Significance
Genome engineering in human pluripotent stem cells holds great promise for biomedical research and regenerative medicine, but it is very challenging. Recently, an RNA-guided nuclease system called clustered, regularly interspaced, short palindromic repeats (CRISPR)/CRISPR-associated (Cas) has been applied to genome engineering, greatly increasing the efficiency of genome editing. Here, using a CRISPR-Cas system identified in Neisseria meningitidis, which is distinct from the commonly used Streptococcus pyogenes system, we demonstrate efficient genome engineering in human pluripotent stem cells. Our study could have a tremendous impact in regenerative medicine.
Keywords: crRNA, tracrRNA, embryonic stem cells, induced pluripotent stem cells
Abstract
Genome engineering in human pluripotent stem cells (hPSCs) holds great promise for biomedical research and regenerative medicine. Recently, an RNA-guided, DNA-cleaving interference pathway from bacteria [the type II clustered, regularly interspaced, short palindromic repeats (CRISPR)-CRISPR-associated (Cas) pathway] has been adapted for use in eukaryotic cells, greatly facilitating genome editing. Only two CRISPR-Cas systems (from Streptococcus pyogenes and Streptococcus thermophilus), each with their own distinct targeting requirements and limitations, have been developed for genome editing thus far. Furthermore, limited information exists about homology-directed repair (HDR)-mediated gene targeting using long donor DNA templates in hPSCs with these systems. Here, using a distinct CRISPR-Cas system from Neisseria meningitidis, we demonstrate efficient targeting of an endogenous gene in three hPSC lines using HDR. The Cas9 RNA-guided endonuclease from N. meningitidis (NmCas9) recognizes a 5′-NNNNGATT-3′ protospacer adjacent motif (PAM) different from those recognized by Cas9 proteins from S. pyogenes and S. thermophilus (SpCas9 and StCas9, respectively). Similar to SpCas9, NmCas9 is able to use a single-guide RNA (sgRNA) to direct its activity. Because of its distinct protospacer adjacent motif, the N. meningitidis CRISPR-Cas machinery increases the sequence contexts amenable to RNA-directed genome editing.
Human pluripotent stem cells (hPSCs) can proliferate indefinitely while maintaining the potential to give rise to virtually all human cell types (1). These cells are therefore invaluable for regenerative medicine, drug screening, and biomedical research. However, to realize the full potential of hPSCs, it will be necessary to manipulate their genomes in a precise, efficient manner. Historically, gene targeting in hPSCs has been extremely difficult (2). The development of zinc-finger nucleases (ZFNs) and transcription activator-like endonucleases (TALENs) (reviewed in refs. 3 and 4) has facilitated gene targeting in hPSCs (5–7). Nonetheless, both approaches require the design, expression, and validation of a new pair of proteins for every targeted locus, rendering both of these platforms time-consuming and labor-intensive (8–10).
Clustered, regularly interspaced, short palindromic repeat (CRISPR) loci, along with CRISPR-associated (cas) genes, underlie an adaptive immune system of bacteria and archaea that defends against bacteriophages (11) and limits horizontal gene transfer (12–14). “Protospacer” sequences from invading nucleic acids are incorporated as “spacers” within CRISPRs, conferring immunity and providing a genomic memory of past invasions. CRISPR-Cas systems have been classified into three types (types I, II, and III) and numerous subtypes (15). All of these types and subtypes use short CRISPR RNAs (crRNAs) (16, 17) to specify genetic interference via the destruction of invading nucleic acids (18). The target nucleic acids are recognized by crRNA Watson–Crick pairing. Importantly, most CRISPR-Cas subtypes target DNA directly (13, 19, 20), suggesting the possibility of engineered, RNA-directed gene-targeting/editing systems. The use of RNA guides for gene targeting would confer many advantages over ZFNs and TALENs, especially by obviating the need for repeated protein design/optimization. Recently, this vision has become a reality (21–31).
Type II CRISPR-Cas systems are noteworthy in that the essential targeting activities—crRNA binding, target DNA binding, R-loop formation, and double-stranded DNA cleavage—are all executed by a single polypeptide, Cas9 (32–35). In addition to crRNA and Cas9, an additional RNA, transacting CRISPR RNA (tracrRNA), is essential for interference in bacteria (14, 32, 36) and in vitro (34, 36). The tracrRNA is partially complementary to pre-crRNA repeats, leading to the formation of duplexes that are cleaved by the host factor ribonuclease III (RNase III) (32). The type II crRNA maturation pathway was originally characterized in strains of Streptococcus pyogenes (32) and Streptococcus thermophilus (35, 36), and RNase III-catalyzed pre-crRNA processing is essential for interference in both native systems. Recent studies of a type II CRISPR-Cas locus from Neisseria meningitidis revealed an intrinsically RNase III- and processing-independent system, which nonetheless requires tracrRNA (14). Importantly, crRNA-directed DNA cleavage was reconstituted in vitro with recombinant S. pyogenes Cas9 (SpCas9) (34) or S. thermophilus Cas9 (StCas9) (33, 36). The SpCas9 in vitro system enabled the development of fused crRNA-tracrRNA chimeras called single-guide RNAs (sgRNAs) that bypass processing (34). Subsequent development of eukaryotic genome editing applications has focused on sgRNAs (21–30), although separately encoded pre-crRNAs and tracrRNAs are also effective (21).
