Significance
Chromatin, the functional form of DNA inside the cell nucleus, has been heavily studied using biochemical and genetic methods, but we still know little about its large-scale organization and even less about its dynamics. We present a unique method, displacement correlation spectroscopy (DCS), which allows us to map interphase chromatin dynamics simultaneously across the entire nucleus and follow the temporal evolution of the global chromatin dynamics in vivo. Using DCS we discovered that chromatin moves coherently across micron-scale regions for a few seconds, which implies a transient mechanical coupling between chromatin on different chromosomes. This coupling might allow different regions of the nucleus to communicate by a unique, mechanochemical mechanism, e.g., to coordinate responses to DNA damage.
Keywords: self-organization, active materials, soft matter
Abstract
Chromatin structure and dynamics control all aspects of DNA biology yet are poorly understood, especially at large length scales. We developed an approach, displacement correlation spectroscopy based on time-resolved image correlation analysis, to map chromatin dynamics simultaneously across the whole nucleus in cultured human cells. This method revealed that chromatin movement was coherent across large regions (4–5 µm) for several seconds. Regions of coherent motion extended beyond the boundaries of single-chromosome territories, suggesting elastic coupling of motion over length scales much larger than those of genes. These large-scale, coupled motions were ATP dependent and unidirectional for several seconds, perhaps accounting for ATP-dependent directed movement of single genes. Perturbation of major nuclear ATPases such as DNA polymerase, RNA polymerase II, and topoisomerase II eliminated micron-scale coherence, while causing rapid, local movement to increase; i.e., local motions accelerated but became uncoupled from their neighbors. We observe similar trends in chromatin dynamics upon inducing a direct DNA damage; thus we hypothesize that this may be due to DNA damage responses that physically relax chromatin and block long-distance communication of forces.
The structure and dynamics of interphase chromatin are major unsolved questions in the biology of eukaryotic cells. Chromatin is composed of DNA wrapped around nucleosomes made of core histones (H2A, H2B, H3, and H4), but its structure at larger length scales is controversial (1). Despite the lack of physical compartmentalization of the nucleus, chromatin appears to be highly organized. Single chromosomes occupy distinct territories in the 3D landscape of the nucleus (2–4). Chromatin cross-linking is starting to reveal complex patterns of genome folding with 1-Mbp resolution (5), but this provides a static picture, and the organization of interphase chromatin is highly dynamic in living cells (6–9): Chromosomes with active genes tend to locate in the nuclear interior, whereas chromosomes with inactive genes reside mostly at the nuclear periphery. Further, active genes tend to locate toward the surface of chromosome territories, whereas the silenced genes tend to remain inside territories. Moreover, in the case of gene coregulation two distinct genes share the same transcription factor, which requires them to be in close proximity (6). In general, it is not clear to what extent gene activity influences gene position or gene position dictates the gene activity. This question is complicated by gene expression and gene localization changes as a function of cell type, cell cycle state, and response to exogenous stimuli (6–11). Because all these factors act on gene dynamics, it is likely dynamic measurements methods will be required to resolve the activity–position problem for genes.
In addition to gene repositioning, chromatin remodeling, i.e., local dynamic modifications of chromatin architecture, contributes to chromatin’s complex dynamics (6, 7). Chromatin remodeling plays an epigenetic regulatory role in all DNA manipulations, e.g., gene transcription, replication, DNA repair, apoptosis, or development (12), which are again all dynamic processes of their own, powered by different ATP-dependent nuclear enzymes (e.g., RNA polymerase, helicase, etc.). Clearly, chromatin dynamics are very complex and contain many layers of convoluted dynamic processes operating at different timescales and length scales.
To date, studies of chromatin dynamics consist largely of photobleaching (13–16) and photoactivation techniques (17, 18), which visualize turnover of nuclear proteins and movement of whole sections of the nucleus, and tracking of chromatin-attached single particles to quantify movement dynamics (19–30). Single particles within chromatin that have been tracked include fluorescently labeled single-DNA sites (19–22), nuclear proteins (23–26), chromosome territories (27, 28) and subchromosomal foci (29, 30). These studies revealed dynamics that can be largely described as diffusive or subdiffusive with constraint, with few instances showing an apparent directional movement for single genes (19, 20). How to reconcile these static and dynamic pictures to provide a physical picture of the interphase nucleus is unclear. The physical nature of a bulk material cannot be understood from tracking single points within it, but approaches for probing bulk chromatin dynamics in vivo have lagged (14).
