Abstract
The assignment of the two substrate water sites of the tetra-manganese penta-oxygen calcium (Mn4O5Ca) cluster of photosystem II is essential for the elucidation of the mechanism of biological O-O bond formation and the subsequent design of bio-inspired water-splitting catalysts. We recently demonstrated using pulsed EPR spectroscopy that one of the five oxygen bridges (μ-oxo) exchanges unusually rapidly with bulk water and is thus a likely candidate for one of the substrates. Ammonia, a water analog, was previously shown to bind to the Mn4O5Ca cluster, potentially displacing a water/substrate ligand [Britt RD, et al. (1989) J Am Chem Soc 111(10):3522–3532]. Here we show by a combination of EPR and time-resolved membrane inlet mass spectrometry that the binding of ammonia perturbs the exchangeable μ-oxo bridge without drastically altering the binding/exchange kinetics of the two substrates. In combination with broken-symmetry density functional theory, our results show that (i) the exchangable μ-oxo bridge is O5 {using the labeling of the current crystal structure [Umena Y, et al. (2011) Nature 473(7345):55–60]}; (ii) ammonia displaces a water ligand to the outer manganese (MnA4-W1); and (iii) as W1 is trans to O5, ammonia binding elongates the MnA4-O5 bond, leading to the perturbation of the μ-oxo bridge resonance and to a small change in the water exchange rates. These experimental results support O-O bond formation between O5 and possibly an oxyl radical as proposed by Siegbahn and exclude W1 as the second substrate water.
Keywords: PSII, OEC, water oxidizing complex, water-oxidation, Mn cluster
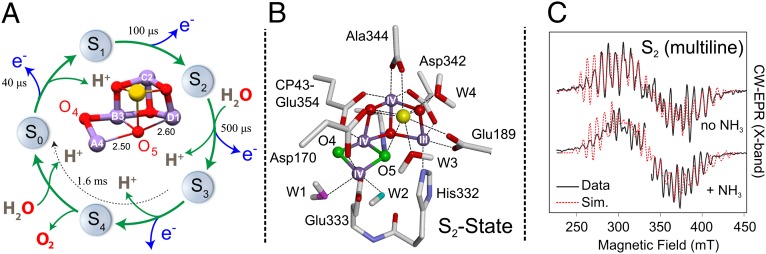
In oxygenic photosynthesis, light-driven water splitting is catalyzed by the oxygen-evolving complex (OEC) of the membrane bound, pigment-protein complex photosystem II (PSII). The OEC consists of an inorganic tetra-manganese penta-oxygen calcium (Mn4O5Ca) cluster (1–3) and the nearby redox-active tyrosine residue YZ (D1-Tyr161) that couples electron transfer from the Mn4O5Ca cluster to P680, the photo-oxidant of PSII. The cluster resembles a “distorted chair”, where the base is formed by an oxygen-bridged (μ-oxo) cuboidal Mn3O4Ca unit (1) (Fig. 1A). The fourth Mn (MnA4) is located outside of the cuboidal unit and is linked via a μ-oxo–bridged ligation (O4) to one of its corners (MnB3). A second linkage between the outer Mn and the cube is provided by a fifth oxygen O5. The Mn4O5Ca cluster is also held together by six carboxylate ligands and has only one directly coordinating nitrogen ligand, D1-His332 (Fig. 1B).
Fig. 1.
(A) The S-state cycle of the OEC. The crystal structure of the manganese tetramer is also shown, indicating the unusual ligation of O5, equidistant between MnA4 and MnD1 (1). (B) A representative, unified DFT model of the OEC in the S2 state (10). The oxygen ligands W1 (pink), W2 (cyan), and O4/O5 (green) were assigned as sites exchangeable with solvent water in the S1 state (17). (C) The effect of ammonia on the CW-EPR (multiline) signal of the S2 state [11,29] (Fig. S1 and Table S1).
The OEC cycles through a series of five intermediate states that are known as S states (4) (Fig. 1A): S0, S1 (dark stable), S2, S3, and S4 (not yet isolated), where the subscript refers to the number of oxidizing equivalents stored in the OEC through successive electron withdrawals by YZ•. In the 1.9-Å resolution structure, the S state of the cluster was assigned to be S1 (1). However, this is unlikely as all Mn-Mn, Mn-Ca, and Mn-O/N distances of the crystal structure are ∼0.1 Å longer compared with those determined by extended X-ray absorption fine structure (EXAFS) spectroscopy (5–7). Moreover, the central O5 has unusually long bonds to three Mn ions and to the Ca ion, outside the range seen for model complexes. All these structural details suggest that the Mn ions of the cluster were photoreduced during X-ray data collection, and as such, the X-ray structure represents a nonphysiological, overreduced S state (8, 9). This structural ambiguity can be eliminated by combining the X-ray data with spectroscopic constraints and the introduction of computational modeling. In these unified models, O5 is generally considered to be a μ-oxo bridge between MnA4 and MnB3 in the S1 and S2 states, rendering this unit bis–μ-oxo bridged, and MnD1 as five coordinate (10–13) (Fig. 1B).
The S2 state is readily observed using EPR spectroscopy and related techniques. In this state, the four Mn ions of the OEC are coupled together, resulting in a ground electronic state with one unpaired electron, i.e., effective spin Seff = 1/2 (14). A distinctive “multiline” EPR spectrum is observed at liquid helium temperature, where the line splittings reflect the coupling of the four 55Mn magnetic nuclei to the unpaired electron spin (hyperfine interaction) (Fig. 1C). The unpaired electron of the Mn4CaO5 cluster also couples to other magnetic nuclei in the vicinity of the OEC (e.g., 17O, 14N/15N, 1H/2H), such as those that coordinate the Mn ions, e.g., 17O, 14N/15N, 1H/2H. These hyperfine couplings are sufficiently small so that the interactions are not directly observed by continuous wave (CW)-EPR spectroscopy. Such interactions can instead be detected using pulse magnetic resonance techniques that probe NMR transitions (15). Such techniques include electron spin echo envelope modulation (ESEEM), electron nuclear double resonance (ENDOR), and electron–electron double-resonance–detected NMR (EDNMR). Each technique is suited to probe specific electron–nuclear interactions of the OEC. For example, exchangeable oxygen sites of the OEC, which are potential substrate sites (16), have been recently studied with W-band EDNMR using 17O isotopic labeling (17). This methodology is particularly useful as it allows all water-exchangeable sites, including fully deprotonated Mn–μ-oxo bridges, to be observed. It is known from time-resolved membrane inlet mass spectrometry (TR-MIMS) that at least one substrate is bound in all S states and exchanges with bulk water on a seconds timescale (16, 18–20). In the equivalent EDNMR experiment performed in the S1 state, rapid mixing of PSII with 17O-labeled water led to the uptake of the 17O label at three different Mn-ligand sites: (i) as one μ-oxo bridge, most likely O4 or O5; (ii) as a terminal hydroxide ligand, most likely W2, a ligand of MnA4; and (iii) as a terminal water ligand, most likely W1, also a ligand of MnA4 (17). This latter species dominates the weakly coupled “matrix” envelope, which also has contributions from the Ca-bound waters (W3/W4) and second coordination shell H2O ligands. These assignments are based on comparison with model compounds and the recent 1.9-Å resolution PSII crystal structure in conjunction with density functional theory (DFT) models (10).