Target cleavage by many CRISPR-Cas systems, including those from type II, require proximity to a 2- to 5-nt sequence called a protospacer adjacent motif (PAM) (37–40). Genome-editing applications reported thus far have focused almost exclusively on SpCas9, which has a 5′-NGG-3′ PAM. StCas9 (from the CRISPR1 locus of strain LMD-9) has also been used in eukaryotes (21), and that system has a 5′-NNAGAAW-3′ PAM (W = A or T). Eukaryotic editing capabilities will benefit from the increased frequency of target sites stemming from the development of additional Cas9s with distinct PAMs.
Targeting by sgRNAs usually relies on either of two approaches. First, double-strand break (DSB) repair by nonhomologous end joining (NHEJ) can be used to generate insertions or deletions (indels) that induce frame-shifts. Second, the addition of a homologous repair template can allow Cas9-induced DSBs or nicks to be repaired by homology-directed repair (HDR). The latter strategy is useful for making precise changes such as repairing mutations or inserting transgenes. Most studies thus far have relied on either NHEJ, or on HDR using short DNA fragments or oligos (24–26, 29, 31). Currently there is very limited information available on gene targeting using long DNA donor templates in hPSCs (23).
Here, we report the development of N. meningitidis Cas9 (NmCas9) (14) as a genome-editing platform, and its application to high-efficiency targeting of an endogenous gene in hPSCs. This system uses a 24-nt proto-spacer for targeting and requires a PAM that is different from those of SpCas9 or StCas9. We have achieved ∼60% targeting efficiency with two human embryonic stem cell (hESC) lines and one human induced pluripotent stem (iPS) cell line. Our work demonstrates the feasibility of using the N. meningitidis CRISPR-Cas system in genome editing in hPSCs using long DNA donor templates. This work also provides an alternative to the S. pyogenes and S. thermophilus CRISPR-Cas system and expands the genomic contexts that are amenable to RNA-directed genome editing in eukaryotes.
Results
Functional Expression of NmCas9 in Mammalian Cells.
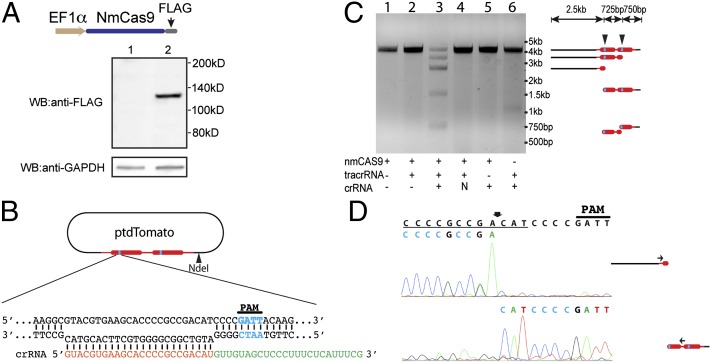
Our recent work has shown that N. meningitidis strain 8013 has a functional type II-C CRISPR-Cas system (14), and that Cas9 is the only Cas protein required for interference activity. We set out to test whether this system could be used for efficient gene targeting in hPSCs. We cloned the ORF from the 3.25-kb cas9 gene, along with a C-terminal FLAG tag, into a mammalian expression plasmid under the control of an elongation factor-1α (EF1α) promoter (Fig. 1A). This NmCas9-containing vector was transfected into 293FT cells and the expression of NmCas9 protein was analyzed by anti-FLAG Western blot. As shown in Fig. 1A, full-length NmCas9 was efficiently expressed in 293FT cells. We then assayed the nuclease activity of NmCas9 expressed in mammalian cells by in vitro plasmid cleavage. Cell extract was prepared from 293FT cells 2 d after transfection with the NmCas9-containing vector (the same one as in Fig. 1A). We assembled cleavage reactions using cell extract, various in vitro-synthesized small RNAs, and the plasmid ptdTomato prelinearized by NdeI (Fig. 1B). tdTomato is a fusion of two copies of the dTomato gene, each of which has one consensus PAM sequence (5′-NNNNGATT-3′) (in blue in Fig. 1B). As shown in Fig. 1C, we achieved efficient plasmid cleavage only in the presence of both tracrRNA and a cognate crRNA (Fig. 1C, lane 3). The pattern of the cleavage products was consistent with two predicted cleavage sites in the PAM-proximal regions (Fig. 1C, Right). Importantly, a noncognate crRNA (N), which contains sequences from EGFP, did not direct NmCas9-mediated cleavage (Fig. 1C, lane 4), indicating that the specificity of the NmCas9 nuclease is indeed guided by the spacer-derived sequence in crRNAs. Additionally, plasmid cleavage is deficient when tracrRNA is absent, even in the presence of a cognate crRNA (Fig. 1C, lane 5), suggesting that tracrRNA is necessary for NmCas9 function in vitro. This finding is consistent with the tracrRNA requirement for NmCas9-mediated interference in bacterial cells (14).