Here, we map chromatin dynamics across the entire nucleus and follow the spatiotemporal evolution of the global chromatin dynamics in vivo in nuclei of human HeLa cells. To monitor chromatin dynamics simultaneously across an entire eukaryotic nucleus we imaged GFP-tagged histone H2B, a widely used and reliable reporter of chromatin position (16). Positional fluctuations in the local intensity of H2B-GFP report directly on positional fluctuations of chromatin (Fig. 1A). We recorded 25-s sequences of H2B-GFP imaged by confocal microscopy in a single plane, typically in the middle of the nucleus, at fairly high resolution (∼65 nm in x and y, 250 ms per frame). Our analytic tool, displacement correlation spectroscopy (DCS), uses algorithms of particle image velocimetry (PIV) to measure the direction and magnitude of local movements simultaneously across the entire nucleus and across the entire temporal spectrum of the experiment. Cross-correlation between pairs of images reported on local chromatin displacements between time intervals, and varying their time interval sampled all time intervals in the experiment, typically from 250 ms to 25 s (Materials and Methods).
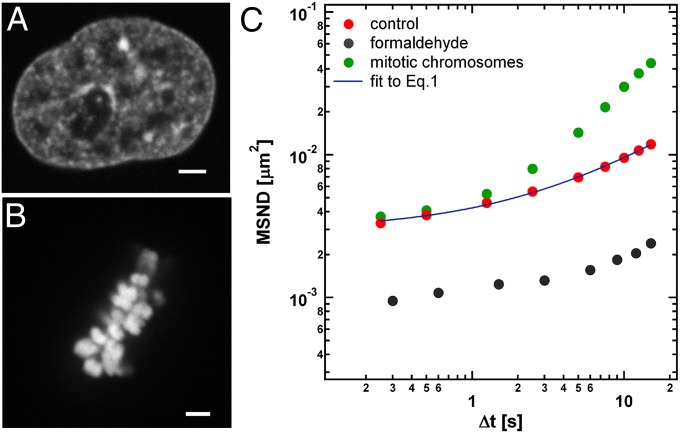
Fig. 1.
Bulk chromatin dynamics. (A) Image of an interphase nucleus expressing H2B-GFP. (B) Mitotic chromosome cluster expressing H2B-GFP. (C) MSND(Δt)= <|d→(r→,Δt)|2>, calculated from displacements d→(r→,Δt) measured by DCS averaging over 16 cells in interphase (red circles). Blue solid line represents the fit of the experimental data to Eq. 1. MSND measured for cells fixed in formaldehyde (gray circles) reveals much lower MSND values, thus confirming that measured values are above the noise floor. MSND measured for mitotic chromosomes (green circles) serves as a positive control. (Scale bar, 2 µm.)
Results and Discussion
To obtain a spatiotemporal evolution of chromatin dynamics across an entire nucleus we used a unique approach we termed DCS (Materials and Methods and Fig. S1). We first calculated mean square network displacement (MSND) as a function of time, which we defined as MSND(Δt) = <|d→(r→,Δt)|2>, where d→(r→,Δt) are displacements measured by DCS, averaging across 16 nuclei (Fig. 1C). We found that MSND follows a power law,
the best fit yielding A = 0.00297 ± 0.00012 µm2, B = 0.00128 ± 0.00011 µm2⋅s−1, and α= 0.71 ± 0.03 (Fig. 1C). Interestingly, our model required a constant A to account for the initial jump of MSND at short times. As a positive control we measured MSND for mitotic chromosomes (Fig. 1B), which are known to undergo larger movements (31), and found consistent with previous observations that MSND values for mitotic chromosomes are much larger than MSND of interphase chromatin (Fig. 1C). MSND measured for cells fixed in formaldehyde revealed much lower MSND values, confirming that measured values were above the noise floor for the instrument and analysis method (Fig. 1C). The constant A might therefore account for another, superimposed type of chromatin fluctuations characterized by shorter timescales. Consistent with previous work, the fit and parameter values were reminiscent of mean square displacement (MSD) for single, tagged genes measured at high temporal resolution (∼30 ms) (19), where the first term was described by another type of movement, constrained fast diffusion, superimposed on the slower motion, which was freely diffusive, unlike subdiffusive in our case. Their model for single-genes movement yielded B = 0.00096 µm2⋅s−1, assuming free diffusion (α= 1) (19). The fast motion at the short times is also consistent with recently observed local nucleosome dynamics (32). Thus, where a direct comparison between DCS and previous methods was possible, the results were similar.