Enzymological studies have indicated that there are at least two independent ammonia-binding sites, SYI and SYII (21, 22) in PSII. Ammonia binding at the SYI site is chloride-concentration dependent, is S-state independent, and results in the inhibition of oxygen-evolving activity (21–23), indicating that SYI likely represents one of the chloride sites identified in the crystal structure (1). In contrast, ammonia binding to SYII is independent of the chloride concentration (21, 22, 24, 25) and does not reduce O2 evolution. It binds only upon formation of the S2 state, to be subsequently released at some later point during the S state cycle (after S3), such that it is not bound upon return to the S1 state (26). SYII exhibits steric selectivity for small Lewis bases and appears to be only accessible to ammonia.
The ESEEM study by Britt et al. (25) demonstrated that SYII represents a Mn coordination site. Interestingly, the bound 14NH3 species displayed a large, rhombic quadrupole coupling (e2Qq/h) of 1.61 MHz, with η = 0.59. From comparison with model compounds, it was suggested that the ammonia is taken up as an amido bridge between either two Mn ions or one Mn ion and the Ca ion, i.e., replacing or modifying one μ-oxo bridge of the complex. Low-frequency FTIR spectroscopy supports this basic hypothesis, identifying a putative Mn-μO-Mn or Mn-μO-Ca vibrational mode (27) lost upon ammonia addition (28).
Here we investigate the binding of ammonia to the OEC, using multiple-pulse EPR techniques and TR-MIMS. It is shown that, although ammonia significantly perturbs all exchangeable Mn-O ligand signals, it only moderately affects the exchange rates of both substrate waters. Instead of it displacing a μ-oxo bridge, our data support a mechanism in which ammonia modifies the μ-oxo bridge by displacing a water ligand trans to the bridge position, specifically the water ligand W1 trans to the μ-oxo bridge O5. Broken symmetry (BS)-DFT calculations, which model this displacement, quantitatively reproduce all spectroscopic observables. Together, our data show that W1 is not a substrate binding site, but instead favor O5 as one of the two substrate waters.
Results and Discussion
Ammonia Binds to the OEC Without Significantly Changing Its Electronic Structure.
PSII isolated from the thermophilic cyanobacteria Thermosynechococcus elongatus was used throughout this study. Ammonia was added to PSII samples in the S1 state, which was advanced before the EPR measurements to the S2 state by low-temperature (180 K) illumination with visible light. In agreement with the literature, this resulted in an unperturbed S2-state multiline EPR signal similar to the “no NH3” spectrum shown in Fig. 1C (26). Subsequent annealing of the sample to 260 K for 30 s led to the induction of the ammonia-modified multiline form (NH3 spectrum, Fig. 1C) (26). No change was observed in the background cytochrome c550/b559 signals upon annealing the sample at 260 K. The ammonia-modified S2-state multiline signal is also centered about g ≈ 2.0, spread over the 250- to 430-mT field range and characteristically contains more hyperfine peaks than the control sample (at least 24 vs. 20; see Fig. 1C) (24, 29). Simulations of the EPR and 55Mn-ENDOR spectra using the spin Hamiltonian formalism are given in Fig. S1 and Table S1.
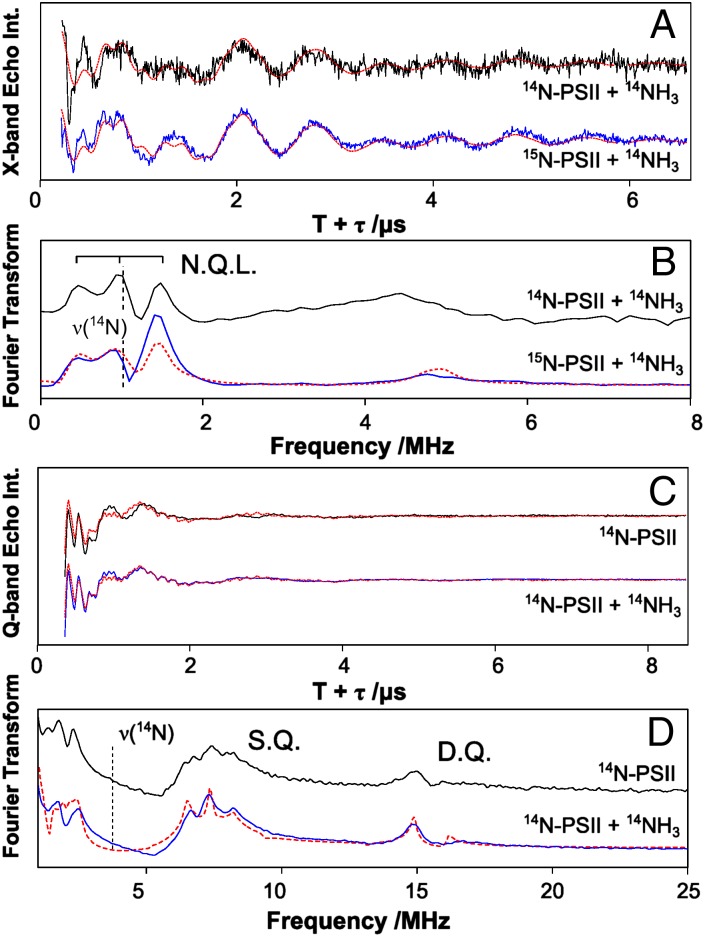
Nitrogen ligands of the OEC can be readily detected using ESEEM. In this type of pulse EPR experiment, the EPR signal intensity (spin echo) is recorded as a function of the time intervals between the successive microwave pulses. Signal intensity modulations arise from the weak coupling of the electron spin with nearby magnetic nuclei such as 14N [I(14N) = 1]. Both native 14N-PSII and universally labeled 15N-PSII [I(15N) = 1/2] were measured. X-band three-pulse ESEEM experiments of 14NH3-containing PSII illuminated at 180 K and subsequently annealed at 260 K (see above) are shown in Fig. 2 A and B. A new modulation, consistent with 14NH3 binding to the Mn4O5Ca cluster, is observed in the light-minus-dark difference spectra only after the 260-K annealing step (25). The bound 14NH3 species displays three sharp nuclear-quadrupole lines (N.Q.L.) at 0.5, 1.0, and 1.5 MHz in the Fourier-transformed spectrum (Fig. 2B). Spin Hamiltonian simulations of the lineshape are shown in Fig. 2 A and B as dashed red lines and the fitted parameters (Table 1, Fig. S2, and Table S2) for T. elongatus are similar to those reported in the earlier higher-plant study (25). The relatively small magnitude of the hyperfine coupling supports the assignment of 14NH3 as a ligand to one of the MnIV ions as opposed to the MnIII ion of the S2 state. This is because the MnIV ions carry a lower spin density (spin projection) than the MnIII ion and thus their ligands are expected to display smaller effective hyperfine couplings (30).