Fig. 1.
Functional NmCas9 can be expressed in mammalian cells. (A) Western blot analysis demonstrates that FLAG-tagged NmCas9 is expressed in 293FT cells. (Lane 1) Untransfected 293FT cells. (Lane 2) 293FT cells transfected with FLAG-tagged NmCas9 expressing plasmid. (Upper) Anti-FLAG tag Western. (Lower) Anti-GAPDH Western as loading control. (B) Design of the crRNA that targets the tdTomato coding sequence. Blue: PAM sequence; orange: crRNA spacer; green: crRNA repeat. (C) The plasmid containing the tdTomato coding sequence (see B) was linearized with NdeI and mixed with different combinations of tracrRNA, crRNA, and cell lysate prepared from NmCas9-expressing 293FT cells. After incubation at 37 °C, DNA was purified and analyzed by agarose gel electrophoresis. The diagram on the right shows the expected cleavage products and their predicted sizes. “N” indicates inclusion of a nonspecific crRNA that does not target tdTomato. (D) Cleavage product (see C) was extracted from the gel and analyzed by Sanger sequencing using the primers indicated in the Right panel. The cleavage site, indicated by the arrow, was inferred from the sequencing.
Two Cas9 orthologs, SpCas9 and StCas9, were previously demonstrated to induce blunt DSBs in their DNA targets, between the third and fourth nucleotide counting from the PAM-proximal end of protospacers (19, 33, 34). We hypothesized that NmCas9 cleaves the DNA target in a similar way, and we tested this by mapping the NmCas9 cleavage site on ptdTomato by Sanger sequencing. Two cleavage products in Fig. 1C (the 1.5- and 2.5-kb fragments) were gel-extracted and sequenced to identify the NmCas9 cleavage sites on the sense strand and the antisense strand, respectively. As expected, NmCas9 induced a blunt-end DSB between the third and fourth nucleotides counting from the PAM-proximal end of the proto-spacer (Fig. 1D).
NmCas9 Functions in RNA-Directed Gene Disruption in hPSCs.
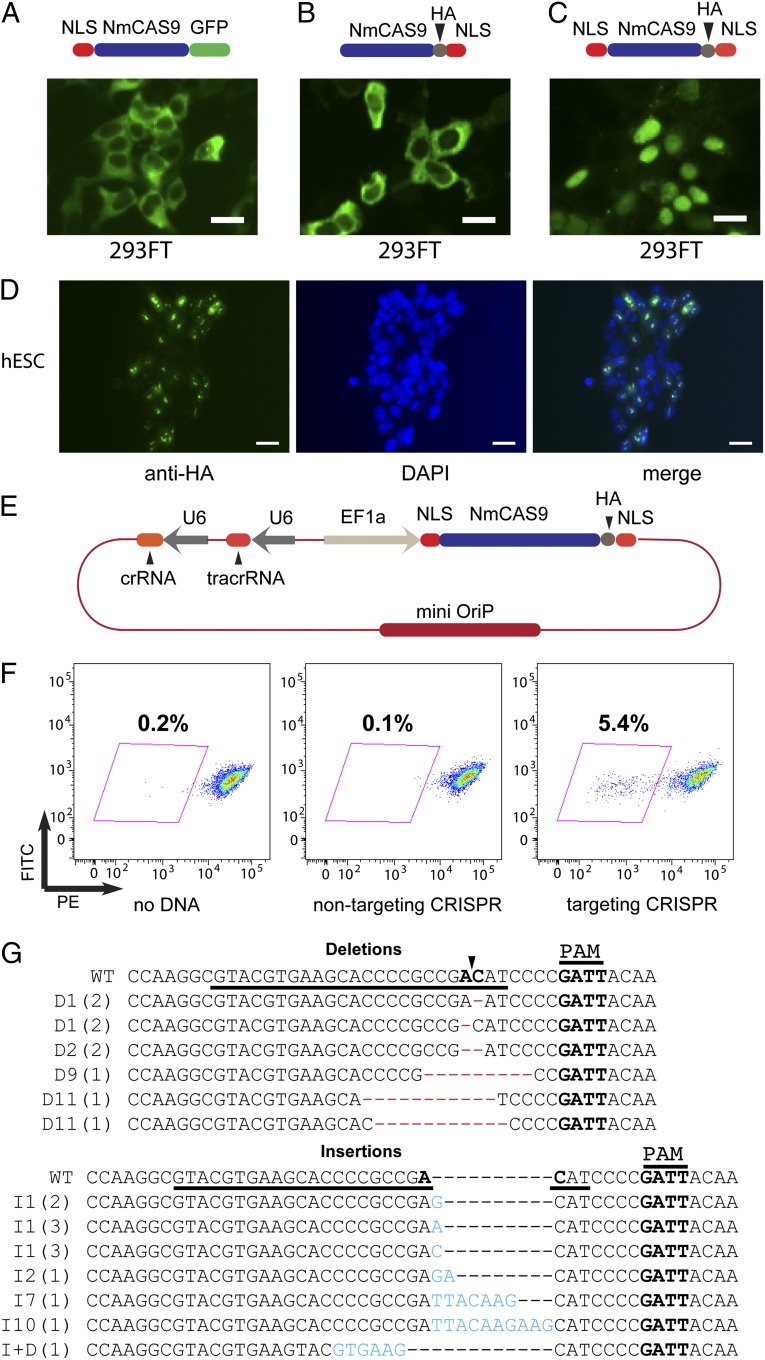
Knowing that NmCas9, without any codon optimization, can be efficiently expressed in mammalian cells and is functional in vitro, we next tested its utility in genome editing in hPSCs. We first monitored its localization. We transfected 293FT cells with several NmCas9 constructs with various nuclear localization signal (NLS) arrangements, and analyzed NmCas9 protein localization by either GFP fluorescence or anti-HA immunostaining. NmCas9 with NLSs on both N and C termini localized efficiently to the nucleus (Fig. 2C), but NmCas9 constructs with just one NLS did not (Fig. 2 A and B). In addition, the same NmCas9 construct with two NLSs also localized to the nucleus of hESCs (Fig. 2D). We noticed that in hESCs, NmCas9, without any crRNA/tracrRNA, displayed a punctate pattern similar to the organization of the nucleolus in hESCs. It is not yet clear if this phenomenon is related to the organization of the double NLS on the protein.