Unlike previous methods, DCS tracked chromatin simultaneously in the whole nucleus with subpixel spatial resolution, so we were able to ask whether chromatin motion was spatially correlated over any time interval in our data. We first plotted d→(r→,Δt) maps of movement for different intervals, color coding displacement vectors by direction to reveal directional correlation. At time intervals <0.5 s the maps contained mostly uncorrelated displacements (Fig. 2A). Small patches of displacements of the same directions were caused partly by overlap of the interrogation windows for local image correlation in our technique (Materials and Methods). At intervals of ∼1–10 s the maps repeatedly revealed large regions of similar color, i.e., locally correlated motions (Fig. 2 B–D). These regions typically spanned several micrometers. Importantly, the appearance of these regions was independent of the size and overlap of the interrogation windows used by DCS (Materials and Methods), thus ruling out a measurement artifact. Different regions in the same nucleus underwent correlated movement in different directions (Fig. 2 B–D), so the maps were not reporting translation or rotation of the entire nucleus, but rather relative displacements of regions within a single nucleus. Sequential maps of the same nucleus showed that regions of correlated movement did not persist (Fig. 2D). Instead, over tens of seconds they broke up, reversed, or in other ways appeared to randomize. Thus, motion was not coherent or unidirectional over long timescales, but it was strikingly correlated over microns and tens of seconds.
Fig. 2.
Local coherence in interphase chromatin dynamics. Shown are DCS maps of the same nucleus calculated for different time intervals Δt: (A) Δt = 0.25 s, (B) Δt = 2.5 s, and (C) Δt = 5 s. Displacement vectors are color coded by their direction to reveal directional correlation. In A the motion is uncorrelated, whereas in B and C regions of correlated motion can be observed. (D) Sequential DCS maps calculated at different times t for time interval Δt = 10 s. The regions of correlated motion seem to disintegrate over tens of seconds, while new regions of correlated motions are formed. Figs. S2 and S3 show corresponding confocal microscopy images and high-resolution images of A–D, respectively. (E) We calculate average spatial displacement autocorrelation function <cdx(Δr,Δt)> for a Δt by averaging over all cdx(Δr,Δt) calculated for single DCS fields at given Δt. The plot shows <cdx(Δr,Δt)> for different Δt. First, an increase in correlation can be observed for Δt = 0.25–5 s, then the correlation reaches its maximum at Δt = 5–10 s, and then it decreases again. (F) Examples of cdx(Δr,Δt) calculated for single fields for Δt = 2.5 s, 5 s, and 10 s. The log-log plots of cdx(Δr,Δt) show an initial power law behavior followed by an abrupt fall consistent with a power law with an exponential cutoff. We fit cdx(Δr,Δt) of single fields to Eq. 2 to obtain the scaling exponent n and the correlation length ξ (yellow solid line). (G) Plot of the average correlation length ξ and scaling exponent n as a function of Δt. We obtain ξ and n by fitting cdx(Δr,Δt) of single DCS fields to Eq. 2 and then averaging over all fields at a Δt and all cells (n = 16) . (Scale bar, 2 µm.)
To quantify spatial correlation of motions we computed the spatial autocorrelation function cdx(Δr→,Δt) =<dx(r→,Δt), dx(r→ + Δr→,Δt)> for the x-component dx of the measured displacements d→(r→,Δt). Fig. 2E shows a plot of radially averaged <cdx(Δr,Δt)> for different Δt, where < > denotes the average over all cdx(Δr,Δt) obtained for all DCS fields at given Δt. These traces quantify an unprecedented type of dynamics. For time intervals between 0.25 s and 5 s, spatial correlation in movement increases with increasing time. Correlation reaches a maximum at 5–10 s and then decreases again. These curves are consistent with visual inspection of maps (Fig. 2 A–D). Regions of correlated motion were most evident for 5- to 10-s intervals and became less evident for shorter and longer intervals. To investigate the time-dependent behavior of <cdx(Δr,Δt)>, we analyzed every cdx(Δr,Δt) separately (Fig. 2F). We found that cdx(Δr,Δt) of a single DCS field shows an initial power law behavior followed by an abrupt fall consistent with a power law with an exponential cutoff. We fitted cdx(Δr,Δt) to
This fit yielded an average exponent n ≈ −0.5 for all times and ξ having a time-dependent behavior: increasing for 0.25–5 s, reaching its maximum ξ ≈ 5 µm at 5–10 s, and then decreasing again (Fig. 2G). Time-dependent behavior of ξ provides a direct measure of the lifetime of the correlated motion τ∼ 10 s. Both the correlation length ξ and the lifetime of the correlated motion τ agree with visual inspection of vector maps for 2.5- to 10-s correlation intervals, where regions of correlated motion are of the same order.
