Abstract
Direct polymerase chain reaction (PCR) detection of insertion/deletion (indel) polymorphisms requires sample homozygosity. For the indel polymorphisms that have the deletion allele with a relatively low frequency in the autosomal regions, direct PCR detection becomes difficult or impossible. The present study is, to our knowledge, the first designed to directly detect indel polymorphisms in a human autosomal region (i.e., the immunoglobulin VH region), through use of single haploid sperm cells as subjects. Unique marker sequences (n=32), spaced at ∼5-kb intervals, were selected near the 3′ end of the VH region. A two-round multiplex PCR protocol was used to amplify these sequences from single sperm samples from nine unrelated healthy donors. The parental haplotypes of the donors were determined by examining the presence or absence of these markers. Seven clustered markers in 6 of the 18 haplotypes were missing and likely represented a 35–40-kb indel polymorphism. The genotypes of the donors, with respect to this polymorphism, perfectly matched the expectation under Hardy-Weinberg equilibrium. Three VH gene segments, of which two are functional, are affected by this polymorphism. According to these results, >10% of individuals in the human population may not have these gene segments in their genome, and ∼44% may have only one copy of these gene segments. The biological impact of this polymorphism would be very interesting to study. The approach used in the present study could be applied to understand the physical structure and diversity of all other autosomal regions.
Introduction
Because of the diploidy of the human autosomes, direct detection of insertion/deletion (indel) polymorphisms by PCR amplification requires the presence of the deletion allele in a homozygous state. When the deletion allele frequency is relatively low, homozygous individuals in the human population become very rare. For example, if the allele frequency is 0.2, which is relatively common, then only 1 homozygous individual can be found among 25 individuals. If the allele frequency is 0.01, then 10,000 individuals need to be examined in order to find one homozygous individual. In these cases, it becomes difficult or impossible to detect indel polymorphisms on the autosomes. In the present study, we describe a method that can be used to directly detect indel polymorphism on the autosomes. The method uses single haploid sperm cells and DNA sequence markers of a high density (5 kb each). Amplification by PCR with multiple sperm for each parental haplotype in each donor allowed us to detect an indel polymorphism without ambiguity. The strength of the analysis was enhanced by the use of markers of a high density, since, in the haplotype with the deletion allele, absent markers were found in a cluster that could be clearly distinguished from markers that were not amplified successfully by PCR. The method was used to detect the indel polymorphisms in the human immunoglobulin VH region. It may also be used for detecting indel polymorphisms in any other regions of the human genome.
During the vertebrate immune response, immunoglobulin and T cell receptors play a key role in antigen recognition. The basic unit of an immunoglobulin or antibody molecule (Ig) consists of two light chains (L) and two heavy chains (H). The light chains are encoded by two independent gene loci—namely, Igκ and Igλ gene complexes. The heavy chains are encoded by the IgH gene complex, located on chromosome 14 (Croce et al. 1979). The amino terminal end of each chain is called the “variable” (V) region, because of its diverse amino acid sequence, and it is responsible for interaction with a wide spectrum of antigens. The V region of an IgH molecule is encoded by three gene segments: variable (VH), diversity (D), and joining (JH). The carboxyl terminal end of each chain is called the constant (C) region, because of its conserved sequence. In the germline configuration, each of these gene segments is present in multiple copies. The nucleotide sequences of the copies for each gene segment differ from each other, although some may share a high degree of sequence identity. Generation of Ig genes from various combinations of gene segments is one of the major mechanisms underlying antibody diversity.
Although human VH gene segments have been mapped to three chromosomes—14q32.3, 15q11, and 16p11 (Croce et al. 1979; Cox et al. 1982; Kirsch et al. 1982; Cherif and Berger 1990; Nagaoka et al. 1994; Tomlinson et al. 1994)—only the segments on chromosome 14 are expressed. VH sequences in each family share >80% identity (Lee et al. 1987; Schroeder et al. 1987; Berman et al. 1988; Buluwela and Rabbitts 1988; Humphries et al. 1988; Tomlinson et al. 1992; Willems van Dijk et al. 1993). Many studies suggest that the VH region is highly diversified in gene segment number and composition (Chen and Yang 1990; Shin et al. 1991; Sasso et al. 1992, 1993, 1995; Rubinstein et al. 1993; Walter et al. 1993; Milner et al. 1995; Cui and Li 1997, 1998). Further diversity is achieved by the allelic variation in VH and intergenic sequences (Souroujon et al. 1989; Willems van Dijk et al. 1989, 1991; Guillaume et al. 1990; Sasso et al. 1990; Walter et al. 1990; Rubinstein et al. 1993).
Efforts have been made to obtain an overview of the physical structure of the VH region. Matsuda et al. (1993) constructed a physical map of the 3′ 0.8-Mb region of the human VH locus, with 64 segments identified in the mapped region. The general organization of the VH segments indicated recent dynamic reshuffling of the VH locus. By determining the VH gene segment organization in the most 3′ end of the VH locus and by integrating the map information from Matsuda et al. (1993) and other data (Shin et al. 1991; Tomlinson et al. 1992), Cook et al. (1994) constructed a composite map for the entire VH locus. The map contained 87 VH segments, of which 46 were functional. The VH segments were classified into seven different families (VH1–VH7). Recently, the complete nucleotide sequence of the 957 kb of DNA of the VH region on chromosome 14q32.3 was determined by Matsuda et al. (1998). Results from analysis of the sequence indicate that the VH region contains 123 gene segments, of which 79 are pseudogenes. Of the 44 VH segments with ORFs, 39 are expressed as heavy-chain proteins and 1 is expressed as mRNA, whereas the remaining 4 are not found in Ig cDNAs. Matsuda et al. (1998) also observed that the promoter and recombination signal sequences were more conserved in functional VH segments than in pseudogenes. The existence of many VH pseudogenes is indicative of the fact that frequent gene conversions took place during the evolution of the VH locus (Haino et al. 1994).