Fig. 2.
NmCas9 functions in gene disruption in human ES cells. (A–C) The localization of NmCas9 with an NLS at the N terminus (A), C terminus (B), or both termini (C) was analyzed by either EGFP fluorescence (A) or anti-HA immunofluorescence (B and C) in 293FT cells. (Scale bars, 20 µm.) (D) The localization of NmCas9 with the double NLS (see C) was analyzed by anti-HA immunofluorescence in hESCs. (Scale bars, 20 µm.) (E) Design of a single plasmid used for gene editing in hESCs. (F) FACS analysis of tdTomato reporter hESC lines after electroporation of the indicated crRNA/tracrRNA/NmCas9 constructs. The number in the plot indicates the percentage of tdTomato-fluorescence-negative cells 5 d after electroporation. (G) Indels introduced by the targeting CRISPR in the tdTomato negative population (see F) were analyzed by targeted PCR amplification and sequencing. The protospacer sequence is underlined. The numbers in parentheses indicate the number of sequenced clones containing that specific indel.
To test the genome-editing activity of NmCas9, we used an hESC cell reporter line that has a single copy of the tdTomato fluorescent protein gene knocked into the highly expressed DNMT3b locus (H9 DNMT3b-tdTomato), leading to tdTomato fluorescence. If NmCas9 is able to introduce a DSB in the tdTomato sequence in the genome, repair by NHEJ would likely lead to indels that disrupt tdTomato expression. Accordingly, the appearance of tdTomato-negative cells would be predicted to reflect genome-editing activity.
hESCs are known to have low transfection efficiencies. To achieve maximum genome-editing efficiency in hPSCs, we assembled expression cassettes of all of the necessary components (NmCas9, tracrRNA, and crRNA) onto one single plasmid that contains an OriP sequence (Fig. 2E). OriP was reported to increase the transfection efficiency and plasmid stability in hPSCs if cotransfected with an RNA expressing the EBNA protein (41). The encoded tracrRNA and crRNA both corresponded to the mature, processed forms as they exist in N. meningitidis cells (14). The resulting all-in-one plasmids were electroporated into H9 DNMT3b-tdTomato cells, and tdTomato fluorescence was monitored by FACS 4–6 d after electroporation. As shown in Fig. 2F, a subpopulation (5.4%) of tdTomato-negative cells became detectable only when a tdTomato-targeting crRNA was encoded on the plasmid. Importantly, for the control plasmid expressing nontargeting crRNA, only background levels (∼0.1%) of tdTomato-negative cells appeared (Fig. 2F), likely because of the low level of spontaneous differentiation in the culture, leading to repression of the DNMT3b promoter. The increased frequency of nonfluorescent cells in the presence of the cognate crRNA suggests successful genome editing by NmCas9.
