Significance
Many proteins are modified by the covalent attachment of a small polypeptide called ubiquitin to their lysine residues. Lysines in ubiquitin itself are further ubiquitinated, leading to formation of ubiquitin chains. Single ubiquitins and ubiquitin chains attached to integral membrane proteins, such as receptors, transporters, and channels, serve as molecular signals mediating endocytosis of these proteins and their subsequent targeting to lysosomes for degradation. This work uses quantitative mass spectrometry to show that activated EGF receptor is ubiquitinated by one to two short polyubiquitin chains linked via ubiquitin lysine 63 or conjugated with a single monoubiquitin. It is demonstrated that these Lys63-linked polyubiquitin chains are necessary for efficient targeting of EGF receptor to the lysosomal degradation pathway.
Abstract
Ubiquitination mediates endocytosis and endosomal sorting of various signaling receptors, transporters, and channels. However, the relative importance of mono- versus polyubiquitination and the role of specific types of polyubiquitin linkages in endocytic trafficking remain controversial. We used mass spectrometry-based targeted proteomics to show that activated epidermal growth factor receptor (EGFR) is ubiquitinated by one to two short (two to three ubiquitins) polyubiquitin chains mainly linked via lysine 63 (K63) or conjugated with a single monoubiquitin. Multimonoubiquitinated EGFR species were not found. To directly test whether K63 polyubiquitination is necessary for endocytosis and post-endocytic sorting of EGFR, a chimeric protein, in which the K63 linkage-specific deubiquitination enzyme AMSH [associated molecule with the Src homology 3 domain of signal transducing adaptor molecule (STAM)] was fused to the carboxyl terminus of EGFR, was generated. MS analysis of EGFR-AMSH ubiquitination demonstrated that the fraction of K63 linkages was substantially reduced, whereas relative amounts of monoubiquitin and K48 linkages increased, compared with that of wild-type EGFR. EGFR-AMSH was efficiently internalized into early endosomes, but, importantly, the rates of ligand-induced sorting to late endosomes and degradation of EGFR-AMSH were dramatically decreased. The slow degradation of EGFR-AMSH resulted in the sustained signaling activity of this chimeric receptor. Ubiquitination patterns, rate of endosomal sorting, and signaling kinetics of EGFR fused with the catalytically inactive mutant of AMSH were reversed to normal. Altogether, the data are consistent with the model whereby short K63-linked polyubiquitin chains but not multimonoubiquitin provide an increased avidity for EGFR interactions with ubiquitin adaptors, thus allowing rapid sorting of activated EGFR to the lysosomal degradation pathway.
Ubiquitination, a posttranslational modification of proteins by attachment of the ubiquitin (Ub) polypeptide, is an important molecular signal that regulates endocytosis and post-endocytic sorting of membrane proteins (1–3). Ubiquitination is carried out by the sequential activity of E1, E2, and E3 enzymes; the latter, E3 ligases, typically determine the substrate specificity of Ub conjugation (4). Deubiquitinating enzymes (DUBs), a group of proteases capable of cleaving Ub from conjugates with target proteins, counteract the activity of the ubiquitination system (5). Ub is predominantly conjugated to lysine residues and much more rarely to the amino-terminal methionine or other amino acids in the substrate. Lysines and the amino-terminal methionine in the Ub molecule can also be conjugated to another molecule of Ub, leading to the formation of polyUb chains (6). Depending on the specific residue that links Ubs into a chain, polyUb chains have different molecular folding, are recognized by specific Ub-binding domains (UBDs) and have distinct functions (7). The structure and interaction mechanisms of lysine 48 (K48)- and K63-linked chains are most well-characterized (8–12). Crystal and NMR structures of K63 di-Ubs revealed extended, open conformation of two Ubs with high conformational freedom, as opposed to closed conformation of K48-polyUb linkages (reviewed in ref. 11). Therefore, ubiquitination substrates including endocytic cargo can be mono- and polyubiquitinated by different chains, but the role of these diverse types of ubiquitination in the regulation of endocytic trafficking remains incompletely understood.
Epidermal growth factor (EGF) receptor (EGFR) was one of the first endocytic cargos in mammalian cells that were found to be ubiquitinated (13). This receptor has the profound role in eukaryotic development, regulation of various tissues in adult organisms, and pathogenesis of cancer (14). Therefore, EGFR has been a prototypic model for studying the mechanisms of endocytosis and endocytosis-relevant ubiquitination. EGFR is ubiquitinated by Cbl E3 ligases at the cell surface and after internalization in endosomes (15–17). The internalization step of EGFR trafficking is regulated by multiple redundant mechanisms, including ubiquitination, and is not significantly inhibited in the absence of receptor ubiquitination (18). By contrast, sorting of the internalized receptor in multivesicular bodies (MVBs), which leads to its incorporation into intraluminal vesicles of MVB and degradation in lysosomes, is highly sensitive to the extent of EGFR ubiquitination (15, 19).
Based on differential recognition by Ub antibodies, EGFR was proposed to be conjugated with multiple monoUbs (20). Moreover, replacement of the cytoplasmic domain of EGFR with the Ub mutant incapable of polyubiquitination resulted in EGF-independent endocytosis and degradation of such chimeric receptor, thus suggesting that monoubiquitination is sufficient for EGFR endocytosis and MVB sorting (20). Subsequently, mass spectrometric (MS) analysis demonstrated a significant amount of EGFR polyubiquitination, mainly by K63-linked chains (19, 21, 22). However, whether K63 polyubiquitination is necessary for EGFR endocytic trafficking remains unknown.
The role of cargo ubiquitination by K63-linked chains has been proposed in studies of endocytosis and MVB sorting of yeast permeases (23–27). These studies, however, used an approach of global elimination of K63 polyubiquitination in the cell to demonstrate the importance of these chains in endocytic trafficking. Because numerous proteins, including ESCRT components mediating MVB sorting are polyubiquitinated with K63 linkages, the inhibitory effects of the blockade of K63-linked polyubiquitination on endocytosis and MVB sorting observed in these studies may be indirect [e.g., not related to cargo ubiquitination (25)]. By contrast, an alternative approach based on the analysis of genetically engineered chimeric cargo molecules fused to Ub or a DUB demonstrated that monoubiquitination is fully sufficient for endocytosis and sorting of several membrane proteins to the vacuole in yeast (28).