The presence of a large number of polymorphisms in the VH region is indicated by a number of studies (Matthyssens and Rabbitts 1980; Brodeur and Riblet 1984; Turnbull et al. 1987; Walter et al. 1987; Pincus 1988; Sam et al. 1988; Walter and Cox 1988, 1991; Willems van Dijk et al. 1989, 1991, 1993; Guillaume et al. 1990; Sasso et al. 1990; Weng et al. 1992; Adderson et al. 1993). Shin et al. (1991) compared the VH segments and restriction map in the 3′ portion of the VH region in a clone from a YAC library with those in the clones from a cosmid library, constructed from different genomic DNA. It was shown that one of the haplotypes contained a VH segment, 1-4.1b, that was not present in the other haplotype. Indel polymorphisms were also observed in a 300–370-kb region immediately 5′ to the region analyzed by Shin et al. (1991). Results from our laboratory indicated that this region was completely missing in one haplotype (Cui and Li 1998). A deletion/duplication polymorphism involving two gene segments, 3-30 and 4-28, was also found in this region (Chen and Yang 1990; Yang et al. 1990; Sasso et al. 1992). The duplicates in this polymorphic region were shown to be in a tandem array, with numbers varying among individuals. Each human may have as many as three duplicates (Sasso et al. 1992). In some haplotypes, this duplicated region may contain a pseudogene, 3-29p, whereas this gene segment is missing in others (Milner et al. 1995). The 5′ end of this region contains an indel polymorphism involving two VH3 and three VH4 gene segments with a length of ∼60–65 kb (Walter et al. 1993). Approximately 60 kb downstream of this polymorphism, a VH3 gene segment, 3-23, was found duplicated, with the number of duplications varying among the subjects (Rubinstein et al. 1993; Sasso et al. 1995).
Before our recent study (Cui and Li 1998), very little was known about the degree of diversification for the 5′ part of the VH region. This region occupies ∼210 kb and contains ∼20–22 VH segments. This region was left unmapped in the first detailed map for the VH region (Matsuda et al. 1993), presumably because of the high degree of diversity. We showed that among the seven VH loci included in our analysis in this region, five contained null alleles (Cui and Li 1998). Sasso et al. (1993) showed that a VH1 gene segment, 1-69, located ∼120 kb from the most 5′ end of the VH region, was present in a very complicated fashion. This gene segment may have been duplicated into two loci and may contain 13 alleles with minor differences.
Although significant progress has been made in the study of VH region diversity, it has been difficult to obtain precise information about the extent of the diversity, simply because the basic units carrying the genetic information are single chromosomes but human somatic cells are diploid. DNA extracted from somatic cells, as the primary material for studying VH region diversity, is a mixture of two sets of homologous chromosomes. Our laboratory has been developing approaches to address this issue through the use of single sperm cells. Since sperm are haploid and each sperm contains only one set of chromosomes, VH regions on individual chromosomes can be studied by analyzing single sperm. In our initial effort (Cui and Li 1997), we showed our ability to amplify the VH4 sequences from single sperm through use of family-specific PCR primers. The method was used to determine the parental haplotypes of 13 donors and to distinguish allelic and interlocus differences among the human VH4 gene family sequences. Four of 10 loci were found to contain null alleles that otherwise would have been undetectable, if diploid materials had been used. Through use of family-specific primers and DNA fragments, which are prepared by lysing single sperm and by aliquoting the sperm lysate (Cui and Li 1998), we were able to determine the gene organization of the VH1 and VH4 sequences in the parental haplotypes of a sperm donor. By comparing our haplotype maps and the composite map by Cook et al. (1994), we found that the VH region on chromosome 14 could be subdivided into four portions. The numbers and compositions of the VH gene segments differed considerably among the maps in two subregions but were highly conserved in the other two. Success in this study demonstrated that high-resolution haplotype maps could be constructed without the help of molecular cloning. The present study, to our knowledge, is the first to identify indel polymorphisms by comparing a panel of selected marker sequences in the haplotypes of single sperm. A region of ∼200 kb near the 3′ end of the VH region was covered in the study.
Material and Methods
Marker Sequence Selection
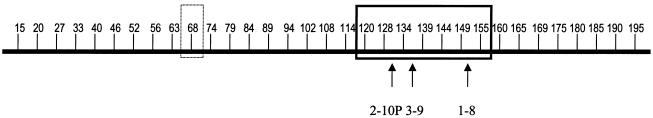
The sequence under GenBank accession number AB019440 (Matsuda et al. 1998) was used to select marker sequences for the present study. This sequence is ∼200 kb in length and is next to the 3′ end of the VH region. Thirty-three unique sequences, spaced at ∼5-kb intervals, were chosen from the intergenic regions and were used as marker sequences (fig. 1). The uniqueness of these marker sequences was confirmed by subjecting these sequences to the BLAST search program. Marker sequences were designated as 15, 20, 27, 165, and so on, on the basis of their positions from the 5′ end of AB019440.