To confirm that NmCas9 introduced a DSB at the intended genomic site, we FACS-sorted the tdTomato-negative population, PCR-amplified the genomic region flanking the predicted cutting site in the 5′ copy of dTomato, cloned the resulting PCR fragments, and sequenced 22 of the resulting plasmids (selected at random). The sequencing results showed both insertions and deletions in the tdTomato sequence (Fig. 2G; only unique indels are shown) in 95% of the sequenced clones. Most importantly, all of these indels were centered around the NmCas9 cleavage site, indicating that the DSB occurred at the intended position (Fig. 2G).
A Chimeric sgRNA Is Effective for Gene Editing in hPSCs.
To simplify the NmCas9 genome-editing system, we explored the possibility of substituting both crRNA and tracrRNA with a chimeric sgRNA. We fused the 5′ end of the 91-nt processed tracrRNA sequence with the 3′ end of the 48-nt mature crRNA using a 6-nt linker (Fig. S1A). This sgRNA was cloned under the control of the U6 promoter and electroporated into the H9 DNMT3b-tdTomato reporter cell line together with a plasmid expressing NmCas9. FACS analysis showed that this sgRNA indeed resulted in tdTomato-negative cells (Fig. S1B) at a level comparable to that achieved by the all-in-one plasmid expressing separate crRNA and tracrRNA (Fig. 2E). These results indicated that an sgRNA could substitute for separate crRNA and tracrRNA in directing NmCas9-mediated gene editing in hESCs.
Specificity of NmCas9 in hPSCs.
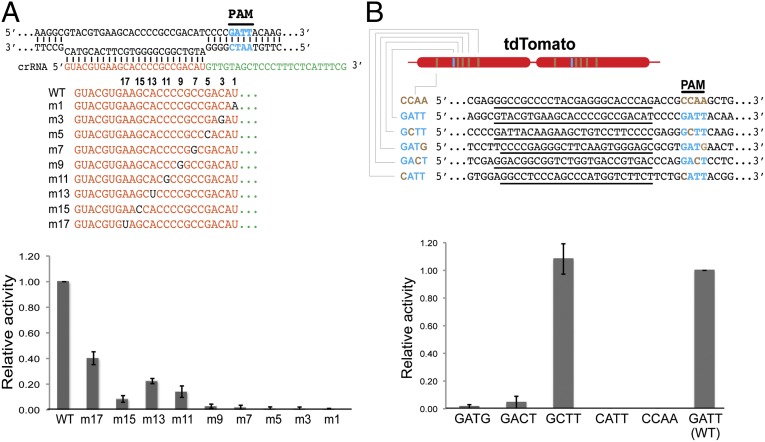
We next tested the specificity of NmCas9 in mammalian cells by mutational analysis. We introduced single-nucleotide mutations at every odd-numbered position from the first to the 17th nucleotide in the PAM-proximal end [spanning the cleavage site (Figs. 1D and 2G) and the functionally critical “seed” sequence] of the spacer in the tdTomato-targeting crRNA construct (Fig. 3A, Upper). We then measured the ability of those constructs to give rise to tdTomato-negative cells in the H9 DNMT3b-tdTomato cell line. As shown in Fig. 3A, mutations at positions 1 through 9 led to background levels of tdTomato-negative cells, indicating that mismatches at these positions in the crRNA/target duplex are not tolerated by NmCas9. As for mutations at positions 11, 13, and 15, some tdTomato-negative cells appeared, but with an efficiency of only 10–25% of that observed with wildtype crRNA (Fig. 3A, Lower). The mismatch at position 17 was ∼40% as efficient as wild-type. These results imply a crRNA/target specificity comparable to that of the SpCas9 system in mammalian cells (21).
Fig. 3.
Specificity screen of NmCas9 system. (A, Upper) The crRNA sequence targeting tdTomato, with locations of single point mutations (black) in the spacer region of the tdTomato targeting crRNA. (Lower) The efficiency of each mutant at disrupting tdTomato expression. All mutants were tested as described in Fig. 2F. The efficiency is defined as percent tdTomato-negative cells (mutant spacer) divided by percent tdTomato-negative cells (wild-type spacer). Error bars: SEM (B, Upper) The locations of different mutant PAMs in the tdTomato sequence. Blue, wild-type PAM; brown, mutant PAM; underlined, spacer sequence. For the bottom-most protospacer, the opposite strand was targeted, and the reverse complement sequence is therefore shown. (Lower) The efficiency of targeting at each site associated with the indicated PAM, as revealed by the loss of tdTomato expression. All targeting experiments were performed as described in Fig. 2F. The efficiency is defined as percent tdTomato-negative cells (mutant PAM) divided by percent tdTomato-negative cells (wild-type PAM). Error bars: SEM.