A number of mammalian endocytic cargo is polyubiquitinated by K63-linked chains (29–31) and, to a lesser extent, with K48 linkages (32–35). Similarly to studies in yeast, the role of these Ub linkages in mammalian cells was mainly examined by overexpressing K63R or K48R Ub mutants incapable of forming corresponding polyUb chains, leading to inhibition of K63 or K48 polyubiquitination of all cellular substrates (31–33). To test whether K63 polyubiquitination is required for EGFR endocytosis and endosomal sorting, we analyzed the stoichiometry of EGFR ubiquitination by MS and generated a chimeric EGFR fused at the carboxyl terminus to a DUB with the specificity toward K63 linkages. Analysis of the internalization and post-endocytic sorting of this chimeric receptor showed that K63 polyUb chains are necessary for the efficient EGF-induced down-regulation of EGFR.
Results and Discussion
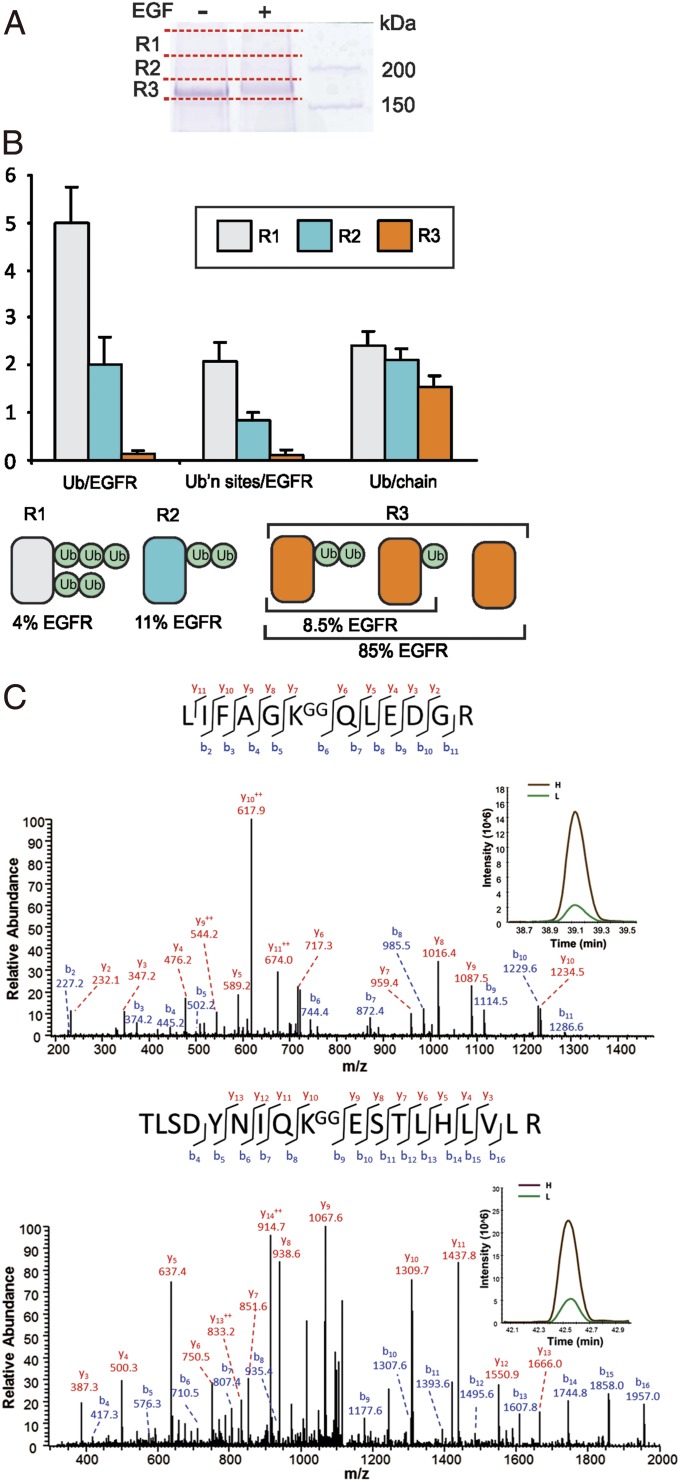
To determine the molar stoichiometry of mono- and polyUb conjugation of EGFR, receptor ubiquitination was analyzed using absolute quantification (AQUA)-based method where heavy isotope-labeled reference peptides standards are used to quantify Ub linkages and EGFR protein (36). Human EGFR constitutively expressed in porcine aortic endothelial (PAE) cells, lacking endogenous EGFR and other ErbB receptors, was immunopurified, resolved by electrophoresis, and visualized by Coomassie blue staining (Fig. 1A). Three gel regions (R1, R2, and R3) containing EGFR (∼175 kDa) and EGFR that is posttranslationally modified upon EGF binding (above 175 kDa) were excised, as shown in Fig. 1A, and digested with trypsin. The presence of EGFR and Ub peptides was confirmed using qualitative MS analysis by liquid chromatography–tandem MS (LC-MS/MS). The absolute amounts of EGFR, monoUb, and two major polyUb chain linkages, K48 and K63 (other Ub linkages represent <3% of total EGFR-associated Ub), in each gel region were quantified using AQUA as described previously (19, 36). Representative MS/MS spectra and selective reaction monitoring (SRM) assays are shown in Fig. 1C and SI Appendix, Fig. S1.
Fig. 1.
Stoichiometry of EGFR ubiquitination. (A) PAE/wtEGFR cells were treated with or without 20 ng/mL EGF for 5 min at 37 °C. EGFR was immunoprecipitated, immunoprecipitates were resolved by SDS/PAGE, and the gels were stained with Coomassie blue. A representative gel is shown, with red lines indicating regions R1, R2, and R3 containing EGFR that were excised and analyzed by AQUA-based targeted proteomics as described in Materials and Methods. (B) Quantification of the number of Ub per EGFR molecule (Ub/EGFR), number of Ub-conjugated sites in EGFR (Ub’n sites/EGFR), and number of Ub per chain conjugated to EGFR (Ub/chain) in gel regions R1, R2 and R3 using equations in SI Appendix. Graph bars represent mean values (±SEM; n = 5). The cartoon illustration below depicts ubiquitinated EGFR kinase domain and summarizes the stoichiometry of EGFR ubiquitination in each of these gel regions calculated on the basis of AQUA analysis. 4%, 11%, and 85% (mean values; n = 5) of total EGFRs are present in R1, R2, and R3, respectively. All receptors present is R1 and R2 are considered to be EGF-activated, whereas in R3, ∼15% of total EGFR are EGF-activated (EGF-occupied before cell solubilization). (C) Representative tandem mass spectra of Ub peptides demonstrating K48-chain and K63-chain ubiquitination. Peaks matching expected singly and doubly charged (++) b and y ions are labeled. Insets show representative mass chromatograms for the unlabeled (L) and isotope-labeled internal standard peptide (H) from the LC-SRM.