Figure 1.
Relative locations of the 32 sequence markers (short vertical lines), the indel polymorphism (boxed with solid lines) and the affected VH gene segments (arrows above gene segment names). Marker 68 (boxed with dashed lines) was excluded from the study, because of its poor amplification efficiency.
Design of PCR Primers
For each marker sequence, forward (F) and reverse (R) primers were designed. A nested (N) primer internal to the F primer was also designed for each marker sequence (see the authors' Web site for the primer sequences). The primer lengths were 20–26 bases. For each marker, sequences of the F and R primers did not match any sequence other than their target sequences; the same was true for the N and R primers. In addition to the specific sequences, each R primer also contained a 5′ universal “tail” sequence (5′-CCCCGCCGCCGCCGCCGCCC-3′). Primers for each marker locus were tested separately, in a pairwise fashion (i.e., F vs. R and N vs. R), to ensure that they could generate a PCR product with good yield and specificity. Primers that did not meet this requirement were replaced, until all primers met the requirement. A two-round multiplex PCR protocol, which was a modification of the protocol described by Lin et al. (1996), was used for marker sequence amplification. All the optimization experiments related to multiplex PCR were performed with human genomic DNA before amplification of the sequences from single sperm.
Single Sperm Sample Preparation
Semen samples used in the present study were the remainder of the specimens used for in vitro fertilization and infertility tests from nine unrelated healthy white donors. Specimens used for infertility tests had sperm counts in the normal range. For each donor, 100 μl of semen was washed with 500 μl of H2O and was passed through a 15-μm mesh. It was then centrifuged at 3,000 rpm for 4 min in an Eppendorf centrifuge (5417C). This process was repeated twice. Sperm were then suspended in 200 μl of water. The suspension was mixed with 200 μl of 70% sucrose.
To fix and stain sperm for flow sorting, we transferred the sperm samples from the above suspension to a 15-ml Falcon tube, and the samples were washed with 1 ml of PBS twice. Centrifugation was performed at 1,200 rpm for 5 min in a Beckman centrifuge. The pellet was suspended in 200 μl of PBS, and 70% ethanol was added to it during vortexing. The suspension was incubated on ice for 20 min and was cetrifuged as described above. The pellet was resuspended in 200 μl of PBS containing 0.3% Triton-X-100. The volume was made up to 2 ml with PBS, followed by addition of 20 μl of propidium iodide (1 mg/ml). The sample was stored at 4°C until further analysis.
Single sperm were sorted, with a fluorescence-activated cell sorter (Beckman Coulter), into the wells of a 96-well V-bottom plate. Each well contained 5 μl of lysis buffer (50 mM dithiothreitol and 200 mM KOH) (Cui et al. 1989). One row in the V-bottom plate was kept empty, to be used later as negative controls. Sperm were lysed by incubating at 65°C for 10 min. After lysis, 5 μl of neutralization buffer (200 mM HCl, 900 mM Tris-HCl [pH 8.3], and 300 mM KCl) was added to each incubated sample. For each donor but D17, 42 sperm were analyzed. Thirty-six sperm were analyzed for donor D17.
Multiplex PCR Amplification
In the first round, all 33 pairs of F and R primers were used to amplify their target sequences in single sperm. Amplification with the R primers resulted in the attachment of the universal tail onto all target sequences. Each sample contained 1× PCR buffer (50 mM KCl, 100 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, and 100 μg/ml gelatin), four dNTPs (50 μM each) (Invitrogen), F and R primers (50 nM each), and 1.2 U of HotStar Taq DNA polymerase (Qiagen). The final volume for each reaction was 50 μl. The samples were first heated to 94°C for 15 min, to activate the Taq DNA polymerase, followed by 40 PCR cycles. Each PCR cycle consisted of 40 s at 94°C for denaturation and 2 min at 55°C, followed by 5 min of ramping from 55°C to 70°C, for annealing and extension. A final extension was performed at 72°C for 3 min at the end of the 40th cycle. All PCR amplifications were performed with the Biometra T3 Thermocycler.
In the second round, 33 marker sequences were subdivided into three groups (groups 1, 2, and 3). Group 1 included eight markers—68, 74, 84, 94, 139, 165, 175, and 190—whereas group 2 included nine: 27, 52, 56, 63, 89, 120, 144, 180, and 185. The remaining 16 markers were placed in group 3, which included 15, 20, 33, 40, 46, 79, 102, 108, 114, 128, 134, 149, 155, 160, 169, and 195. Three small aliquots (1 μl) from the first-round PCR products were reamplified separately with the primers for these three groups. The F primers used in the first round were replaced by their respective N primers (50 nM each), and all R primers were replaced by a universal primer (0.1 μM) that was identical to the universal tail on the R primers. Each sample also contained 1× PCR buffer (same as above), four dNTPs (100 μM each), and 1.2 U of Taq DNA polymerase with a final volume of 30 μl. The same cycling conditions used in the first round were used, except for the ramping step, which was 3 min instead of 5 min, and the number of cycles, which was reduced from 40 to 25. In addition, for the group 3 primers, the annealing step was 60°C for 2 min, followed by 3 min of ramping to 70°C.