Fig. 2.
(A) X-band three-pulse ESEEM traces measured at the center of the S2-state multiline signal (Fig. 1, B0 = 333 mT, microwave frequency = 9.4 GHz). The data represent annealed-minus-dark difference traces collected on ammonia (14NH3)-treated 14N-PSII (black) and (14NH3)-treated 15N-PSII (blue). The traces shown in A were measured with an interpulse spacing τ of 136 ns. Additional data traces using the τ-values 152 ns, 168 ns, and 184 ns are shown in Fig. S2. (B) Fourier transform (FT) of the X-band time domain data. N.Q.L. identifies the nuclear-quadrupole lines caused by the coupling of the OEC with the added 14N (I = 1). The spectrum shown represents the sum of the FT of the four ESEEM traces measured using different τ-values (136–184 ns) to minimize spectral artifacts. (C) Q-band three-pulse ESEEM traces measured at the center of the S2-state multiline signal (B0 = 1.22 T, microwave frequency = 34.0 GHz). The data represent light-minus-dark and annealed-minus-dark difference spectra of native 14N-PSII (black) and ammonia (14NH3)-treated 14N-PSII (blue) respectively. The time domain data were measured using an interpulse spacing τ of 260 ns. Additional data traces using τ-values of 240 ns and 300 ns are shown in Fig. S2. (D) Corresponding FT of the data traces presented in C. S.Q. and D.Q. identify single-quantum and double-quantum transition lines from the coupling with 14N-His332. The red dashed lines superimposing the data represent a simulation using the spin Hamiltonian formalism (SI EPR Theory/Simulations, Fig. S2, and Table S2). The label N.Q.L. identifies the quadrupole lines observed in the X-band 14N-ESEEM spectrum.
Table 1.
Experimentally determined ESEEM and EDNMR spin Hamiltonian parameters: Comparison with calculated magnetic resonance parameters from DFT
| Hyperfine couplings |Aiso|/MHz |
||||
| Exchangable ligands, 14N/17O | ||||
| Experiment/Theory | His332*, 14N* | W1 17O/NH3 14N* | W2 17O* | O5 17O† |
| DFT | ||||
| Native | 4.8 | 1.7 | 5.2 | 17.4 |
| +NH3‡ | 5.2 | 1.5 | 4.3 | 12.2 |
| Δ§ | 0.4 | — | −0.9 | −5.2 |
| Δ/%§ | 8.3 | — | −17 | −30 |
| Experiment | ||||
| Native | 7.2 | 1.4 | 4.5 | 9.7 |
| +NH3* | 7.2 | 2.4 | 3.1 | 6.5 |
| Δ§ | 0.0 | — | 1.4 | 3.2 |
| Δ/%§ | 0.0 | — | −31 | −28 |
Calculated (projected) BS-DFT hyperfine values directly comparable to experiment (/MHz).
Calculated (raw) BS-DFT hyperfine values are not directly comparable to experiment; the percentage change (Δ) due to ammonia binding can, however, be compared.
NH3 replacing W1.
Δ = difference between native and +NH3 samples.
In contrast to the X-band measurements, at Q-band, no difference is seen between the control and the 14NH3-treated sample (Figs. 2C and 2D). Instead, the observed ESEEM modulation is dominated by a 14N hyperfine coupling assigned to the D1-His332 ligand of MnD1 (31). At Q-band the histidine 14N signal is at or near the cancellation condition and as such displays a maximal ESEEM response (30, 31). As a consequence of the D1-His332 14N coupling matching the cancellation condition, the signal of the bound ammonia in comparison is suppressed at Q-band and no direct information on its binding site can be obtained in this way. However, the 14N histidine ESEEM signal, which resolves multiple spectral lines at 0.6, 2.0, and 7.3 MHz, and 14.8 MHz representing single-quantum (SQ) and double-quantum (DQ) transitions, respectively, can be used as spin probe reporting on the electronic structure and the oxidation state of MnD1. Spin Hamiltonian simulations of the lineshape of this signal are shown in Fig. 2 C and D (dashed red lines). The parameters used (Table 1, Fig. S2, and Table S2) are similar to those reported earlier by Stich et al. (31) for PSII purified from Synechocystis sp. 6803. The relatively large magnitude of the D1-His332 14N coupling suggests that it is ligated to the only MnIII ion in the S2 state, i.e., to the Mn ion that carries the largest spin density/spin projection (11, 30, 31). As the D1-His332 signal does not change upon the addition of ammonia, the oxidation state and ligand field of the MnD1 ion cannot change. Thus, the binding site of ammonia at the manganese tetramer is unlikely to be proximal to the MnD1 but instead is distal to it, consistent with NH3 binding to a MnIV ion. It is also noted that protons in the vicinity of the OEC can be readily detected using Q-band 1H-ENDOR (SI EPR Theory/Simulations and Fig. S3A). The addition of NH3 does not change the width of the signal envelope, which has been assigned to the protonated oxygen ligands on MnA4 (32). The absence of a large proton coupling suggests ammonia does not replace one of the μ-oxo bridges of the OEC, excluding this previous suggestion for its binding site (25).
Ammonia Perturbs All Exchangeable Oxygen Ligands of the Manganese Tetramer.