We also investigated PAM sequence requirements for NmCas9 in hESC. We designed five crRNAs that use different sequences as the PAM in the tdTomato coding region (Fig. 3B) and then tested their ability to disrupt tdTomato expression in H9 DNMT3b-tdTomato cells. Four of the sites were associated with a PAM that varied from the 5′-NNNNGATT-3′ consensus by only a single nucleotide. Only a GCTT variant site was efficiently targeted, whereas the other four variants were severely deficient (Fig. 3B, Lower). Our results indicate that an A-to-C mutation at the second nucleotide of the PAM could be tolerated, whereas a G-to-C mutation at the first position, T-to-C at the third, and T-to-G at the fourth likely render the PAM variants nonfunctional. Interestingly, C is the second most-frequent residue at the second nucleotide of the PAM in candidate bacterial protospacers (14), suggesting that GCTT might also be a natural PAM.
NmCas9 Increases Gene-Targeting Efficiency in hPSCs.
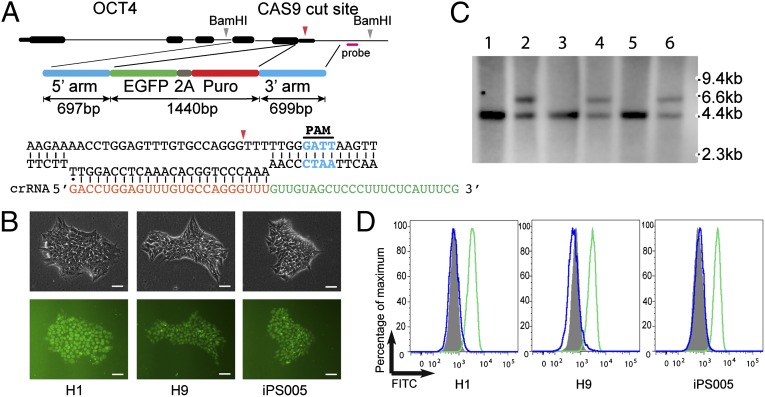
We next explored whether NmCas9 can increase gene-targeting efficiency in hPSCs compared with the traditional method in which no DSB was intentionally introduced at the target site. We used a donor DNA template previously used to target the endogenous POU5F1 (OCT4) gene (6) (Fig. 4A), creating a fusion of OCT4 with EGFP. We designed the crRNA using the consensus PAM sequence located ∼84 bp downstream of the OCT4 stop codon (Fig. 4A). Two hESC lines, H1 and H9, and one human iPS cell line, iPS005 (42), were used in the experiment. After puromycin selection, we were able to obtain clones for all three cell lines when plasmid expressing the OCT4-targeting crRNA was used. Of these clones, ∼60% were correctly targeted with single insertion events (Table 1), comparable to the efficiency obtained using TALENs in a previous report with the same donor DNA (6). Fluorescent images of the targeted clones revealed the expected nuclear localization of EGFP signal because of the fusion with Oct4 protein (Fig. 4B). Southern blots using a probe outside the targeting vector’s homology arm confirmed the correct integration of the donor sequence in the OCT4 locus (Fig. 4C). Most importantly, the EGFP signals respond to differentiation cues as the endogenous Oct4 would (Fig. 4D). In a control experiment with an all-in-one plasmid expressing a nontargeting crRNA, no puromycin-resistant clones were obtained with the H1 ESC line. Only one puromycin-resistant clone each was obtained from H9 ESCs and iPS005 iPSCs, and neither clone was correctly targeted (Table 1). All of the above results indicated that the CRISPR-Cas system from N. meningitidis was able to generate accurately targeted clones in hPSCs with much increased efficiency compared with the traditional method.
Fig. 4.
Gene targeting in hESCs using NmCas9. (A) Donor DNA and crRNA design. The mismatch in the first nucleotide of crRNA is to satisfy the requirement of the U6 promoter for a G residue at the transcription start site. (B) Phase-contrast (Upper) and fluorescent (Lower) images of targeted clones from H1, H9, and iPS005 line. (Scale bars, 50 µM.) (C) Southern blot analysis of H1 WT (lane 1), H9 WT (lane 3), and iPS005 WT (lane 5) and targeted clones of H1 (lane 2), H9 (lane 4), and iPS005 (lane 6) line. Genomic DNA was digested with BamHI. The Southern probe is located outside of donor DNA (see A). The wild-type clone should give one band of 4.2 kb and targeted heterozygous clone should give one additional band of 5.6 kb. (D) Targeted clones (see B) were treated with 10 µM SB431542 and 10 ng/mL BMP4 to initiate differentiation. The EGFP signal was analyzed by FACS 3 d after differentiation. Gray, undifferentiated parental cells before targeting; green, targeted cells before differentiation; blue, targeted cells after differentiation.
Table 1.