Separation of PCR Products by PAGE
PCR products amplified in the second round were resolved in 8% polyacrylamide minigels. DNA bands in the gels were visualized under UV illumination after staining with ethidium bromide or SYBR (Molecular Probes). Amplified fragments for group 1 were 105–163 bp, whereas those for groups 2 and 3 were 100–189 bp and 119–193 bp, respectively. One locus (68) was not amplified well in the multiplex PCR format in the initial study and was excluded from the later analyses.
Results
Determination of Haplotypes of Individual Sperm from Nine Donors
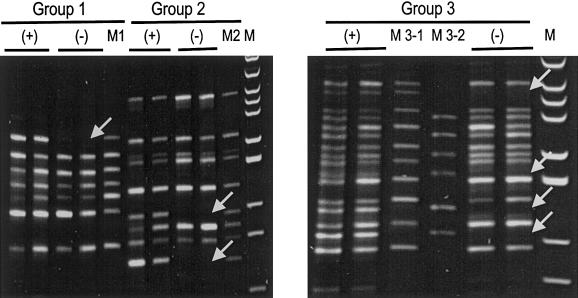
Sperm were purified from semen samples and were fixed and stained with propidium iodide, for fluorescence-activated cell sorting. Single sperm were sorted into 96-well microtiter plates with V bottoms. For each plate, the wells in one row were kept empty and were used as negative controls. After lysis and neutralization, the 32 marker sequences were amplified simultaneously, from single sperm, with a two-round multiplex PCR protocol. In the first round, F and R primers for all 32 markers were used. Amplification with the R primers attached the universal tail sequence to all the amplified products. The lengthy annealing-extension step coupled with ramping helped all the primers with different melting temperatures to anneal to their respective target sequences and to be well extended. In the second round, the markers were amplified in three groups, with 74, 84, 94, 139, 165, 175, and 190 (seven markers) in group 1; 27, 52, 56, 63, 89, 120, 144, 180, and 185 (nine markers) in group 2; and 15, 20, 33, 40, 46, 79, 102, 108, 114, 128, 134, 149, 155, 160, 169, and 195 (16 markers) in group 3. The amplification was achieved by reamplifying three small aliquots (1 μl each) of the first-round PCR products with the respective primers for each group. In the second round, all the F primers were replaced by their respective N primers. N primers were internal and in the same direction with respect to the F primers, to enhance the amplification yield and efficiency. All the R primers were replaced by a universal primer whose sequence was identical to the universal tail at the 5′ ends of the R primers. The final PCR products were resolved in 8% polyacrylamide minigels. Some of the results are shown in figure 2.
Figure 2.
Results from the analysis of two sperm with the insertion (+) haplotype and two with the deletion (−) haplotype. The three groups of primers used in the second round are indicated. M1, M2, and M3-1/M3-2 are mixtures of PCR products amplified with a single pair of primers that belong to the three groups. M is pBR322 DNA digested with MspI and is used as a molecular size marker. Missing bands are indicated with arrows.
Haplotypes Identified among the Nine Donors
Each sperm was scored for the presence or absence of bands corresponding to all 32 marker sequences. On the basis of the banding patterns (fig. 2), the majority of the sperm could be assigned to two groups. One group had sperm with all the expected bands, and the other group had sperm with all but seven bands. These seven bands represented markers 120, 128, 134, 139, 144, 149, and 155, which were clustered. It is very likely that they represented an indel polymorphism that was 35–40 kb in size. Table 1 shows the banding patterns of sperm from one individual, AB027. As shown in table 1, the majority of the single sperm fell into two categories. Within each of these two categories, a few sperm had banding patterns slightly different from the typical ones. These included some sperm with one or very few bands that were not amplified, presumably because of incomplete lysis or amplification failure. Six sperm had one of the seven polymorphic bands amplified, but not the other six. It is likely that this could be due to a low level of contamination. Therefore, these sperm were placed into the group with the deletion haplotype. No bands were amplified from the other six sperm that were placed in a separate group.
Table 1.
Haplotypes of Individual Sperm from Donor AB027[Note]
|
Marker |
||||||||||||||||||||||||||||||||
| Haplotypeand Sperm | 15 | 20 | 27 | 33 | 40 | 46 | 52 | 56 | 63 | 74 | 79 | 84 | 89 | 94 | 102 | 108 | 114 | 120 | 128 | 134 | 139 | 144 | 149 | 155 | 160 | 165 | 169 | 175 | 180 | 185 | 190 | 195 |
| Haplotype 1: | ||||||||||||||||||||||||||||||||
| AB027-1 | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + |
| AB027-4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| AB027-8 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| AB027-12 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| AB027-17 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| AB027-19 | + | + | + | + | + | + | − | + | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + |
| AB027-20 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + |
| AB027-21 | + | + | + | + | − | + | + | + | − | + | + | + | + | + | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| AB027-23 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| AB027-27 | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| AB027-30 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + |
| AB027-31 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + |
| AB027-35 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| AB027-41 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Haplotype 2: | ||||||||||||||||||||||||||||||||
| AB027-2 | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-5 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + |
| AB027-9 | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | − | + | + | + | + | + | + |
| AB027-10 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-11 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-14 | + | + | − | + | − | + | − | + | + | + | + | + | + | + | + | + | + | − | − | − | − | + | − | − | + | + | + | + | − | + | + | + |
| AB027-15 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | + | + | + | + | − | + | + | + |
| AB027-16 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | + | − | − | + | + | + | + | + | + | + | + |
| AB027-18 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-22 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-24 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | − | + | + | + |
| AB027-28 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-29 | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-32 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-33 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | + | − | + | + | + | + | + | + | + | + |
| AB027-36 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-38 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-39 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-40 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-42 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| AB027-6 | − | + | − | − | − | − | + | − | + | + | − | + | + | − | − | + | − | − | − | − | − | − | − | − | + | − | + | − | − | − | + | − |
| AB027-37 | + | − | + | + | − | + | − | − | + | − | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | − | + | + | + |
Note.— No band was amplified from sperm AB027-3, AB027-7, AB027-13, AB027-25, AB027-26, and AB027-34.