Analysis of EGFR from unstimulated cells revealed a low background level of Ub. Following stimulation of cells with 20 ng/mL EGF for 5 min at 37 °C, conditions of the maximum high EGFR ubiquitination, the total amount of Ub in EGFR immunoprecipitates increased 20- to 40-fold. Quantification of EGFR using AQUA showed that in cells treated with EGF, 4%, 11%, and 85% of EGFR protein were detected in R1, R2, and R3, respectively (Fig. 1B). Calculations of Ub stoichiometry using AQUA equations (SI Appendix) (36) revealed that highly ubiquitinated EGFRs in R1 were conjugated with two short (2-3Ubs) chains (Fig. 1B). R2 contained EGFRs that are mostly conjugated at a single site with the di-Ub. The number of ubiquitinated sites per EGFR in R3 was 0.1, indicating that only 10% of EGFRs in this gel region (8.5% of total EGFR) were conjugated to either monoUb or di-Ub. [125I]EGF-binding analysis revealed that 30% of total EGFR were occupied by EGF under conditions of these experiments. Because EGFRs in R1 and R2 (15% of total EGFR) were considered to be EGF-activated (EGF-occupied before cell lysis), the remaining ∼15% (of total EGFR) EGF-activated EGFRs were in R3. Therefore, more than half of these EGF-activated EGFR in R3 were ubiquitinated (8.5% Ub-EGFR compared with total 15% EGFR-activated), which, in turn, leads to the conclusion that more than 75% of total EGF-activated EGFRs were ubiquitinated (Fig. 1B). Altogether, the data of the quantitative MS analysis demonstrated that EGFR is mostly polyubiquitinated with short chains or monoubiquitinated. It should be noted that these experiments provide an estimation of an average Ub:EGFR stoichiometry. It is possible that longer chains can be conjugated to EGFR, although the population of such Ub-EGFR species is predicted to be small. Importantly, multimonoubiquitinated EGFR species were not detected.
Because the affinity of monomeric interactions of Ub with UBD is low, the general notion is that ubiquitin-dependent sorting processes are mediated by multivalent interactions of ubiquitinated substrates with adaptor proteins containing either multiple UBDs or an UBD with two ubiquitin-binding sites (37–39). The multivalency and high avidity of interactions have been proposed to be achieved through either multimonoubiquitination or polyubiquitination of the endocytic cargo. The Ub:EGFR stoichiometry data in Fig. 1 support the model whereby the predominant mode of cargo (EGFR) interaction with UBD-containing adaptors involves short polyUb chains rather than multiple monoUbs.
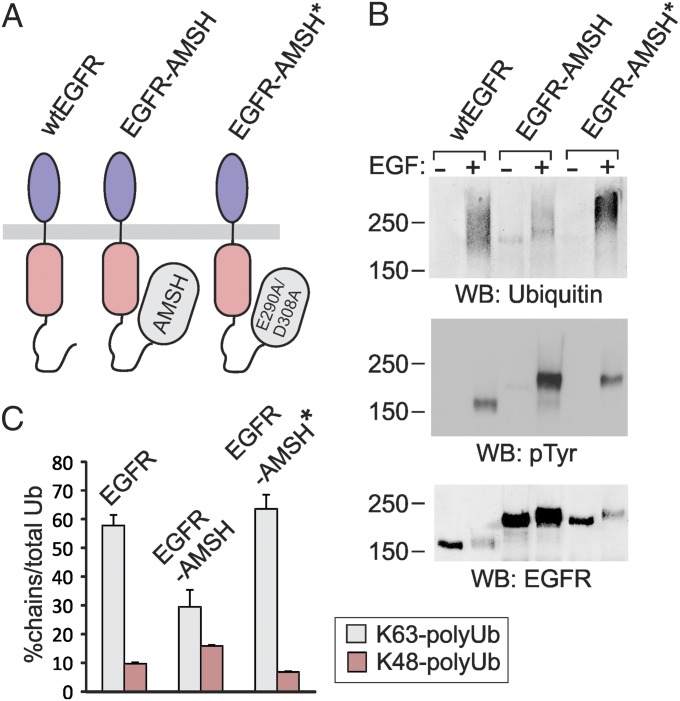
The bulk of polyUb conjugated to the kinase domain of the EGF-activated EGFR is represented by K63-linked chains (19). Consistent with the data in Fig. 1 demonstrating predominant conjugation of EGFR by di-Ub, K63-linked di-Ub was implicated in endocytosis and sorting of yeast permeases (23). To directly test the role of K63-linked polyubiquitination of EGFR in its endocytosis, we used an approach based on the hypothesis that attachment of a K63-polyubiquitin–specific DUB to the cytoplasmic domain of EGFR may result in the cleavage of these chains conjugated to the receptor. Therefore, we generated a fusion protein, in which AMSH, a DUB that displays strong preference toward K63-linked polyubiquitin chains (40), was attached to the carboxyl terminus of EGFR (EGFR-AMSH; Fig. 2A). To control for additional, deubiquitination-independent effects of AMSH attachment to EGFR, catalytically inactive AMSH mutant was substituted for the wild-type (wt) AMSH in the EGFR fusion protein. In previous studies such AMSH mutant (D348N) was used to examine cellular functions of AMSH (22). However, EGFR chimeric proteins containing the D348N AMSH mutant were found to be highly unstable as revealed by the presence of multiple truncated forms of this EGFR chimera. Therefore, an EGFR fusion with the double mutant of AMSH, E280A/D309A, which displayed normal protein stability, was generated (EGFR-AMSH*; Fig. 2A). E280 is important for normal coordination of Zn ions in the catalytic center of AMSH. However, D309 has the flexibility to turn in the absence of E280 and bind H2O, which results in normal coordination of Zn ions (41). Thus, combination of these two mutations was necessary for inactivating AMSH. EGFR-AMSH and EGFR-AMSH* were constitutively expressed in PAE cells. Several independent single-cell clones expressing different levels of wt and chimeric EGFRs were used in subsequent experiments to account for clonal variability.