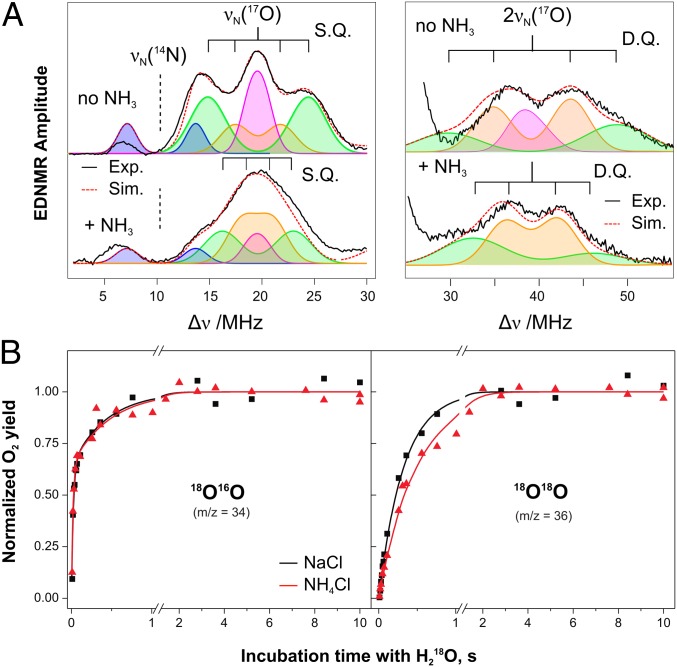
EDNMR (32), a pump–probe technique, which employs two independent microwave pulses, has recently been shown to be the magnetic resonance method of choice for the detection of (17O) ligands of metallocofactors, such as the Mn4O5Ca cluster of the OEC. In this experiment, the EPR signal is monitored at a fixed microwave frequency matched to the resonator (probe pulse). Before the detection sequence, a microwave pulse of varying frequency, termed the high turning-angle (HTA) pulse, is applied (pump pulse). The pumping (HTA) pulse drives spin-forbidden transitions where both the electron spin and the nuclear spin state change (|Δms| = 1, |ΔmI| = 1). Magnetic nuclei appear as doublets centered about their characteristic (Larmor) frequencies; i.e., νN(14N) = 10.46 MHz and νN(17O) = 19.6 MHz at 3.4 T. As described in Rapatskiy et al. (17), in S2-state PSII samples resuspended in H217O-containing buffer, two structured signal envelopes are observed centered at the Larmor frequency and at twice the Larmor frequency of 17O. These two signal envelopes correspond to SQ and DQ transitions of exchangeable oxygen ligands of the manganese tetramer (Fig. 3A). Three components were identified (17): (i) a large coupling, assigned to a μ-oxo bridge from comparison with model complexes, most likely O4 or O5; (ii) an intermediate coupling, assigned to the terminal oxygen ligand of MnA4 (W2); and (iii) a weak coupling (unsplit matrix line), representing the second terminal oxygen ligand of MnA4 (W1) but also including contributions from W3 and W4. The couplings of W1 and W2 are proposed to differ due to their protonation state. In DFT models, W2 is preferentially a hydroxo ligand in the S2 state, whereas W1 represents a water ligand (10, 13). In comparison with the hydroxo ligand (W2), the water ligand (W1) is expected to have a much smaller coupling, owing to its additional covalent bond to hydrogen, which weakens its bond to the MnIV ion.
Fig. 3.
(A) W-band 17O-EDNMR spectra of native and 14NH3-treated 14N-PSII samples (17). The black line represents the data; the red dashed line represents the total simulation. Fitted isotropic hyperfine values are listed in Table 1. A complete list of parameters is given in Table S3. The colored traces represent the four components of the fit: the 14N of D1-His332, blue; the strongly coupled 17O species, green; the intermediately coupled 17O species, orange; and the weakly coupled 17O species, pink. (B) TR-MIMS traces monitoring substrate exchange in the S2 state at pH 7.6 in the presence of either 100 mM NH4Cl (red triangles) or 100 mM NaCl (black squares). The lines represent biexponential (34O2, Left) and monoexponential (36O2, Right) fits. NH4Cl: kf = 52 s−1, ks = 2 s−1. NaCl: kf = 38 s−1, ks = 3 s−1.
Ammonia binding to the OEC modifies the 17O signal profile (17) (Fig. 3A and Fig. S4). The widths of the 17O single- and double-quantum envelopes narrow by ∼30%, and the splitting of the two outer single-quantum satellite peaks, which corresponds to the large coupling (μ-oxo bridge), becomes unresolved. Additionally, the sharp central matrix line (W1) appears to be of lower intensity.
The intermediate coupling is best resolved in the double-quantum region, owing to spectral congestion in the single-quantum region. Ammonia binding modifies the intermediate-coupling feature, narrowing it by ∼1–2 MHz. Furthermore, the whole double-quantum region becomes more symmetric compared with the spectra of the control sample (Fig. 3A, Fig. S4, and Table S3); this asymmetry was previously thought to be due to the matrix signal (17). The reduced asymmetry in the double-quantum region is taken as additional evidence that the matrix component is reduced by ammonia binding to the OEC, which is further supported by considering the power dependence of the EDNMR signal (for further details see SI EPR Theory/Simulations and Fig. S4). Thus, ammonia likely displaces W1, perturbing W2 and the μ-oxo bridge signal. It is also noted that the water ligands of the Ca2+ ion (W3, W4) were measured independently using 17O-Mims ENDOR, and no change was observed; ergo, W3 and W4 are not displaced by ammonia (Fig. S3B).
The Site of Ammonia Binding: A Mechanism for the Perturbation of the μ-oxo Bridge.
The binding of ammonia as a terminal ligand to MnA4 instead of W1 could potentially modify the hyperfine coupling of the μ-oxo bridge O5 via the trans effect. To test whether this rationale can quantitatively explain the observed spectral changes, DFT calculations were performed using previously reported S2-state OEC models consistent with geometric, thermodynamic, and spectroscopic parameters (10, 17). Calculated EPR parameters (33, 34) of both the W1- and the NH3-containing structure are shown in Table 1 and Tables S2–S4. This single-ligand substitution quantitatively reproduces all experimental observables, including the 14N hyperfine and quadrupole couplings of the bound ammonia, the ∼1-MHz decrease in the 17O hyperfine coupling of the terminal hydroxide (W2), and the 14N-His332 hyperfine coupling and its insensitivity to ammonia addition. Although it is currently not possible to reliably calculate projected hyperfine coupling constants for bridging ligands, as this is yet to be calibrated in model systems, it is possible to compare the raw BS-DFT values to ascertain the effect of the ammonia ligand. The calculations show that ammonia binding at the W1 site selectively perturbs the O5 μ-oxo bridge. The observed change in coupling is again quantitatively reproduced, with a decrease in the hyperfine coupling of O5 by 30%, the same as seen for the μ-oxo bridge species using EDNMR. All other μ-oxo bridge couplings are calculated as being very similar for the H2O- and the NH3-containing structure, including the O4, which actually increases upon NH3 binding, excluding it as the exchangeable bridge. The only exception is O1, where the calculated raw BS-DFT hyperfine has a large percentage change; however, the absolute magnitude of the O1 hyperfine coupling is small and the absolute change is only 0.55 MHz (Table S4).