Confirmation of the Presence of the Deletion Haplotype
To further confirm the absence of the seven bands, aliquots of the first-round PCR products from a number of single sperm with the missing bands were reamplified separately with the universal primer and single N primers. A few sperm with the insertion haplotype were also included, as positive controls. These experiments were performed for donors AC11, AB027, D17, AB012, and AC09. No bands that were undetectable with multiplex PCR could be detected from the amplification with a single pair of primers.
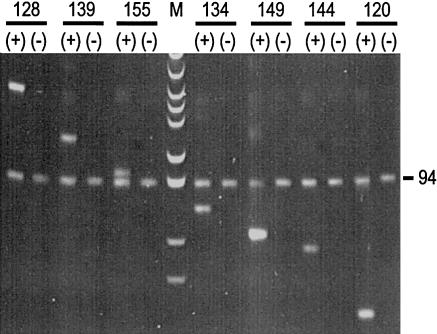
Two-marker PCR was performed for two sperm from donor AB027 with both haplotypes. The so-called “two-marker PCR” was a PCR with one marker in the polymorphic region and another outside the region. PCR products from two sperm were used for the test. One of the two sperm had the deletion haplotype, and the other had the insertion haplotype. Marker 94, which was outside the deletion region, was used as the positive marker of the two markers, and one of the seven polymorphic markers was used as the other marker. This experiment was repeated with all seven polymorphic markers (fig. 3). Both markers could be amplified from the sperm with the insertion haplotype; however, none of the seven polymorphic markers could be amplified from the sperm with the deletion haplotype. Only the marker outside the deletion region could be amplified from this sperm.
Figure 3.
Two-marker PCR, with one marker (94) outside and the other (one of the markers 128, 139, 155, 134, 149, 144 and 120) inside the deletion region. The markers were amplified from one sperm containing the insertion haplotype (+) and another sperm containing the deletion haplotype (−). M is pBR322 DNA digested with MspI and is used as a molecular size marker.
A further confirmation was performed with the donor AC09, both of whose haplotypes had the deletion. Two-marker PCR was performed, starting from the first round from six sperm. This experiment was also performed for all seven polymorphic markers. Only the marker that was outside the deletion region could be amplified, and none of the seven loci that were within the deletion region could be amplified.
Haplotypes of the Nine Donors
The derived haplotypes for the nine donors are summarized in table 2. Of these nine individuals, four (D18, D20, AB005, and 002) had sperm with only the insertion haplotype, and another four (AC11, AB027, D17, and AB012) had both insertion and deletion haplotypes. One individual (AC09) had all sperm with the deletion haplotype and was, therefore, homozygous for the deletion haplotype. The ratio (2:1, insertion:deletion) of the two haplotypes and the ratio (4:4:1) of the genotypes for the nine individuals perfectly matched the expected ratios, if one assumes that the haplotypes are in the Hardy-Weinberg equilibrium.
Table 2.
Haplotypes of the Nine Donors
|
Marker |
||||||||||||||||||||||||||||||||
| IndividualandHaplotype | 15 | 20 | 27 | 33 | 40 | 46 | 52 | 56 | 63 | 74 | 79 | 84 | 89 | 94 | 102 | 108 | 114 | 120 | 128 | 134 | 139 | 144 | 149 | 155 | 160 | 165 | 169 | 175 | 180 | 185 | 190 | 195 |
| I-D18: | ||||||||||||||||||||||||||||||||
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I-D20: | ||||||||||||||||||||||||||||||||
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I-AB005: | ||||||||||||||||||||||||||||||||
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I-002: | ||||||||||||||||||||||||||||||||
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I-AC11: | ||||||||||||||||||||||||||||||||
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| I-AB027: | ||||||||||||||||||||||||||||||||
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| I-D17: | ||||||||||||||||||||||||||||||||
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| I-AB012: | ||||||||||||||||||||||||||||||||
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| I-AC09: | ||||||||||||||||||||||||||||||||
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
Discussion
A large number of polymorphisms have been detected by Southern analyses in the VH region (Turnbull et al. 1987; Walter et al. 1987; Sam et al. 1988; Walter and Cox 1988; Willems van Dijk et al. 1989, 1991, 1993; Guillaume et al. 1990; Sasso et al. 1990; Weng et al. 1992; Adderson et al. 1993). The polymorphic band accounted for a large fraction (30%–88%) of the total hybridization bands. These results cannot be explained by the polymorphisms consisting of minor sequence variations, such as single-base substitutions and small indels, because these types of sequence variations rarely affect the restriction fragments resolved by Southern analyses. Although the presence of relatively large indel polymorphisms in the human Ig VH region have been implicated in many studies, it is difficult to obtain precise information about the extent, locations, and impact of these polymorphisms, since they are mostly present in a heterozygous state and, so far, most of the studies of the VH region have been performed using diploid materials. The present study is, to our knowledge, the first to show the ability to identify indel polymorphisms by analysis of single sperm. Because single sperm are haploid, homologous chromosomes of different parental origins in each sperm donor can be studied separately, and complications with diploid materials can be avoided. Through use of this method, we identified an indel polymorphism with a precise size and location. Three VH gene segments (1-8, 3-9, and 2-10P), of which two (3-9 and 1-8) are functional, are affected by this indel polymorphism (fig. 1). It would be very interesting to learn the biological impact of this polymorphism, since ∼10% of individuals in the human population would be homozygous for the deletion haplotype and 44% would have only one copy of the two affected VH gene segments.