Fig. 2.
AMSH attachment to EGFR results in impaired K63 ubiquitination of the receptor. (A) Schematic representation of EGFR-AMSH chimeric proteins. (B) PAE cells expressing wtEGFR, EGFR-AMSH (clone 10), or EGFR-AMSH* (clone 27) were treated or not with 20 ng/mL EGF for 5 min at 37 °C and lysed, and EGFRs were immunoprecipitated. Immunoprecipitates were resolved by SDS/PAGE and probed with Ub, phosphotyrosine, and EGFR antibodies. Note that Ab1005 poorly recognize activated EGFR. (C) PAE cells expressing wtEGFR, EGFR-AMSH (clone 10), or EGFR-AMSH* (clone 27) were treated with 20 ng/mL EGF for 5 min at 37 °C and lysed, and EGFRs were immunoprecipitated. Absolute amounts (femtomole quantities) of total Ub and K48 and K63 polyUb-chain linkages were measured by SRM-based MS. The data are presented as percentages of polyUb linkages to the total amount of Ub in the combined three gel regions R1, R2, and R3 of EGFR staining that was excised similarly to Fig.1A.
To analyze ubiquitination of EGFR-AMSH fusions, Western blotting and parallel quantitative MS analysis of EGFR immunoprecipitates were performed as in Fig. 1. Because a reduced level of EGFR-AMSH ubiquitination was expected, the PAE cell clone expressing a high amount of EGFR-AMSH mutant receptors was used in these experiments. Western blot analysis of wtEGFR and chimeric EGFR immunoprecipitates demonstrated abolished ubiquitination of EGFR-AMSH (if normalized to the total EGFR), whereas ubiquitination of EGFR-AMSH* was moderately but consistently increased compared with wtEGFR (Fig. 2B). Interestingly, distribution of the Ub immunoreactivity in the EGFR-AMSH lane was different from that in wtEGFR and EGFR-AMSH* lanes. The Ub signal overlapping with the major EGFR-AMSH band (∼230–240 kDa) (presumably, mostly monoubiquitinated EGFR-AMSH) was increased, whereas the high-molecular-mass smear (presumably highly ubiquitinated EGFR-AMSH) was decreased (Fig. 2B). Consistent with this observation, label-free MS-based relative quantification of ubiquitinated EGFR peptides (listed in SI Appendix, Fig. S2A) indicated that the amount of Ub adjusted for the amount of ubiquitinated EGFR is lowest in EGFR-AMSH, followed by wtEGFR and EGFR-AMSH* (SI Appendix, Fig. S2B), suggesting that the number of Ubs per ubiquitinated EGFR is lowest in EGFR-AMSH. The extent of tyrosine phosphorylation of wtEGFR, EGFR-AMSH, and EGFR-AMSH* was comparable, indicative of a normal kinase activity of the chimeric receptors (Fig. 2B; also see Fig. 5 below).
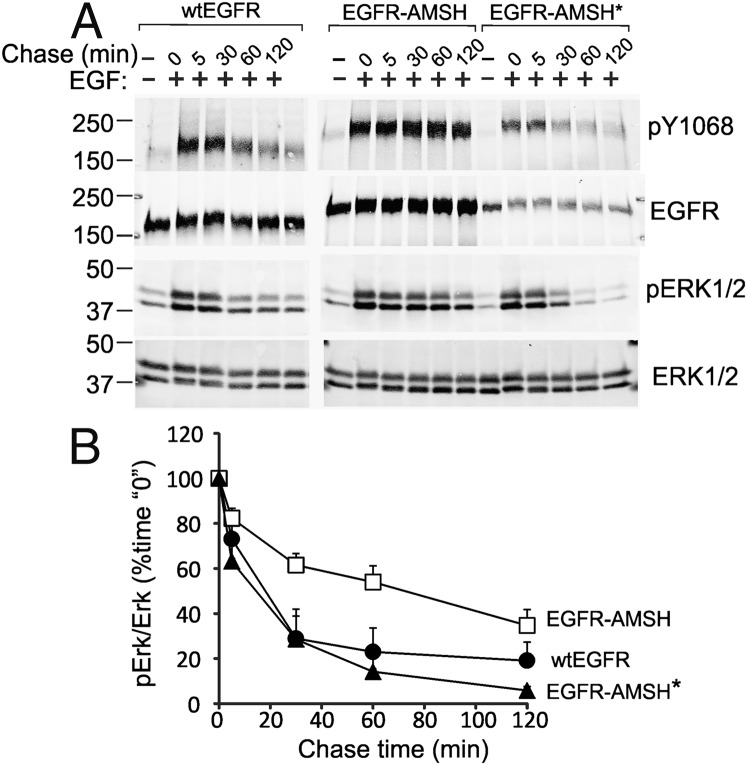
Fig. 5.
Signaling by EGFR-AMSH chimeric proteins. (A) Cells expressing wtEGFR, EGFR-AMSH, or EGFR-AMSH* were serum-starved, treated with 10 ng/mL EGF for 5 min at 37 °C, washed, and further incubated for the indicated times (Chase Time) without EGF. The cells were lysed in the presence of OV and NEM. The cell lysates were probed for active EGFR (antibody pY1068), total EGFR, phosphorylated ERK1/2, and total ERK1/2. The experiment is representative of three independent experiments. (B) Mean amounts of active ERK1/2 normalized to total ERK1/2 from three experiments (± SEM) plotted against chase time are presented on the graphs.
SRM-based quantitative MS revealed that about 55–60% of Ub in wtEGFR immunoprecipitates was present in the form of K63-linked chains (Fig. 2C). The percentage of K63-linked polyubiquitination of EGFR-AMSH was decreased two times but not changed in the control chimera containing inactive AMSH. In contrast, the relative amount of Lys48 polyUb chains was higher in EGFR-AMSH than in wtEGFR and EGFR-AMSH* (Fig. 2C). Correspondingly, the proportion of monoUb was the highest in EGFR-AMSH. These data suggest that AMSH attached to the EGFR carboxyl terminus efficiently and specifically removes K63 polyUb linkages conjugated to the kinase domain of EGFR.
