LP1 paralog, whereas testis-specific expression originates from both paralogs. Orange arrows depict segmental duplications. Lower depicts the coverage of mRNA-Seq reads mapping uniquely and perfectly (without mismatch) onto the ancestral paralog (Lower Left) or the derived duplicate (Lower Right). Numbers at the outer ends of the mRNA-Seq plots indicate maximum coverage height (average read density over a window size of 25 bp) in the respective tissue.
We next investigated the effects of gene duplications on expression in different tissues. To this end, we examined the relationship of 113 fixed whole-gene duplications with patterns of tissue-specific expression (37) across six tissues in humans and nonhuman primates (SI Appendix). We identified genes that showed gene expression in a tissue (normalized gene expression value ≥ 0.2) in one species but were not expressed (normalized gene expression value = 0) in the same tissue in other species. We then evaluated these data jointly with our set of interspecies gene duplications. Ultimately, we identified 13 (11.5%) whole-gene duplications associated with expression in a new tissue (Dataset S5), a notable enrichment compared with partial gene duplications (∼3.5-fold enrichment; P = 0.001139, two-sided Fisher's test) and nonduplicated genes (approximately fourfold enrichment; P = 0.001213, two-sided Fisher's exact test) (Fig. 4B), which remained significant when assuming different thresholds and scenarios for considering genes as expressed (SI Appendix, Fig. S9 C–E). Hence, our findings suggest that, in primates, newly emerged gene duplicates frequently coincide with gene expression in new tissues.
Analyzing these 13 duplications in detail, we observed that 5 duplications coincided with acquisition of gene expression in brain tissues, a finding of potential interest in the light of the proposed role of gene duplications in primate brain evolution (38). Furthermore, four duplications were related to processes linked with interactions with the environment, including immune response (IGLL1, and LYG2) and xenobiotic metabolism (CYP2A13 and UGT2B7). Lysozyme G-like protein 2 (LYG2) encodes a bacterial cell wall-degrading lysozyme with a role in innate immunity, which in humans, is expressed in eye and testis (39). Our analyses showed that LYG2 is additionally expressed in human brain, where it may participate in brain-specific innate immunity (Fig. 4C and SI Appendix, Fig. S9F). Importantly, in conjunction with whole-gene duplication, LYG2 acquired expression in the liver of orang-utans (Fig. 4C and SI Appendix, Fig. S9F), suggesting a potential acquired functional role in orang-utan livers.
The incomplete nature of nonhuman primate reference assemblies (8) hampered detailed analyses of LYG2 paralogs. We were, however, able to pursue such analysis with the putative cystatin 9-like protein 1 gene (CST9LP1) (an evolutionarily conserved gene of unknown function with homology to the cystatin 9 gene, which encodes a protein with endopeptidase inhibitory activity) displaying whole-gene duplication in macaque. Although CST9LP1 showed little or no expression across examined tissues in the great apes (37), we detected appreciable expression levels in macaque heart (Fig. 4D). Although only one copy of CST9LP1 has been annotated in the macaque genome, we identified two intact coding sequences separated by 200 kb on chromosome 10 occurring in an inverted orientation (Fig. 4D). Although both copies showed similar levels of sequence identity (∼94%) with the human ortholog, preserved synteny allowed us to distinguish the ancestral and derived locus. We reasoned that, if the striking increase in expression in heart results from cis regulatory context changes, then the mRNA-Seq reads would exclusively map to the derived paralog, and indeed, expressed mRNAs in heart that could be confidently mapped originated exclusively from this paralog (which additionally showed enhanced expression in macaque testis) (Fig. 4D). Hence, our findings link CST9LP1 duplication with the gain of expression in heart, a gain that may be explained by exposure of the derived gene locus to a different cis regulatory context.
Discussion
Here, we have provided comprehensive SV maps in different nonhuman primates and shown that the activity of SV formation mechanisms, specifically of MEI and NAHR, is subject to rapid evolutionary change visible at timescales less than 25 million years ago. By generating MPS-based genome-scale sets of polymorphic reference and nonreference MEIs in several primates, we observed a notable excess of Alu activity in macaque compared with chimpanzee and orang-utan. Because Alu represents the most active human mobile element (3, 40), our findings suggest a rapid turnover of active transposable DNA sequences, leading to a divergent set of species-specific MEIs.