Because of the complications with diploid materials, the identification of chromosomal deletions by PCR has so far been used for the Y chromosome (Bor et al. 2001; Yao et al. 2001; Peterlin et al. 2002), because all males are haploid with respect to the Y chromosome, although it shares a certain amount of sequence identity with the X chromosome. In this case, an unlimited amount of DNA from somatic cells can be used for the study. In fact, in most of the studies, the amount (50–500 ng) of genomic DNA used was relatively large. In the present study, the subjects—that is, single sperm—contained only one copy of the sequences that needed to be analyzed. Our group has already developed a highly efficient multiplex PCR protocol that can be used to amplify a large number of loci that could be resolved either by conventional PAGE or by denaturing gradient gel electrophoresis (Lin et al. 1996; Cui and Li 1998; Cui et al. 2000). Very recently, we showed that 10 independent sequences may be amplified simultaneously from single diploid cells (Cui et al. 2002). In the present study, we used a subgrouping strategy in the second round of amplification and successfully amplified 32 marker sequences from single sperm, compared with 5 or 6 in most studies with large amounts of DNA. Such a capacity and sensitivity allowed us to examine a region with a high marker density. Further increases in marker density may allow us not only to locate the borders of the deletion but also to determine the sequences at these borders. The results could be very informative for understanding the mechanisms underlying VH region diversity.
When studying indel polymorphisms by PCR, extra precautions need to be taken. Conclusions cannot be drawn unless real deletions and failures in PCR amplification can be confidently distinguished from each other. In the present study, we confirmed our results by reamplifying the first-round PCR product with a single pair of primers or with two pairs of primers, of which one pair was used to amplify a sequence outside the deleted region, as an internal positive control. To further confirm the deletion, we reamplified sperm from a donor who was homozygous for the deletion, through use of the two-marker PCR protocol. All results were consistent with those from the multiplex amplification. The indel polymorphism detected in the present study is further confirmed by the fact that the ratio of the sperm donor genotypes perfectly matched the expectation based on Hardy-Weinberg equilibrium. In fact, multiplex amplification itself is a powerful method for confirmation. When a group of clustering markers is missing, it is a strong indication of deletion, since the chance of failing to amplify seven clustered yet independent markers would be very, very low.
The ability to analyze indel polymorphisms could have a significant impact on our understanding of human genome diversity. Many chromosomal regions, such as gene complexes encoding Ig light chains, T cell receptors, and human leukocyte antigen, may have a structure similar to that of the VH region and therefore may be analyzed by the method used in the present study. The method may also be used to understand the physical structure of the entire human genome. Although the Human Genome Project has been completed, the extent of human genome diversity remains unclear. Large or relatively large indel polymorphisms may be abundant in the human genome, and at least a portion of these polymorphisms may contain genes. On the one hand, the affected genes would be completely missing in the homozygous state of the deletion allele, and it may have a significant physiological impact. On the other hand, the hemizygous state of the deletion may confer constitutional cancer susceptibility if some tumor suppressor genes are located in the deleted regions, since the only copy of the affected genes could be lost or inactivated in some cells, by mechanisms such as mutation or loss of heterozygosity.
Acknowledgments
The authors would like to thank Dr. Chimgee Nyam-Osor and Xiangfeng Cui for their experimental help and suggestions. This work was supported by National Institute of Allergy and Infectious Diseases grant R01 AI45956 (to H.L.).