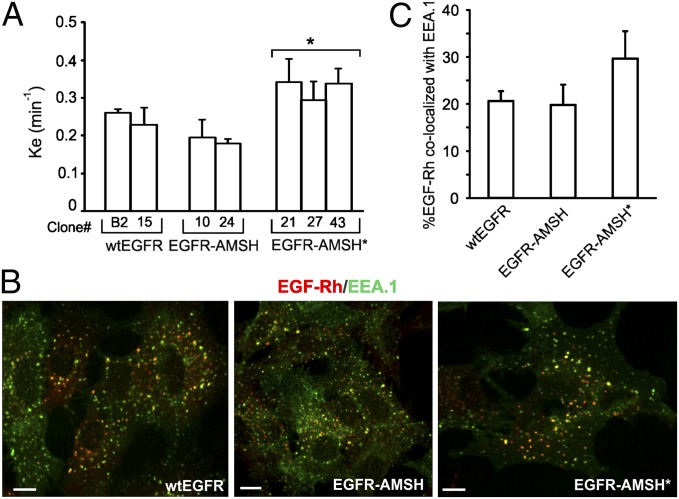
EGFR ubiquitination is implicated in the regulation of two steps of the EGFR traffic: (i) internalization from the cell surface to endosomes; and (ii) sorting from endosomes to lysosomes. Therefore, first, internalization rates of wtEGFR and chimeric proteins were compared. When low concentration of [125I]EGF was used, conditions that favor EGFR internalization via clathrin-mediated endocytic pathway, the rates of wtEGFR and EGFR-AMSH were not statistically different (Fig. 3A). Internalization of EGFR-AMSH* was slightly increased compared with that of wtEGFR and EGFR-AMSH (Fig. 3A), likely because of low expression levels of EGFR-AMSH* in all clones generated. These data are consistent with the previous studies demonstrating that ubiquitination is not essential for clathrin-mediated internalization of EGFR and represents one of the redundant internalization mechanisms (18). When saturating concentration of EGF is used, both clathrin-dependent and -independent pathways contribute to EGFR internalization. Incubation of cells with 100 ng/mL EGF conjugated to rhodamine (EGF-Rh) for 10 min resulted in accumulation of wt and chimeric EGFRs in early endosomes containing EEA.1 (Fig. 3B). The extent of endosomal accumulation of wtEGFR and EGFR-AMSH in early endosomes was similar, whereas internalization of EGFR-AMSH* was slightly increased (Fig. 3C). These data suggest that reduced K63 polyubiquitination of EGFR does not significantly affect the internalization step of receptor trafficking.
Fig. 3.
Internalization of EGFR-AMSH chimeric proteins. (A) Internalization rate constants (ke) of [125I]EGF (1 ng/mL) were measured in several single cell clones of cells stably expressing wtEGFR, EGFR-AMSH, or EGFR-AMSH*. *P < 0.05 (EGFR-AMSH* relative to EGFR-AMSH). (B) Cells stably expressing similar levels of wtEGFR, EGFR-AMSH, or EGFR-AMSH* were incubated with EGF-Rh (100 ng/mL) for 10 min at 37 °C. After fixation, the cells were stained with antibody to EEA.1 followed by secondary Cy5-conjugated antibody. A z stack of confocal images were acquired although 561 nm (EGF-Rh, red) and 640 nm (EEA.1, green) channels. Confocal sections through the middle of the cell are shown. “Yellow” signifies the overlap of rhodamine and Cy5 fluorescence. (Scale bars: 10 μm.) (C) The relative amount of EGF-Rh in EEA.1-containing endosomes was calculated from two independent experiments performed as in A). Data represent values averaged from 10 cells (± SEM).
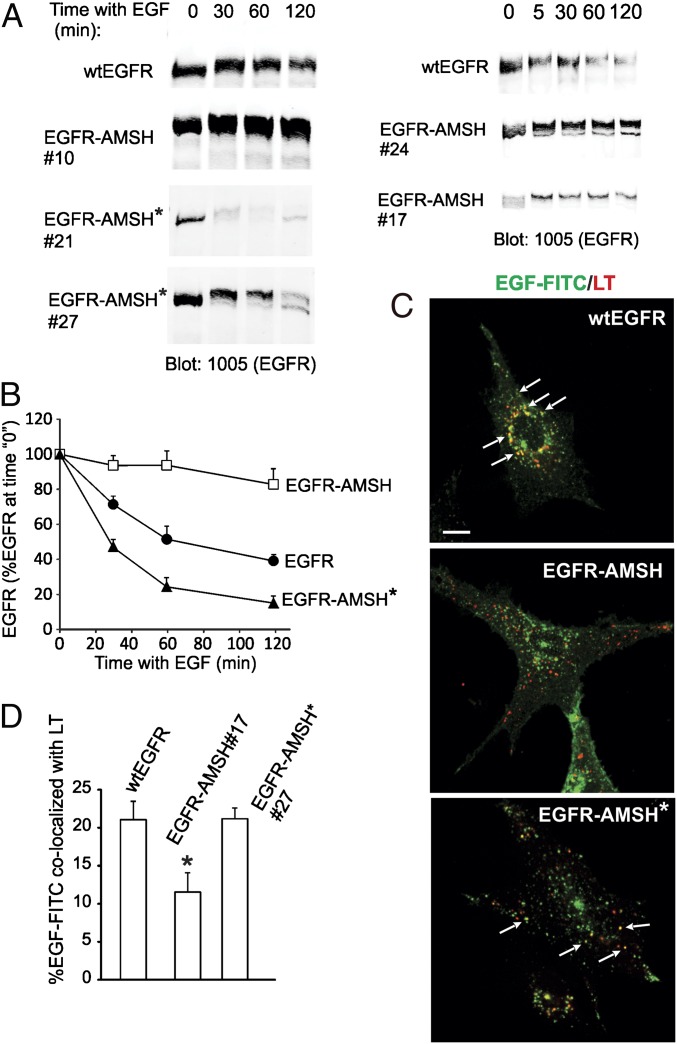
To examine the role of K63 polyubiquitination of EGFR in ligand-induced receptor degradation, several single-cell clones expressing wtEGFR or chimeric EGFRs were incubated with EGF at a saturating concentration, and the amounts of the EGFR protein were measured by Western blotting. As shown in Fig. 4, activated wtEGFR was degraded with the half-life time of about 1 h. By contrast, EGF-activated EGFR-AMSH was very slowly degraded regardless of its expression level in several single cell clones. Interestingly, EGFR-AMSH* was degraded even more rapidly than wtEGFR, presumably because of increased ubiquitination and relatively low expression levels of this chimera. Altogether, these data indicated that K63 polyubiquitination is essential for the maximally rapid EGF-induced degradation of EGFR.