From this, we can confidently assign the site of NH3 binding to the W1 coordination site of MnA4. A comparison of the different geometries of the two BS-DFT structures (with and without NH3) shows a small elongation of the MnA4-O5 bond of 0.02 Å upon NH3 substitution, as expected. This bond lengthening reduces the MnA4 to O5 spin polarization and consequently the overall spin density on O5, resulting in the 30% decrease in the observed 17O hyperfine value. This change should also modify the vibrational mode of the O5 bridge, consistent with low-frequency IR spectroscopic results reported in ref. 28. Indeed, vibrational frequencies computed for the optimized structures of the two models indicate that a MnA4–O5 stretching mode along the MnA1-MnD1 vector at 644 cm−1 shifts upon NH3 binding to 617 cm−1 with concomitant ∼50% loss in intensity, consistent with experimental observations.
W1 Is Not a Substrate Water.
TR-MIMS, a mass spectrometric pump–probe technique, employing H218O labeling, provides important information regarding the binding of the substrate to the catalyst during the S-state cycle (16). This experiment involves poising the OEC in the desired S state with light flashes and the subsequent rapid injection (t1/2 = 3 ms) of isotopically labeled water (H218O), followed by successive light flashes to release the product O2. By varying the incubation time of the sample in labeled water, the extent to which 18O is incorporated into the product O2 is varied, allowing the determination of substrate water exchange rates with the bulk solvent. These experiments have established that the two substrate waters exchange with different rates that also vary independently with the S states. Thus, the two substrates bind at chemically distinct sites. The slowly exchanging substrate (Ws) is bound throughout the S-state cycle, whereas the fast-exchanging substrate (Wf) is bound latest in the S2 state (16, 18, 20, 35, 36).
TR-MIMS data monitoring the fast and slow substrate exchange in the S2 state at pH 7.6 in the presence of 100 mM NH4Cl (red) or 100 mM NaCl (black) are shown in Fig. 3B. If ammonia displaces a substrate, a major slowing or even abolishment of one exchange rate is expected. This is not observed experimentally: The exchange rates of Ws and Wf with bulk water lie within factors of 1.5 in the presence and absence of NH4Cl. This demonstrates that ammonia does not displace a substrate water, but instead slightly modifies exchange rates by binding in their vicinity. Thus, the combined EPR and TR-MIMS data exclude W1 as a substrate site. Importantly, these results exclude O-O bond mechanisms that involve both terminal Mn oxygen ligands on MnA4, i.e., the Kusunoki-type mechanism (37).
This model also provides a simple rationale for ammonia binding/release during the S-state cycle (24, 26). In the lower S states (S0, S1), MnA4 is usually considered to be in the MnIII oxidation state and is thus potentially five-coordinate, with W1 being only a weakly associated ligand. It is noted that DFT calculations support assigning the Jahn–Teller axis of MnA4III along the W1/O5 axis (13, 38, 39). As such, ammonia does not bind in these S states as its nominal binding site is preferentially unoccupied. Upon formation of the S2 state, the MnA4 is oxidized to +IV and is required to be six-coordinate, thus allowing ammonia to bind to the OEC. As NH3 is a better (more tightly bound) ligand to MnIV than water in the S3 and presumably the S4 states, ammonia is unlikely to be released until after the O-O bond formation step, at which point MnA4 returns to its +III oxidation state and is again five-coordinate.
O5 Represents a Substrate Site.
The slow rate of exchange of Ws and the observation that the rate is S-state (i.e., Mn oxidation state) dependent suggest that Ws represents a Mn–oxygen ligand (16, 18, 20, 35). In Rapatskiy et al. (17), three exchangeable Mn-O ligands were identified, and thus, all three potentially represent WS: W1, W2, and a μ-oxo bridge, either O4 or O5. As described above, the ammonia effect excludes W1 and demonstrates that O5 (and not O4) represents the exchangeable bridge. Thus, we can now reduce the number of possible candidates for Ws to only two: W2 and O5.
A series of studies are converging with regard to the role of O5 instead of W2 as the Ws substrate site. Critical to this assignment has been the recent demonstration that one of the μ-oxo bridges (shown here to be O5) exchanges rapidly with bulk water (17), with an exchange rate consistent with mass spectrometry measurements (16, 18–20) and over 1,000 times faster than that seen in synthetic model systems (40). A rationale for this enhanced exchange rate was recently provided by the theoretical study of Pantazis et al. (13), where it was shown that O5 has a flexible coordination, acting as either a μ-oxo linkage to the outer Mn (MnA4) or a vertex of the cuboidal unit proper. Similarly, the OEC appears to contain several pathways for internal oxygen exchange between terminal water ligands to Ca or Mn, which may allow a calcium-ligated bridge such as O5 to exchange rapidly (41).
Site-selective perturbations such as protein mutagenesis provide further support for the assignment of O5 over W2 as Ws. The replacement of Ca with Sr strongly enhances the exchange rate of Ws (36). As O5 (not W2 or O4)) is a ligand to Ca/Sr (1), this result is readily understood (36, 41). Similarly, the mutation of the D1-Glu189 (bridge between MnD1 and Ca), the D1-Asp170 (bridge between MnA4 and Ca), and the CP43-Glu354 (bridge between MnB3 and MnC2) all enhance the rate of Ws exchange (20, 42, 43). As O5 is a ligand to MnA4, MnB3, and MnD1 (owing to its two isoenergetic forms in the S2 state and potentially the S3 state) (1, 13), the observed perturbation in the exchange rate seen in these mutants is again readily explained.
An O-O Bond Formation Mechanism Involving O5.
The O-O bond reaction can proceed via either (i) a nucleophilic attack of O5 by a nearby substrate, i.e., between the μ-oxo bridge (O5) and a terminal hydroxide/Ca2+-bound water (W3), or (ii) an oxo/oxyl radical coupling of O5 and an as yet unidentified water (possibly previously bound to Ca/MnA4) that is located proximal to O5 in the S3/S4 states, as proposed by Siegbahn (12) (see also refs. 41, 44).