Summary of gene targeting efficiency using NmCas9 in hPSCs
| Cell line | crRNA | Clone analyzed | Nontargeted | Targeted with additional insertions | Targeted | Targeting efficiency (%) |
| H1 (ES) | Nontargeting | 0 | 0 | 0 | 0 | 0 |
| Targeting | 20 | 5 | 3 | 12 | 60 | |
| H9 (ES) | Nontargeting | 1 | 1 | 0 | 0 | 0 |
| Targeting | 39 | 9 | 7 | 23 | 59 | |
| iPS005 (iPS) | Nontargeting | 1 | 1 | 0 | 0 | 0 |
| Targeting | 10 | 1 | 3 | 6 | 60 |
The gene targeting efficiencies using the donor construct shown in Fig. 4A were calculated as the ratio between the number of correctly targeted clones and the total number of analyzed clones. For H1 cells, no puromycin-resistant clones were obtained using nontargeting crRNA.
Discussion
Genome Editing by N. meningitidis Cas9.
In this report, we have successfully used the type II-C CRISPR-Cas system from N. meningitidis to achieve both NHEJ-mediated gene editing and long DNA donor-directed gene targeting of an endogenous locus in hPSCs. The targeting efficiency we obtain with NmCas9 is comparable to that achieved with TALENs. Using the same donor construct, we were able to get ∼60% targeting efficiency in all three different hPSC lines tested (Table 1), whereas the targeting efficiency of a TALEN was 48% in the one hESC line tested (6). A previous report using SpCas9 in human iPSCs achieved a targeting efficiency of 43%, close to what we observed with NmCas9 (6). However, that report only identified seven clones and did not perform further analysis to confirm the correct integration of the donor DNA sequence only at the intended site. Therefore, additional work will be needed to compare the efficiency of mammalian gene targeting using these two CRISPR-Cas systems.
CrRNA/Target Mismatch Tolerance by NmCas9 in Mammalian Cells.
One potential advantage of NmCas9, relative to SpCas9, is that it might offer better targeting specificity by virtue of its longer crRNA spacer (24 vs. 20 nts) and its longer PAM (14). We chose 24 nt as the crRNA spacer length for NmCas9 because that is the length of the crRNA spacer in N. meningitidis. CrRNA-target mismatches distant from the PAM were tolerated to various extents for both NmCas9 (Fig. 3B) and SpCas9 (21) in mammalian cells. However, NmCas9 was more sensitive than SpCas9 to mismatches at the 13th, 15th, and 17th nts (counting from the PAM-proximal end of the proto-spacer). NmCas9 gene-editing efficiencies with mismatches at those positions were no higher than 10–40% of those observed with the perfectly matched crRNA (Fig. 3B), whereas with SpCas9, mismatches at equivalent positions retained 60–90% of the nonmismatched efficiency (21).
PAM Requirements in Mammalian Cells.
One hallmark of type II CRISPR-Cas systems is the requirement of a nearby PAM on the target sequence. This sequence varies between different Cas9 orthologs. Among Cas9 proteins validated for mammalian genome editing, PAM functional requirements have been defined for three: those from S. pyogenes SF370 (21–23, 32, 34), S. thermophilus LMD-9 (the CRISPR1 locus) (19, 21, 38), and N. meningitidis 8013 (Fig. 3B) (14). On the one hand, the PAM requirement adds a second layer of specificity for gene targeting, beyond that afforded by spacer/protospacer complementarity. For longer PAMs (such as the NmCas9 PAM, 5′-NNNNGATT-3′) the frequency of off-target cutting events should potentially drop significantly compared with SpCas9, which requires a 5′-NGG-3′ PAM. On the other hand, longer PAM requirements also constrain the frequency of targetable sites. By developing genome-editing systems using a range of Cas9 proteins with distinct PAM requirements, the genomic regions that can be targeted by CRISPR-Cas editing would expand significantly.
The results in Fig. 3B show that NmCas9 does allow limited deviation from the 5′-NNNNGATT-3′ PAM. Having a variable PAM can potentially increase the flexibility during the design of targeting construct. However, it also increases the potential of off-target cleavage. Because of the limited options afforded by the sequence of tdTomato, we only tested one nucleotide substitution in each position of the PAM. It is possible that additional nucleotide substitutions will also be tolerated. A detailed mutational analysis will be needed to fully understand the PAM requirements of NmCas9 in mammalian cells.
Editing the Genomes of hPSCs.
Compared with two other widely used systems for enhancing gene targeting efficiency (ZFNs and TALENs), the CRISPR-Cas system offers a much simpler and more user-friendly design. For each different genomic locus to be targeted, one only needs to design a small RNA by applying simple Watson–Crick base-pairing rules. This system’s ease of use will make gene targeting in hPSCs, once considered a difficult project, a routine laboratory technique. This simple and high-efficiency gene-targeting system for hPSC will also have a tremendous impact on personalized regenerative medicine. One concern with using CRISPR-Cas in human genome editing is off-target cleavage. Our work (Fig. 3A) and that of others (21, 43) has shown that the CRISPR-Cas system can tolerate mismatches within the crRNA, especially in the PAM-distal region. This finding raises concerns that other regions in the genome might be cleaved unintentionally. Indeed, recent work has shown various off-target cleavage rates in the human genome using SpCas9 with different sgRNAs (43). To fully understand this issue, whole-genome sequencing of cells targeted by different Cas9 proteins with different crRNA/sgRNA constructs will be needed. A potential way to get around this problem is to use a nickase, a Cas9 variant in which one nuclease domain is inactivated by a mutation (21, 34), so that off-target cleavage will have a much lower chance of generating unwanted mutations in the genome but HDR will still be stimulated.