By comparison, our analyses showed a marked increase in NAHR-formed SVs in the great apes. Because NAHR-mediated SVs are usually larger in size, often intersect genes, and have been implicated in numerous genomic disorders (26, 35), these results are relevant to the generation of evolutionary novelty by gene duplication and the formation of pathogenic SVs. The markedly increased number of segmental duplications observed in great ape genomes most likely contributes to the activity of NAHR in these species, implying a direct link between genomic architecture and SV formation mechanism landscapes (12).
The burst of segmental duplications in the great ape lineage (12), linked to the NAHR mechanism, and an abundance of MEIs in the Old World monkey lineage compared with the great ape lineage (11) have been previously reported. We now furthered this observation by providing strong evidence for present day lineage-specific activities of NAHR and retrotransposition influencing within-species polymorphism landscapes at genome-wide scale. These mechanistic differences have two interrelated implications. First, fixed NAHR and MEI differences between great ape and Old World monkey lineages will likely further accumulate differentially in these lineages, thereby promoting additional diversification. Second, the likelihood of an adaptive variant to form through NAHR is higher in great apes than Old World monkeys, whereas the likelihood of an adaptive variant to be formed by retrotransposition is higher in Old World monkeys compared with great apes. Therefore, differences in rates of SV formation can predispose great apes and Old World monkeys to disparate evolutionary trajectories.
Our study also uncovered hundreds of fixed whole- and partial gene duplications, which we related to gene expression data to investigate their evolutionary impact. A possible explanation for the imperfect correlation between gene duplications and transcript-level increases that we observed is that gene duplicates are frequently regulated by an altered cis regulatory program. Furthermore, fixed partial gene duplications only rarely coincided with an increase in gene expression in keeping with recent studies reporting complex relationships between partial gene duplication and gene expression (41, 42).
Evolution of genes and gene families by duplication has been proposed to constitute a major driving force in evolution (27). Duplicated mammalian genes evolve rapidly after gene duplication (43, 44), and we have observed gene expression patterns of certain duplicated genes, implying diversification and function acquisition. Indeed, different possible fates of gene duplications have been proposed, which are referred to as neofunctionalization (i.e., a gene duplicate or paralog acquires a unique function) and subfunctionalization [specialization of both copies (parent and duplicate), each of which retain different subfunctions of the ancestral gene] (45). Roles of neofunctionalization and subfunctionalization have previously been studied in different organisms (45, 46), including humans, in which ancient gene duplications occurring after the human–mouse split (>90 Mya) were evaluated for tissue expression (47). Our duplication map enabled us to associate gene duplication across recent primate evolution and revealed 13 recently duplicated genes that are candidates for neofunctionalization (i.e., gene duplication coinciding with newly acquired tissue expression.) One possible explanation is that the newly emerged gene duplicate is located in a unique cis regulatory environment, facilitating the acquisition of expression in a new tissue. Irrespective of the mechanism involved, our results underscore the presumption that gene duplications can have a major influence on gene expression patterns.
Materials and Methods
Primate fibroblast-derived cell lines were obtained from Coriell (Dataset S1). MPS DNA and RNA libraries were prepared according to the vendor’s protocols. Sequence variants were detected using different algorithms as detailed in SI Appendix. To infer fixed gene duplications, we used paralog-specific and aggregate read mapping approaches, making use of eggNOG database v. 3 (28). Variants were validated by aCGH, PCR, qPCR, and FISH (SI Appendix).
Supplementary Material
Acknowledgments
We thank Kalliopi Trachana for enabling access to the latest eggNOG release; Andreas Schlattl, Tobias Rausch, Benjamin Raeder, Alejandro Reyes, and Wolfgang Huber for support with the gene expression analyses; and David Garfield for valuable discussions. We also thank David Radke, Sunita Setlur, and Psalm Haseley for technical assistance and Anita Hawkins for support in FISH, and we acknowledge the European Molecular Biology Laboratory Genomics Core Facility and Information Technology Unit for support. V.T. was supported by European Molecular Biology Organization Short-Term Fellowship ASTF 150-2013. R.C.I. was supported by National Institutes of Health Fellowship F32 AG 039979. C.L. was supported by National Institutes of Health Grants National Institute of Allergy and Infectious Diseases 5 R01 AI 089246-01A1 and The National Institute of General Medical Sciences 5 R01 GM081533. J.O.K. received support from an Emmy Noether Fellowship from the Deutsche Forschungsgemeinschaft (KO 4037/1-1).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequencing and aCGH data reported in this paper have been deposited in the European Nucleotide Archive, www.ebi.ac.uk/ena/ (accession no. ERP002376) and the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE45741), respectively. In addition, all the callsets are available at http://www.korbel.embl.de/primate_sv/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305904110/-/DCSupplemental.