Of the two pathways to O-O bond formation, only the nucleophilic attack mechanism has been previously observed in Mn model systems, albeit with a much slower rate than seen for the OEC (45, 46). In contrast, the radical coupling mechanism has no precedence in Mn model chemistry, but has been demonstrated as an efficient O-O bond formation pathway in second-row transition metal catalysts; see, for example, the ruthenium (Ru-Hbpp) dimer complex (47). This latter mechanistic route has been demonstrated in silico by Siegbahn as the most efficient O-O bond formation pathway (12).
A unique feature of the oxo/oxyl mechanism proposed by Siegbahn is that the second, fast-exchanging water substrate (Wf) binds to the OEC late in the S-state cycle, a conclusion supported by FTIR difference spectroscopy (48). This additional substrate from the bulk binds to the open coordination site of MnD1 as a water/hydroxide ion in the S3 state, forming an oxyl radical in the S4 state (Fig. 4) (12). Superficially, this appears to be in disagreement with TR-MIMS measurements, which suggest that Wf has a similar affinity in the S2 state to that in the S3 state, requiring it to be in a chemically similar environment in both states. The inherent structural flexibility of the OEC provides a rationale for this problem, suggesting a second binding sequence for Wf, reconciling the oxo/oxyl mechanism with the observation that Wf is already bound in the S2 state. Instead, of binding directly to MnD1, the second substrate could bind to the solvent-accessible outer MnA4 ion, as the open coordination site of the complex can exist at either MnA4 or MnD1 via the facile movement of the O5/Ws bridge. In this instance, the terminal hydroxide ligands of MnA4 in the S3 state (W2 and Wf) would be indistinguishable, owing to rapid interchange, and could be considered to represent the same species. O5/Ws, which upon proton movement from Wf returns to the putative S3 state proposed by Siegbahn, represents in this tautomeric structure a terminal hydroxide ion bound to a MnIV ion. This ligand motif is considered to exchange with bulk solvent on a seconds timescale in Mn model complexes. The MnD1-bound oxygen is, however, within a more hydrophobic pocket compared with the MnA4-bound oxygen, which explains why two exchange rates are still observed for the two putative MnIV-O(H) substrate ligands in the S3 state. The hydrophobic region about MnD1 potentially acts to stabilize the subsequent ligand oxidation of the MnD1-bound oxygen to an oxyl radical upon advancement to the S4 state.
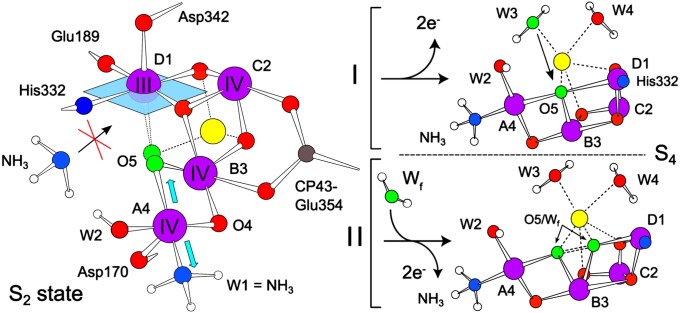
Fig. 4.
(Left) Site for NH3 binding to the OEC poised in the S2 state. NH3 displaces W1, a water ligand of the outer MnA4 (a MnIV ion in the S2 state), which slightly affects the binding strength of the oxo-bridge O5, which is trans to this position. (Right) O-O bond formation mechanisms consistent with this study (see main text): (I) a nucleophilic attack of O5 by a nearby substrate; (II) an oxo/oxyl radical coupling of O5 and an as yet unidentified additional water marked Wf (possibly W2). Mn, purple; Ca, yellow; N, blue; O, red; and substrate O, green.
Thus, a concerted tetramer mechanism involving O5, which uses the unique geometry of the Mn4O5Ca cluster to bind and position the two substrates, provides a rationale for the substrate exchange phenomenology described in the literature. The sequential uptake of the two substrates ensures that simultaneous binding of both substrates does not occur in the resting states (S0, S1) of the catalyst, which is likely critical for efficient (high turnover frequency) and highly selective O2 product formation.
Materials and Methods
14N- and 15N-PSII core complex preparations from T. elongatus were isolated as described earlier (49, 50) with modifications described in SI Materials and Methods. The S2 state was generated by short, white-light illumination (5 s) with a tungsten lamp at 185–200 K.
EPR measurements were performed at X-band using Bruker ELEXSYS 500 and 580 spectrometers, at Q-band using a Bruker ELEXSYS E580 spectrometer, and at W-band using a Bruker ELEXSYS E680 spectrometer. X-band CW and pulse EPR measurements were performed at 8.6 K and 4.2 K, respectively. Q- and W-band pulsed EPR measurements were performed at 4.8–5.2 K. Experimental settings were as reported in refs. 11 and 17 and in Figs. S1–S3.
TR-MIMS experiments were performed at 20 °C using a modified membrane-inlet cell connected to a magnetic sector field isotope ratio mass spectrometer. Further details regarding experimental procedures and data analysis are described in SI Materials and Methods and refs. 16, 19, and 35.
Density functional theory calculations of geometries, exchange coupling constants, vibrational frequencies, and EPR parameters were performed similarly to those described in refs. 10 and 17. Computational details and Cartesian coordinates of the optimized structures are given in SI Materials and Methods and Table S5, respectively.
Supplementary Material
Acknowledgments
We thank A. Savitsky (Max-Planck-Institut für Chemische Energiekonversion) and J. Martínez (Consejo Superior de Investigaciones Científicas, Universidad de Zaragoza) for their assistance. This work was supported by The Max-Planck-Gesellschaft, the “Bioénergie” program from the Commissariat à l’Énergie Atomique et aux Énergies Alternatives, the European Union SOLAR-H2 project (FP7 Contract 212508), Vetenskapsrådet, Strong Research Environment Solar Fuels (Umeå University), The Artificial Leaf Project (K&A Wallenberg Foundation), the Swedish Energy Agency, and the Kempe Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304334110/-/DCSupplemental.