Materials and Methods
Cell Culture.
hESCs and iPS cells were cultured in E8 medium (42) on Matrigel-coated tissue culture plates with daily media change at 37 °C with 5% (vol/vol) CO2. Cells were split every 4–5 d with 0.5 mM EDTA in 1× PBS. 293FT cells were cultured similarly in DMEM/F12 media supplemented with 10% (vol/vol) FBS.
NmCas9 DNA Transfection and in Vitro Plasmid Digestion.
All transfections with 293FT cells were done using Fugene HD (Promega) following the manufacturer’s instructions. Cell lysate was prepared 2 d after transfection. Plasmid digestion using cell lysate was carried out at 37 °C for 1–4 h in digestion buffer (1× PBS with 10 mM MgCl2). See SI Materials and Methods for a detailed procedure. To map the cleavage site of NmCas9, the digested plasmid DNA was excised from the agarose gel and purified using Gel Extraction Kit (Qiagen). The purified fragments were then sequenced to map the cleavage site.
Gene Editing in hPSCs.
All plasmids used in this experiment were purified using the MaxiPrep Kit from Qiagen. hPSCs were passaged 2 or 3 d before the experiments. Immediately before the experiment, hPSCs were individualized by Accutase treatment, washed once with E8 medium, and resuspended at densities of 2.5–6.2 × 106 cells/mL in E8 medium with 10 mM Hepes buffer (pH 7.2–7.5) (Life Technologies). For electroporation, 400 µL of cell suspension, 15 µg of pSimple-Cas9-Tracr-CrRNA plasmid, 5 µg of EBNA RNA, and (for those experiments involving gene targeting by HDR) 5 µg of linearized DNA template plasmid (Addgene 31939) were mixed in a 4-mm cuvette (Bio-Rad) and immediately electroporated with a Bio-Rad Gene Pulser. Electroporation parameters were 250 V, 500 µF, and infinite resistance. Cells were then plated into appropriate Matrigel-coated culture dishes in E8 supplemented with 10 µM ROCK inhibitor Y-27632. Media was changed the next day to E8. For those experiments involving gene editing by HDR, puromycin selection was started 4 d after electroporation. Surviving colonies were picked 4–6 d after selection and expanded in E8 medium.
For lists of plasmids, crRNA and tracrRNA sequences and primers used in this study, see Tables S1–S4. For the sequence of NmCas9, see Dataset S1. For the comparison of NmCas9 with SpCas9 and StCas9, see Datasets S2 and S3.
Supplementary Material
Acknowledgments
We thank Krista Eastman for editorial assistance. This work was supported by The Charlotte Geyer Foundation; Morgridge Institute for Research; The Wynn Foundation; and National Institutes of Health Grants 1UH2TR000506-01 (to J.A.T.) and R01 GM093769 (to E.J.S.).
Footnotes
Conflict of interest statement: Northwestern University (Y.Z. and E.J.S.) and Wisconsin Alumni Research Foundation (Z.H. and J.A.T.) have filed a related patent: DNA cleavage and genome editing using Cas9 from Neisseria meningitidis.
See Commentary on page 15514.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313587110/-/DCSupplemental.
References
- 1.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21(3):319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 3.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 4.Joung JK, Sander JD. TALENs: A widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14(1):49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hockemeyer D, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27(9):851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou J, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5(1):97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, et al. Synthetic zinc finger transcription factor action at an endogenous chromosomal site. Activation of the human erythropoietin gene. J Biol Chem. 2000;275(43):33850–33860. doi: 10.1074/jbc.M005341200. [DOI] [PubMed] [Google Scholar]
- 9.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol Ther. 2006;13(2):438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 12.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12(2):177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(5909):1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, et al. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50(4):488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14(12):2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 19.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 20.Westra ER, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46(5):595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 26.Chang N, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23(4):465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiCarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41(7):4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013 doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao A, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Q, et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12(4):393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39(21):9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karvelis T, et al. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 2013;10(5) doi: 10.4161/rna.24203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah SA, Erdmann S, Mojica FJ, Garrett RA. Protospacer recognition motifs: Mixed identities and functional diversity. RNA Biol. 2013;10(5):891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190(4):1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horvath P, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190(4):1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 41.Kameda T, Smuga-Otto K, Thomson JA. A severe de novo methylation of episomal vectors by human ES cells. Biochem Biophys Res Commun. 2006;349(4):1269–1277. doi: 10.1016/j.bbrc.2006.08.175. [DOI] [PubMed] [Google Scholar]
- 42.Chen G, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8(5):424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013 doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.