Fig. 4.
Slow degradation and lysosomal targeting of EGFR-AMSH. (A) Several individual single-cell clones of cells expressing wtEGFR, EGFR-AMSH, and EGFR-AMSH* were serum-starved and incubated with EGF (100 ng/mL) for the indicated times before lysis in the absence of OV and NEM. EGFR was detected with antibodies 1005. (B) The amount of EGFR immunoreactivity was quantitated from five to six experiments for each variant, and the mean values (± SEM) for each type of EGFR/mutant-expressing cells were plotted against time. (C) Cells were preincubated with leupeptin and then incubated with EGF-FITC (100 ng/mL) for 2 h at 37 °C, including with LysoTrackerRed for the last 30 min of this incubation with EGF-FITC. After fixation, a z stack of images were acquired although 561-nm (LysoTrackerRed) and 488-nm (EGF-FITC) channels. Confocal sections through the middle of the cell are shown. “Yellow” signifies the overlap of red and green fluorescence. Arrows indicate examples of colocalization of EGF-FITC and LysoTracker. (Scale bars: 10 μm.) (D) The percentage of EGF-FITC located in vesicles containing LysoTrackerRed relative to the total cell-associated EGF-FITC was calculated from two experiments performed as in C. Each data represents value averaged from 10 cells (± SEM). *P < 0.05 (EGFR-AMSH relative to wtEGFR and EGFR-AMSH*).
After 2 h of cell incubation with labeled EGF (EGF-FITC), wtEGFR could be readily detected in compartments with increased acidity (late endosomes and lysosomes) labeled with LysoTrackerRed (Fig. 4). By contrast, a very low level of EGF-FITC colocalization with LysoTracker was observed in clone 17 of cells expressing EGFR-AMSH at a similar level as wtEGFR clone 15. Significant amount of this chimera was detected in EEA.1 containing early endosomes after 2 h of continuous endocytosis (SI Appendix, Fig. S3). The EGFR-AMSH* protein was efficiently targeted to acidic compartments (Fig. 4). These data indicate that sorting of EGFR to late endosomes/lysosomes correlates with the extent of K63-linked polyubiquitination. Sorting of EGFR in MVB involves interaction of ubiquitinated receptors with hepatocyte growth factor-regulated substrate (Hrs) and, possibly, signal-transducing adaptor molecule (STAM), which have been proposed to be regulated themselves by ubiquitination (42, 43) (reviewed in ref. 3). However, expression of EGFR-AMSH did not decrease Hrs-associated ubiquitination or affected cellular concentrations of Hrs, STAM1, and STAM2 compared with that in wtEGFR- and EGFR-AMSH*–expressing cells (SI Appendix, Fig. S4), suggesting that slow lysosomal targeting of EGFR-AMSH is not attributable to decreased ubiquitination of these adaptor proteins.
Because EGFR-AMSH chimera was very slowly down-regulated, we hypothesized that the signaling activity of this receptor is prolonged. To compare the kinetics of EGFR activation, cells were stimulated by supraphysiological concentration of EGF (10 ng/mL) for 5 min, washed, and then incubated without EGF to monitor the decay of the receptor activity. Under these conditions, only a pool of EGFR was occupied by EGF, and, therefore, no significant down-regulation of the total EGFR protein was observed. The amount of tyrosine phosphorylated wtEGFR and EGFR-AMSH* was rapidly decreased during the chase incubation, whereas EGFR-AMSH remained to be highly phosphorylated for at least 2 h (Fig. 5). Similarly, EGF-induced activity of extracellular growth factor regulated kinase (ERK) 1/2 was prolonged in cells expressing EGFR-AMSH compared with cells expressing wtEGFR or EGFR-AMSH* (Fig. 5). Some decay in ERK1/2 phosphorylation in EGFR-AMSH–expressing cells could be attributable to EGF-induced up-regulation of ERK1/2 phosphatases (44).
In summary, we propose that K63-linked polyUb chains are necessary for rapid lysosomal degradation of EGF-activated EGFR based on (i) observation of the predominant ubiquitination of the receptor by short K63 linkages and absence of multimonoubiquitinated receptors; and (ii) slow degradation of the EGFR-AMSH chimeric protein with impaired K63-linked polyubiquitination. This model appears to disagree with studies using linear cargo-Ub fusion proteins in yeast and similar EGFR-Ub fusions in mammalian cells, which proposed that monoUb is the fully sufficient sorting signal (20, 28, 45). It should be noted, however, that studies of cargo-Ub chimeras in yeast typically measured the end point of the sorting process, and it is possible that the kinetics of endocytosis and/or vacuole targeting of these chimeric proteins is slower than that of natively polyubiquitinated transporters and receptors. Likewise, the rates of endocytosis and degradation of EGFR-Ub chimera were two times lower than those rates of wtEGFR (20). Another important consideration is that all molecules of engineered cargo-Ub fusions bear Ub at all steps of endocytosis and intracellular sorting, thus maintaining constantly high concentration of cargo capable of weak interactions with Ub adaptors. By contrast, the extent of ubiquitination of endogenous cargo depends on the opposing activities of ubiquitination and deubiquitination systems. Therefore, only a limited pool of total cellular cargo protein is ubiquitinated and capable of interaction with UBD adaptors at a given stage of endocytosis. We postulate that in higher eukaryotic cells with evolving of the large family of ubiquitinated cargo, such as receptor tyrosine kinases, K63 chains provide an advantage for the efficient sorting of these signaling receptors over monoUb in competing for binding to Ub adaptors in endosomes, thus allowing physiological down-regulation of these receptors upon their activation.
Materials and Methods
Reagents.