References
- 1.Umena Y, Kawakami K, Shen J-R, Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature. 2011;473(7345):55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004;303(5665):1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 3.Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature. 2005;438(7070):1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- 4.Kok B, Forbush B, McGloin M. Cooperation of charges in photosynthetic O2 evolution-I. A linear four step mechanism. Photochem Photobiol. 1970;11(6):457–475. doi: 10.1111/j.1751-1097.1970.tb06017.x. [DOI] [PubMed] [Google Scholar]
- 5.Yano J, et al. Where water is oxidized to dioxygen: Structure of the photosynthetic Mn4Ca cluster. Science. 2006;314(5800):821–825. doi: 10.1126/science.1128186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pushkar YL, Yano J, Sauer K, Boussac A, Yachandra VK. Structural changes in the Mn4Ca cluster and the mechanism of photosynthetic water splitting. Proc Natl Acad Sci USA. 2008;105(6):1879–1884. doi: 10.1073/pnas.0707092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dau H, Grundmeier A, Loja P, Haumann M. On the structure of the manganese complex of photosystem II: Extended-range EXAFS data and specific atomic-resolution models for four S-states. Philos Trans R Soc Lond B Biol Sci. 2008;363(1494):1237–1243, discussion 1243–1244. doi: 10.1098/rstb.2007.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yano J, et al. X-ray damage to the Mn4Ca complex in single crystals of photosystem II: A case study for metalloprotein crystallography. Proc Natl Acad Sci USA. 2005;102(34):12047–12052. doi: 10.1073/pnas.0505207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grabolle M, Haumann M, Müller C, Liebisch P, Dau H. Rapid loss of structural motifs in the manganese complex of oxygenic photosynthesis by X-ray irradiation at 10-300 K. J Biol Chem. 2006;281(8):4580–4588. doi: 10.1074/jbc.M509724200. [DOI] [PubMed] [Google Scholar]
- 10.Ames W, et al. Theoretical evaluation of structural models of the S2 state in the oxygen evolving complex of Photosystem II: Protonation states and magnetic interactions. J Am Chem Soc. 2011;133(49):19743–19757. doi: 10.1021/ja2041805. [DOI] [PubMed] [Google Scholar]
- 11.Cox N, et al. Effect of Ca2+/Sr2+ substitution on the electronic structure of the oxygen-evolving complex of photosystem II: A combined multifrequency EPR, 55Mn-ENDOR, and DFT study of the S2 state. J Am Chem Soc. 2011;133(10):3635–3648. doi: 10.1021/ja110145v. [DOI] [PubMed] [Google Scholar]
- 12.Siegbahn PEM. Structures and energetics for O2 formation in photosystem II. Acc Chem Res. 2009;42(12):1871–1880. doi: 10.1021/ar900117k. [DOI] [PubMed] [Google Scholar]
- 13.Pantazis DA, Ames W, Cox N, Lubitz W, Neese F. Two interconvertible structures that explain the spectroscopic properties of the oxygen-evolving complex of photosystem II in the S2 state. Angew Chem Int Ed Engl. 2012;51(39):9935–9940. doi: 10.1002/anie.201204705. [DOI] [PubMed] [Google Scholar]
- 14.Dismukes GC, Siderer Y. Intermediates of a polynuclear manganese center involved in photosynthetic oxidation of water. Proc Natl Acad Sci USA. 1981;78(1):274–278. doi: 10.1073/pnas.78.1.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schweiger A, Jeschke G. Principles of Pulse Electron Paramagnetic Resonance. Oxford: Oxford Univ Press; 2001. [Google Scholar]
- 16.Messinger J, Badger M, Wydrzynski T. Detection of one slowly exchanging substrate water molecule in the S3 state of photosystem II. Proc Natl Acad Sci USA. 1995;92(8):3209–3213. doi: 10.1073/pnas.92.8.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rapatskiy L, et al. Detection of the water binding sites of the oxygen-evolving complex of photosystem II using W-band 17O ELDOR-detected NMR spectroscopy. J Am Chem Soc. 2012;134(40):16619–16634. doi: 10.1021/ja3053267. [DOI] [PubMed] [Google Scholar]
- 18.Hillier W, Wydrzynski T. The affinities for the two substrate water binding sites in the O2 evolving complex of photosystem II vary independently during S-state turnover. Biochemistry. 2000;39(15):4399–4405. doi: 10.1021/bi992318d. [DOI] [PubMed] [Google Scholar]
- 19.Hillier W, Wydrzynski T. Substrate water interactions within the Photosystem II oxygen evolving complex. Phys Chem Chem Phys. 2004;6(20):4882–4889. [Google Scholar]
- 20.Cox N, Messinger J. Reflections on substrate water and dioxygen formation. Biochim Biophys Acta. 2013;1827(8–9):1020–1030. doi: 10.1016/j.bbabio.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Sandusky PO, Yocum CF. The chloride requirement for photosynthetic oxygen evolution. Analysis of the effects of chloride and other anions on amine inhibition of the oxygen-evolving complex. Biochim Biophys Acta. 1984;766(3):603–611. [Google Scholar]
- 22.Sandusky PO, Yocum CF. The chloride requirement for photosynthetic oxygen evolution: Factors affecting nucleophilic displacement of chloride from the oxygen-evolving complex. Biochim Biophys Acta. 1986;849(1):85–93. [Google Scholar]
- 23.Beck WF, Brudvig GW. Binding of amines to the O2-evolving center of photosystem II. Biochemistry. 1986;25(21):6479–6486. doi: 10.1021/bi00369a021. [DOI] [PubMed] [Google Scholar]
- 24.Beck WF, de Paula JC, Brudvig GW. Ammonia binds to the manganese site of the oxygen-evolving complex of photosystem II in the S2 state. J Am Chem Soc. 1986;108(14):4018–4022. [Google Scholar]
- 25.Britt RD, Zimmermann JL, Sauer K, Klein MP. Ammonia binds to the catalytic manganese of the oxygen-evolving complex of photosystem II. Evidence by electron spin-echo envelope modulation spectroscopy. J Am Chem Soc. 1989;111(10):3522–3532. [Google Scholar]
- 26.Boussac A, Rutherford AW, Styring S. Interaction of ammonia with the water splitting enzyme of photosystem II. Biochemistry. 1990;29(1):24–32. doi: 10.1021/bi00453a003. [DOI] [PubMed] [Google Scholar]
- 27.Chu H-A, Sackett H, Babcock GT. Identification of a Mn-O-Mn cluster vibrational mode of the oxygen-evolving complex in photosystem II by low-frequency FTIR spectroscopy. Biochemistry. 2000;39(47):14371–14376. doi: 10.1021/bi001751g. [DOI] [PubMed] [Google Scholar]