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- Authors' Web site, http://www2.umdnj.edu/lilabweb/SupplementaryData.htm (for primer sequences)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for accession number AB019440)
References
- Adderson EE, Azmi FH, Wilson PM, Shackelford PG, Carroll WL (1993) The human VH3b gene subfamily is highly polymorphic. J Immunol 151:800–809 [PubMed] [Google Scholar]
- Berman JE, Mellis SJ, Pollock R, Smith CL, Suh H, Heinke B, Kowal C, Surti U, Chess L, Cantor CR, Alt FW (1988) Content and organization of the human Ig VH locus: definition of three new VH families and linkage to the Ig CH locus. EMBO J 7:727–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bor P, Hindkjaer J, Ingerslev HJ, Kolvraa S (2001) Multiplex PCR for screening of microdeletions on the Y chromosome. J Assist Reprod Genet 18:291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodeur PH, Riblet R (1984) The immunoglobulin heavy chain variable region (IgH-V) locus in the mouse. I. One hundred IgH-V genes comprise seven families of homologous genes. Eur J Immunol 14:922–930 [DOI] [PubMed] [Google Scholar]
- Buluwela L, Rabbitts TH (1988) A VH gene is located within 95 Kb of the human immunoglobulin heavy chain constant region genes. Eur J Immunol 18:1843–1845 [DOI] [PubMed] [Google Scholar]
- Chen PP, Yang PM (1990) A segment of human VH gene locus is duplicated. Scand J Immunol 31:593–599 [DOI] [PubMed] [Google Scholar]
- Cherif D, Berger R (1990) New localizations of VH sequences by in situ hybridization with biotinylated probes. Genes Chromosomes Cancer 2:103–108 [DOI] [PubMed] [Google Scholar]
- Cook GP, Tomlinson IM, Walter G, Riethman H, Carter NP, Buluwela L, Winter G, Rabbitts TH (1994) A map of the human immunoglobulin VH locus completed by analysis of the telomeric region of chromosome 14q. Nat Genet 7:162–168 [DOI] [PubMed] [Google Scholar]
- Cox DW, Markovic VD, Teshima IE (1982) Genes for immunoglobulin heavy chains and for α1-antitrypsin are localized to specific regions of chromosome 14q. Nature 297:428–430 [DOI] [PubMed] [Google Scholar]
- Croce CM, Shander M, Martinis J, Cicurel L, D'Ancona GG, Dolby TW, Koprowski H (1979) Chromosomal location of the genes for human immunoglobulin heavy chains. Proc Natl Acad Sci USA 76:3416–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Feiner H, Li H (2002) Multiplex loss of heterozygosity analysis by using single or very few cells. J Mol Diagn 4:172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Feiner H, Lin Z, Li H (2000) Multiplex genotype analysis of invasive carcinoma and accompanying proliferative lesions microdissected from breast tissue. J Mol Diagn 2:29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Li H (1997) Discriminating between allelic and interlocus differences among human immunoglobulin VH4 sequences by analyzing single spermatozoa. Hum Genet 100:96–100 [DOI] [PubMed] [Google Scholar]
- ——— (1998) Determination of gene organization in individual haplotypes by analyzing single DNA fragments from single spermatozoa. Proc Natl Acad Sci USA 95:10791–10796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Li H, Goradia TM, Lange K, Kazazian HHJ, Galas D, Arnheim N (1989) Determination of genetic distance between the Gγ globin and parathyroid hormone loci using the polymerase chain reaction and allele specific oligomers. Proc Natl Acad Sci USA 86:9389–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume T, Rubinstein DB, Young F, Tucker L, Logtenberg T, Schwartz RS, Barrett KJ (1990) Individual VH genes detected with oligonucleotide probes from the complementarity-determining regions. J Immunol 145:1934–1945 [PubMed] [Google Scholar]
- Haino M, Hayashida H, Miyata T, Shin EK, Matsuda F, Nagaoka H, Matsumara R, Taka-ishi S, Fukita Y, Fujikura J, Honjo T (1994) Comparison and evolution of human immunoglobulin VH segments located in the 3′ 0.8 megabase region: evidence for unidirectional transfer of segmental gene sequences. J Biol Chem 269:2619–2626 [PubMed] [Google Scholar]
- Humphries CG, Shen A, Kuziel WA, Capra JD, Blattner FR, Tucker PW (1988) A new human immunoglobulin VH family preferentially rearranged in immature B-cell tumors. Nature 331:446–449 [DOI] [PubMed] [Google Scholar]
- Kirsch IR, Morton CC, Nakahara K, Leder P (1982) Human immunoglobulin heavy chain genes map to a region of translocations in malignant B lymphocytes. Science 216:301–303 [DOI] [PubMed] [Google Scholar]
- Lee KH, Matsuda F, Kinashi T, Kodaira M, Honjo T (1987) A novel family of variable region genes of the human immunoglobulin heavy chain. J Mol Biol 195:761–768 [DOI] [PubMed] [Google Scholar]
- Lin Z, Cui X, Li H (1996) Multiplex genotype determination at a large number of gene loci. Proc Natl Acad Sci USA 93:2582–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, Honjo T (1998) The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med 188:2151–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Shin EK, Nagaoka H, Matsumura R, Haino M, Fukita Y, Taka-ishi S, Imai T, Riley JH, Anand R, Soeda E, Honjo T (1993) Structure and physical map of 64 variable segments in the 3′ 0.8-megabase region of the human immunoglobulin heavy-chain locus. Nat Genet 3:88–94 [DOI] [PubMed] [Google Scholar]
- Matthyssens G, Rabbitts TH (1980) Structure and multiplicity of genes for the human immunoglobulin heavy chain variable region. Proc Natl Acad Sci USA 77:6561–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner EC, Hufnagle WO, Glas AM, Suzuki I, Alexander C (1995) Polymorphism and utilization of human VH genes. Ann NY Acad Sci 764:50–61 [DOI] [PubMed] [Google Scholar]