EGF-Rh, EGF-FITC, and LysoTrackerRed were purchased from (Invitrogen). Monoclonal antibody to phosphotyrosine (PY20) conjugated to horseradish peroxidase and monoclonal antibody to EEA.1 were from BD Transduction Laboratories; monoclonal antibody to EGFR (Ab528) was from American Type Culture Collection; mouse monoclonal antibody to Ub (P4D1) and polyclonal antibody to EGFR (Ab1005) were from Santa Cruz Biotechnology; and monoclonal antibody to EGFR phosphotyrosine 1068, polyclonal antibodies to AMSH and ERK1/2, and monoclonal antibody to phosphorylated ERK1/2 were from Cell Signaling Technology. Rabbit polyclonal antibody to actin and GFP were from Sigma and Abcam, respectively. Mouse monoclonal antibody to Hrs and rabbit polyclonal antibodies to STAM1 and STAM2 were from Santa Cruz Biotechnology. Heavy isotope-labeled synthetic peptides for Ub and EGFR were purchased from Cell Signaling Technology and ThermoFisher Scientific, respectively.
Plasmid Constructs and Point Mutations.
The full-length human AMSH cDNA was kindly provided by Daniel Devor (University of Pittsburgh). The EGFR-AMSH chimeric construct was generated by replacing the GFP sequence with the AMSH sequence in the EGFR-GFP construct described previously (46) between SacII and NotI restriction sites. Point mutations in the constructs were generated using the QuikChange mutagenesis kit (Stratagene Cloning Systems) according to the manufacture protocol.
Cell Culture and Transfections.
PAE cells were grown in F12 medium containing 10% (vol/vol) FBS, antibiotics, and glutamine. PAE cell lines stably expressing wt EGFRs were described previously (18, 19). Other cell lines were obtained by transfection of cells with EGFR-AMSH or EGFR-AMSH* plasmids, followed by single cell clone selection with 0.4 mg/mL G418 (Invitrogen).
Mass Spectrometric Analysis of EGFR Ubiquitination.
EGFR immunoprecipitation and MS analysis was performed as described previously (19). Briefly, PAE cells stably expressing wt or mutant EGFR grown in 150-mm dishes were untreated or treated with 20 ng/mL EGF for 5 min at 37 °C, washed and lysed in Triton X-100/glycerol/Hepes solubilization buffer containing 1% sodium deoxycholate, 1 mM ortho-vanadate (OV), and 10 mM N-ethyl-maleimide (NEM). EGFR was immunoprecipitated with antibody Ab528, and the precipitates were washed with 0.5 M NaCl to minimize coprecipitation of other proteins. The precipitates were resolved on 7.5% SDS/PAGE. The gels were stained by Coomassie blue.
Gel regions were excised as indicated and processed for tryptic digestion as described previously (36). Heavy isotope-labeled synthetic peptides were spiked in after digestion, and the resultant peptide mixtures were analyzed using MS. Qualitative LC-MS/MS analysis was carried out using a nano-LC (nanoACQUITY UltraPerformance LC; Waters) coupled with an LTQ Orbitrap Velos mass spectrometer (ThermoFisher Scientific). A more detailed description of the qualitative LC-MS/MS analysis can be found in SI Appendix. AQUA-based quantitative MS analysis presented in this work was a combined effort of the S.G. laboratory at the Harvard Medical School and the Biomedical Mass Spectrometry Center at the University of Pittsburgh (PITT). The LC-SRM analysis by the S.G. laboratory was carried out as described (36). The SRM analysis by PITT was performed on a TSQ quantum ultra (ThermoFisher) coupled with a nanoflow Dionex Ultimate 3000 LC system as described (47). Skyline software (48) was used to facilitate targeted SRM assay method development and data analyses. The transitions of the targeted peptides were preliminarily selected based on human tandem spectrum library downloaded from PeptideAtlas (www.peptideatlas.org) and subsequently optimized on the TSQ. The list of targeted Ub and EGFR peptides and transitions, together with peptide concentrations, is presented in SI Appendix, Table S1.
Immunoprecipitation and Western Blotting.
Western blot analysis of EGFR immunoprecipitates and lysates to examine ubiquitination, tyrosine phosphorylation of EGFR, EGFR degradation, and ERK1/2 phosphorylation was performed as described (19). See SI Appendix for details.
[125I]EGF Internalization.
Mouse receptor-grade EGF (Collaborative Research) was iodinated, [25I]EGF internalization rates were measured, and the specific rate constant for internalization ke was calculated as described previously (18).
Fluorescence Microscopy.
Analysis of EGFR localization in the cells using confocal fluorescence microscopy was performed as described (19) with the modifications listed in SI Appendix.
Supplementary Material
Acknowledgments
We thank Drs. Devor and Stenmark for the gift of reagents and Zhiyuan Sun for assistance with the collection and analysis of the mass spectrometric data. This work was supported by National Cancer Institute (NCI) Grant CA089151 (to F.H., A.S., and W.K.) and the Head-and Neck Cancer Specialized Program of Research Excellence (F.H. and A.S.). This project used the Biomedical Mass Spectrometry Center and University of Pittsburgh Cancer Institute Cancer Biomarker Facility, which are supported, in part, by NCI Grant P30CA047904.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308014110/-/DCSupplemental.