- 28.Hou L-H, Wu C-M, Huang H-H, Chu H-A. Effects of ammonia on the structure of the oxygen-evolving complex in photosystem II as revealed by light-induced FTIR difference spectroscopy. Biochemistry. 2011;50(43):9248–9254. doi: 10.1021/bi200943q. [DOI] [PubMed] [Google Scholar]
- 29.Boussac A, Sugiura M, Inoue Y, Rutherford AW. EPR study of the oxygen evolving complex in His-tagged photosystem II from the cyanobacterium Synechococcus elongatus. Biochemistry. 2000;39(45):13788–13799. doi: 10.1021/bi001159r. [DOI] [PubMed] [Google Scholar]
- 30.Stich TA, Whittaker JW, Britt RD. Multifrequency EPR studies of manganese catalases provide a complete description of proteinaceous nitrogen coordination. J Phys Chem B. 2010;114(45):14178–14188. doi: 10.1021/jp908064y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stich TA, Yeagle GJ, Service RJ, Debus RJ, Britt RD. Ligation of D1-His332 and D1-Asp170 to the manganese cluster of photosystem II from Synechocystis assessed by multifrequency pulse EPR spectroscopy. Biochemistry. 2011;50(34):7390–7404. doi: 10.1021/bi2010703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schosseler P, Wacker T, Schweiger A. Pulsed ELDOR detected NMR. Chem Phys Lett. 1994;224(3–4):319–324. [Google Scholar]
- 33.Sinnecker S, Neese F, Noodleman L, Lubitz W. Calculating the electron paramagnetic resonance parameters of exchange coupled transition metal complexes using broken symmetry density functional theory: Application to a MnIII/MnIV model compound. J Am Chem Soc. 2004;126(8):2613–2622. doi: 10.1021/ja0390202. [DOI] [PubMed] [Google Scholar]
- 34.Pantazis DA, et al. Structure of the oxygen-evolving complex of photosystem II: Information on the S2 state through quantum chemical calculation of its magnetic properties. Phys Chem Chem Phys. 2009;11(31):6788–6798. doi: 10.1039/b907038a. [DOI] [PubMed] [Google Scholar]
- 35.Hillier W, Messinger J, Wydrzynski T. Kinetic determination of the fast exchanging substrate water molecule in the S3 state of photosystem II. Biochemistry. 1998;37(48):16908–16914. doi: 10.1021/bi980756z. [DOI] [PubMed] [Google Scholar]
- 36.Hendry G, Wydrzynski T. 18O isotope exchange measurements reveal that calcium is involved in the binding of one substrate-water molecule to the oxygen-evolving complex in photosystem II. Biochemistry. 2003;42(20):6209–6217. doi: 10.1021/bi034279i. [DOI] [PubMed] [Google Scholar]
- 37.Kusunoki M. Mono-manganese mechanism of the photosystem II water splitting reaction by a unique Mn4Ca cluster. Biochim Biophys Acta. 2007;1767(6):484–492. doi: 10.1016/j.bbabio.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 38.Kusunoki M. S1-state Mn4Ca complex of Photosystem II exists in equilibrium between the two most-stable isomeric substates: XRD and EXAFS evidence. J Photochem Photobiol B. 2011;104(1–2):100–110. doi: 10.1016/j.jphotobiol.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi K, et al. The nature of chemical bonds of the CaMn4O5 cluster in oxygen evolving complex of photosystem II: Jahn-Teller distortion and its suppression by Ca doping in cubane structures. Int J Quantum Chem. 2012;113(4):453–473. [Google Scholar]
- 40.Tagore R, Chen H, Crabtree RH, Brudvig GW. Determination of μ-oxo exchange rates in di-μ-oxo dimanganese complexes by electrospray ionization mass spectrometry. J Am Chem Soc. 2006;128(29):9457–9465. doi: 10.1021/ja061348i. [DOI] [PubMed] [Google Scholar]
- 41.Messinger J. Evaluation of different mechanistic proposals for water oxidation in photosynthesis on the basis of Mn4OxCa structures for the catalytic site and spectroscopic data. Phys Chem Chem Phys. 2004;6(20):4764–4771. [Google Scholar]
- 42.Hillier W, et al. In: Photosynthesis. Energy from the Sun. 14th International Congress on Photosynthesis, vol. I. Allen JF, Gantt E, Golbeck J, Osmond B, editors. Dordrecht: Springer; 2008. pp. 427–430. [Google Scholar]
- 43.Service RJ, et al. Participation of glutamate-354 of the CP43 polypeptide in the ligation of manganese and the binding of substrate water in photosystem II. Biochemistry. 2011;50(1):63–81. doi: 10.1021/bi1015937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamanaka S, et al. Possible mechanisms for the O-O bond formation in oxygen evolution reaction at the CaMn4O5(H2O)4 cluster of PSII refined to 1.9 Å X-ray resolution. Chem Phys Lett. 2011;511(1–3):138–145. [Google Scholar]
- 45.Gao Y, Åkermark T, Liu J, Sun L, Åkermark B. Nucleophilic attack of hydroxide on a MnV oxo complex: A model of the O-O bond formation in the oxygen evolving complex of photosystem II. J Am Chem Soc. 2009;131(25):8726–8727. doi: 10.1021/ja901139r. [DOI] [PubMed] [Google Scholar]
- 46.Privalov T, et al. A computational study of O-O bond formation catalyzed by mono- and bis-MnIV-corrole complexes. Inorg Chem. 2007;46(17):7075–7086. doi: 10.1021/ic700940x. [DOI] [PubMed] [Google Scholar]
- 47.Romain S, Bozoglian F, Sala X, Llobet A. Oxygen-oxygen bond formation by the Ru-Hbpp water oxidation catalyst occurs solely via an intramolecular reaction pathway. J Am Chem Soc. 2009;131(8):2768–2769. doi: 10.1021/ja808166d. [DOI] [PubMed] [Google Scholar]
- 48.Noguchi T. FTIR detection of water reactions in the oxygen-evolving centre of photosystem II. Philos Trans R Soc Lond B Biol Sci. 2008;363(1494):1189–1194, discussion 1194–1195. doi: 10.1098/rstb.2007.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boussac A, et al. Biosynthetic Ca2+/Sr2+ exchange in the photosystem II oxygen-evolving enzyme of Thermosynechococcus elongatus. J Biol Chem. 2004;279(22):22809–22819. doi: 10.1074/jbc.M401677200. [DOI] [PubMed] [Google Scholar]
- 50.Sander J, et al. Functional characterization and quantification of the alternative PsbA copies in Thermosynechococcus elongatus and their role in photoprotection. J Biol Chem. 2010;285(39):29851–29856. doi: 10.1074/jbc.M110.127142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.