- Nagaoka H, Ozawa K, Matsuda F, Hayashida H, Matsumura R, Haino M, Shin EK, Fukita Y, Imai T, Anand R, Yokoyama K, Eki T, Soeda E, Honjo T (1994) Recent translocation of variable and diversity segments of the human immunoglobulin heavy chain from chromosome 14 to chromosomes 15 and 16. Genomics 22:189–197 [DOI] [PubMed] [Google Scholar]
- Peterlin B, Kunej T, Sinkovec J, Gligorievska N, Zorn B (2002) Screening for Y chromosome microdeletions in 226 Slovenian men. Hum Reprod 17:17–24 [DOI] [PubMed] [Google Scholar]
- Pincus SH (1988) Human immunoglobulin heavy-chain-variable (VH) region gene families defined by hybridization with cloned human and murine VH-genes. Hum Immunol 22:199–215 [DOI] [PubMed] [Google Scholar]
- Rubinstein DB, Symann M, Stewart AK, Guillaume T (1993) Restriction fragment length polymorphisms and single germline coding region sequence in VH18/2, a duplicated gene encoding autoantibody. Mol Immunol 30:403–412 [DOI] [PubMed] [Google Scholar]
- Sam M, Walter MA, Cox DW (1988) RsaI polymorphism of a human immunoglobulin VH5 subclass locus. Nucleic Acids Res 16:8748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasso EH, Buckner JH, Suzuki LA (1995) Ethnic differences of polymorphism of an immunoglobulin VH3 gene. J Clin Invest 96:1591–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasso EH, Willems van Dijk K, Bull AP, Milner EC (1993) A fetally expressed immunoglobulin VH1 gene belongs to a complex set of alleles. J Clin Invest 91:2358–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasso EH, Willems van Dijk K, Bull A, van der Maarel SM, Milner EC (1992) VH genes in tandem array comprise a repeated germline motif. J Immunol 149:1230–1236 [PubMed] [Google Scholar]
- Sasso EH, Willems van Dijk K, Milner EC (1990) Prevalence and polymorphism of human VH3 genes. J Immunol 145:2751–2757 [PubMed] [Google Scholar]
- Schroeder HWJ, Hillson JL, Perlmutter RM (1987) Early restriction of the human antibody repertoire. Science 238:791–793 [DOI] [PubMed] [Google Scholar]
- Shin EK, Matsuda F, Nagaoka H, Fukita Y, Imai T, Yokoyama K, Soeda E, Honjo T (1991) Physical map of the 3′ region of the human immunoglobulin heavy chain locus: clustering of autoantibody-related variable segments in one haplotype. EMBO J 10:3641–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souroujon MC, Rubinstein DB, Schwartz RS, Barrett KJ (1989) Polymorphism in human H chain V region genes from the VHIII gene family. J Immunol 143:706–711 [PubMed] [Google Scholar]
- Tomlinson IM, Cook GP, Carter NP, Elaswarapu R, Smith S, Walter G, Buluwela L, Rabbitts TH, Winter G (1994) Human immunoglobulin VH and D segments on chromosomes 15q11.2 and 16p11.2. Hum Mol Genet 3:853–860 [DOI] [PubMed] [Google Scholar]
- Tomlinson IM, Walter G, Marks JD, Llewelyn MB, Winter G (1992) The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J Mol Biol 227:776–798 [DOI] [PubMed] [Google Scholar]
- Turnbull IF, Bernard O, Sriprakash KS, Mathews JD (1987) Human immunoglobulin variable region genes: a new VH sequence used to detect polymorphism. Immunogenetics 25:184–192 [DOI] [PubMed] [Google Scholar]
- Walter G, Tomlinson IM, Cook GP, Winter G, Rabbitts TH, Dear PH (1993) HAPPY mapping of a YAC reveals alternative haplotypes in the human immunoglobulin VH locus. Nucleic Acids Res 21:4524–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MA, Cox DW (1988) Analysis of genetic variation reveals human immunoglobulin VH-region gene organization. Am J Hum Genet 42:446–451 [PMC free article] [PubMed] [Google Scholar]
- ——— (1991) Nonuniform linkage disequilibrium within a 1,500-kb region of the human immunoglobulin heavy-chain complex. Am J Hum Genet 49:917–931 [PMC free article] [PubMed] [Google Scholar]
- Walter MA, Linsley PS, Cox DW (1987) ApaI polymorphism of a human immunoglobulin VH3 subclass locus. Nucleic Acids Res 15:4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MA, Surti U, Hofker MH, Cox DW (1990) The physical organization of the human immunoglobulin heavy chain gene complex. EMBO J 9:3303–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng NP, Snyder JG, Yu-Lee LY, Marcus DM (1992) Polymorphism of human immunoglobulin VH4 germ-line genes. Eur J Immunol 22:1075–1082 [DOI] [PubMed] [Google Scholar]
- Willems van Dijk K, Mortari F, Kirkham PM, Schroeder HWJ, Milner EC (1993) The human immunoglobulin VH7 gene family consists of a small, polymorphic group of six to eight gene segments dispersed throughout the VH locus. Eur J Immunol 23:832–839 [DOI] [PubMed] [Google Scholar]
- Willems van Dijk K, Sasso EH, Milner EC (1991) Polymorphism of the human Ig VH4 gene family. J Immunol 146:3646–3651 [PubMed] [Google Scholar]
- Willems van Dijk K, Schroeder HWJ, Perlmutter RM, Milner EC (1989) Heterogeneity in the human immunoglobulin VH locus. J Immunol 142:2547–2554 [PubMed] [Google Scholar]
- Yang PM, Olsen NJ, Siminovitch KA, Olee T, Kozin F, Carson DA, Chen PP (1990) Possible deletion of a developmentally regulated heavy-chain variable region gene in autoimmune diseases. Proc Natl Acad Sci USA 87:7907–7911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Chen G, Pan T (2001) Study of microdeletions in the Y chromosome of infertile men with idiopathic oligo- or azoospermia. J Assist Reprod Genet 18:612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]