References
- 1.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 2.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 3.Mills RE, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470(7332):59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudmant PH, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330(6004):641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318(5849):420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlattl A, Anders S, Waszak SM, Huber W, Korbel JO. Relating CNVs to transcriptome data at fine resolution: Assessment of the effect of variant size, type, and overlap with functional regions. Genome Res. 2011;21(12):2004–2013. doi: 10.1101/gr.122614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varki A, Geschwind DH, Eichler EE. Explaining human uniqueness: Genome interactions with environment, behaviour and culture. Nat Rev Genet. 2008;9(10):749–763. doi: 10.1038/nrg2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Bleness M, Searles VB, Varki A, Gagneux P, Sikela JM. Evolution of genetic and genomic features unique to the human lineage. Nat Rev Genet. 2012;13(12):853–866. doi: 10.1038/nrg3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437(7055):69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 10.Locke DP, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469(7331):529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316(5822):222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 12.Marques-Bonet T, et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457(7231):877–881. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gazave E, et al. Copy number variation analysis in the great apes reveals species-specific patterns of structural variation. Genome Res. 2011;21(10):1626–1639. doi: 10.1101/gr.117242.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry GH, et al. Copy number variation and evolution in humans and chimpanzees. Genome Res. 2008;18(11):1698–1710. doi: 10.1101/gr.082016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee AS, et al. Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum Mol Genet. 2008;17(8):1127–1136. doi: 10.1093/hmg/ddn002. [DOI] [PubMed] [Google Scholar]
- 16.Gokcumen O, et al. Refinement of primate copy number variation hotspots identifies candidate genomic regions evolving under positive selection. Genome Biol. 2011;12(5):R52. doi: 10.1186/gb-2011-12-5-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan G, et al. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotechnol. 2011;29(11):1019–1023. doi: 10.1038/nbt.1992. [DOI] [PubMed] [Google Scholar]
- 18.Auton A, et al. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336(6078):193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prüfer K, et al. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486(7404):527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee E, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam HY, et al. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat Biotechnol. 2010;28(1):47–55. doi: 10.1038/nbt.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kidd JM, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143(5):837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10(8):551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker JA, et al. Orangutan Alu quiescence reveals possible source element: Support for ancient backseat drivers. Mob DNA. 2012;3:8. doi: 10.1186/1759-8753-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18(2):74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 27.Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- 28.Powell S, et al. eggNOG v3.0: Orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40(Database issue):D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iskow RC, et al. Regulatory element copy number differences shape primate expression profiles. Proc Natl Acad Sci USA. 2012;109(31):12656–12661. doi: 10.1073/pnas.1205199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: A non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40(Web Server issue):W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449(7164):913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ting CT, et al. Gene duplication and speciation in Drosophila: Evidence from the Odysseus locus. Proc Natl Acad Sci USA. 2004;101(33):12232–12235. doi: 10.1073/pnas.0401975101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perry GH, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39(10):1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: Insights from and for human disease. Nat Rev Genet. 2013;14(2):125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 36.Henrichsen CN, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009;41(4):424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 37.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478(7369):343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 38.Sikela JM. The jewels of our genome: The search for the genomic changes underlying the evolutionarily unique capacities of the human brain. PLoS Genet. 2006;2(5):e80. doi: 10.1371/journal.pgen.0020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang P, et al. Characterization and expression of HLysG2, a basic goose-type lysozyme from the human eye and testis. Mol Immunol. 2011;48(4):524–531. doi: 10.1016/j.molimm.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 40.Stewart C, et al. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011;7(8):e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dennis MY, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149(4):912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popesco MC, et al. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313(5791):1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Zhang J. The ortholog conjecture is untestable by the current gene ontology but is supported by RNA sequencing data. PLoS Comput Biol. 2012;8(11):e1002784. doi: 10.1371/journal.pcbi.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pegueroles C, Laurie S, Albà MM. Accelerated evolution after gene duplication: A time-dependent process affecting just one copy. Mol Biol Evol. 2013;30(8):1830–1842. doi: 10.1093/molbev/mst083. [DOI] [PubMed] [Google Scholar]
- 45.He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169(2):1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katju V, Lynch M. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics. 2003;165(4):1793–1803. doi: 10.1093/genetics/165.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huminiecki L, Wolfe KH. Divergence of spatial gene expression profiles following species-specific gene duplications in human and mouse. Genome Res. 2004;14(10A):1870–1879. doi: 10.1101/gr.2705204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.