References
- 1.Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: The network at work. Exp Cell Res. 2009;315(9):1610–1618. doi: 10.1016/j.yexcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Hurley JH, Stenmark H. Molecular mechanisms of ubiquitin-dependent membrane traffic. Annu Rev Biophys. 2011;40:119–142. doi: 10.1146/annurev-biophys-042910-155404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clague MJ, Liu H, Urbé S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev Cell. 2012;23(3):457–467. doi: 10.1016/j.devcel.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 5.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walczak H, Iwai K, Dikic I. Generation and physiological roles of linear ubiquitin chains. BMC Biol. 2012;10:23. doi: 10.1186/1741-7007-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahighi S, Dikic I. Selectivity of the ubiquitin-binding modules. FEBS Lett. 2012;586(17):2705–2710. doi: 10.1016/j.febslet.2012.04.053. [DOI] [PubMed] [Google Scholar]
- 8.Varadan R, et al. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279(8):7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 9.Sekiyama N, et al. NMR analysis of Lys63-linked polyubiquitin recognition by the tandem ubiquitin-interacting motifs of Rap80. J Biomol NMR. 2012;52(4):339–350. doi: 10.1007/s10858-012-9614-9. [DOI] [PubMed] [Google Scholar]
- 10.Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol. 2009;16(12):1328–1330. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- 11.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 12.Ye Y, et al. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature. 2012;492(7428):266–270. doi: 10.1038/nature11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galcheva-Gargova Z, Theroux SJ, Davis RJ. The epidermal growth factor receptor is covalently linked to ubiquitin. Oncogene. 1995;11(12):2649–2655. [PubMed] [Google Scholar]
- 14.Sibilia M, et al. The epidermal growth factor receptor: From development to tumorigenesis. Differentiation. 2007;75(9):770–787. doi: 10.1111/j.1432-0436.2007.00238.x. [DOI] [PubMed] [Google Scholar]
- 15.Levkowitz G, et al. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12(23):3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stang E, Johannessen LE, Knardal SL, Madshus IH. Polyubiquitination of the epidermal growth factor receptor occurs at the plasma membrane upon ligand-induced activation. J Biol Chem. 2000;275(18):13940–13947. doi: 10.1074/jbc.275.18.13940. [DOI] [PubMed] [Google Scholar]
- 17.Umebayashi K, Stenmark H, Yoshimori T. Ubc4/5 and c-Cbl continue to ubiquitinate EGF receptor after internalization to facilitate polyubiquitination and degradation. Mol Biol Cell. 2008;19(8):3454–3462. doi: 10.1091/mbc.E07-10-0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goh LK, Huang F, Kim W, Gygi S, Sorkin A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J Cell Biol. 2010;189(5):871–883. doi: 10.1083/jcb.201001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21(6):737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 20.Haglund K, et al. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5(5):461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 21.Argenzio E, et al. Proteomic snapshot of the EGF-induced ubiquitin network. Mol Syst Biol. 2011;7:462. doi: 10.1038/msb.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meijer IM, van Rotterdam W, van Zoelen EJ, van Leeuwen JE. Recycling of EGFR and ErbB2 is associated with impaired Hrs tyrosine phosphorylation and decreased deubiquitination by AMSH. Cell Signal. 2012;24(11):1981–1988. doi: 10.1016/j.cellsig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Galan JM, Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16(19):5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paiva S, et al. Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter: Role of lysine 63-linked ubiquitin chains. J Biol Chem. 2009;284(29):19228–19236. doi: 10.1074/jbc.M109.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erpapazoglou Z, et al. A dual role for K63-linked ubiquitin chains in multivesicular body biogenesis and cargo sorting. Mol Biol Cell. 2012;23(11):2170–2183. doi: 10.1091/mbc.E11-10-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauwers E, Jacob C, André B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol. 2009;185(3):493–502. doi: 10.1083/jcb.200810114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, André B. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 2010;20(4):196–204. doi: 10.1016/j.tcb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Stringer DK, Piper RC. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J Cell Biol. 2011;192(2):229–242. doi: 10.1083/jcb.201008121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geetha T, Jiang J, Wooten MW. Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol Cell. 2005;20(2):301–312. doi: 10.1016/j.molcel.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Vina-Vilaseca A, Sorkin A. Lysine 63-linked polyubiquitination of the dopamine transporter requires WW3 and WW4 domains of Nedd4-2 and UBE2D ubiquitin-conjugating enzymes. J Biol Chem. 2010;285(10):7645–7656. doi: 10.1074/jbc.M109.058990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duncan LM, et al. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 2006;25(8):1635–1645. doi: 10.1038/sj.emboj.7601056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Xu M, Scotti E, Chen ZJ, Tontonoz P. Both K63 and K48 ubiquitin linkages signal lysosomal degradation of the LDL receptor. J Lipid Res. 2013;54(5):1410–1420. doi: 10.1194/jlr.M035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar KG, et al. Site-specific ubiquitination exposes a linear motif to promote interferon-alpha receptor endocytosis. J Cell Biol. 2007;179(5):935–950. doi: 10.1083/jcb.200706034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao Y, et al. Polyubiquitination of insulin-like growth factor I receptor (IGF-IR) activation loop promotes antibody-induced receptor internalization and down-regulation. J Biol Chem. 2011;286(48):41852–41861. doi: 10.1074/jbc.M111.288514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marx C, Held JM, Gibson BW, Benz CC. ErbB2 trafficking and degradation associated with K48 and K63 polyubiquitination. Cancer Res. 2010;70(9):3709–3717. doi: 10.1158/0008-5472.CAN-09-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8(7):700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 37.Barriere H, Nemes C, Du K, Lukacs GL. Plasticity of polyubiquitin recognition as lysosomal targeting signals by the endosomal sorting machinery. Mol Biol Cell. 2007;18(10):3952–3965. doi: 10.1091/mbc.E07-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hawryluk MJ, et al. Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic. 2006;7(3):262–281. doi: 10.1111/j.1600-0854.2006.00383.x. [DOI] [PubMed] [Google Scholar]
- 39.Ren X, Hurley JH. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 2010;29(6):1045–1054. doi: 10.1038/emboj.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato Y, et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455(7211):358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 41.Davies CW, Paul LN, Kim MI, Das C. Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: Nearly identical fold but different stability. J Mol Biol. 2011;413(2):416–429. doi: 10.1016/j.jmb.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polo S, et al. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416(6879):451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 43.Urbé S, et al. The UIM domain of Hrs couples receptor sorting to vesicle formation. J Cell Sci. 2003;116(Pt 20):4169–4179. doi: 10.1242/jcs.00723. [DOI] [PubMed] [Google Scholar]
- 44.Caunt CJ, Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs): Shaping the outcome of MAP kinase signalling. FEBS J. 2013;280(2):489–504. doi: 10.1111/j.1742-4658.2012.08716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mosesson Y, et al. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J Biol Chem. 2003;278(24):21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- 46.Carter RE, Sorkin A. Endocytosis of functional epidermal growth factor receptor-green fluorescent protein chimera. J Biol Chem. 1998;273(52):35000–35007. doi: 10.1074/jbc.273.52.35000. [DOI] [PubMed] [Google Scholar]
- 47.Zeng X, et al. Lung cancer serum biomarker discovery using label-free liquid chromatography-tandem mass spectrometry. J Thorac Oncol. 2011;6(4):725–734. doi: 10.1097/JTO.0b013e31820c312e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacLean B, et al. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.