One potential explanation of correlated motion on the micron scale is that single chromosomes in their interphase territories tend to move as a single unit. The observed correlation length ξ ≈ 5 µm is comparable to the size of single-chromosome territories seen previously in fixed cells by fluorescence in situ hybridization (FISH) (2). To test this, we visualized territories, using a published method (33) in which DNA strands were labeled during replication with Cy3-dCTP and then diluted with unlabeled strands by allowing cell division without label (Fig. 3B). We then overlaid DCS-derived maps of histone H2B-GFP movement with Cy3-dCTP marked territories (Fig. 3D). We found instances where boundaries of regions of correlated motion corresponded to boundaries between labeled and unlabeled territories (Fig. 3D, Inset A) but also many examples where regions of correlated motion spanned across several territories (Fig. 3D, Inset B). We conclude that regions of correlated motion are not chromosome territories in most cases. Fig. 3 E and F provide a cartoon view of our data; yellow arrows denote motion within the regions of coherent motion (red or blue box), and green and blue circles denote neighboring territories.
Fig. 3.
Regions of coherent motion vs. chromatin territories. (A) A fluorescent image of an interphase nucleus expressing H2B-GFP. (B) Chromosome territories visualized by incorporation of Cy3-dCTP. (C) An overlay of A and B. (D) An overlay of B and DCS vector map. There are instances where boundaries of regions of correlated motion correspond to boundaries between labeled and unlabeled territories (Inset A). However, we also find many examples where regions of correlated motion span across several territories (Inset B). Therefore, we conclude that regions of correlated motion are not chromosome territories in most cases. Fig. S4 shows a high-resolution image (D). (E and F) A model for the two observed regimes of the chromatin dynamics. Blue and green chains represent distinct chromosomes. In E, regions of coherent motion correspond to chromosome territories; in F, regions of coherent motion span beyond the chromosome territories; i.e., parts of different chromosomes are moving coherently (red or blue box). (Scale bar, 2 µm.)
To explore the origin and organization underlying the correlated motion we investigated its ATP dependence. ATP depletion blocked correlation in bulk chromatin dynamics completely. It also strongly reduced all movement. MSND(t) shows a strong reduction in measured displacements, yielding A = 0.00199 ± 0.00014 µm2 and α = 0.32 ± 0.03, when fitted to Eq. 1 (Fig. 4G), averaging across 10 nuclei. The exponent α is strongly reduced. In addition to a reduction in the extent of the motion, we observed an apparent chromatin condensation in images (Fig. 4B), which concurs with overall higher compaction reported earlier by fluorescence lifetime imaging microscopy (FLIM)-FRET studies (34). Thus, chromatin requires ATP to preserve its spread out, dynamic state.
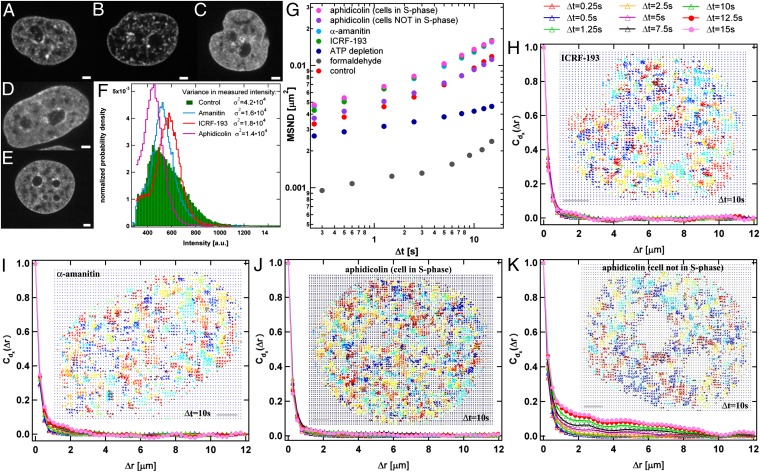
Fig. 4.
Chromatin perturbations. Shown are changes in phenotype upon perturbations: (A–E) control (A); after ATP depletion (B); and upon addition of ICRF-193 (topoisomerase II inhibitor) (C), α-amanitin (polymerase II inhibitor) (D), and aphidicolin (DNA polymerase inhibitor) (E). (F) Histograms of normalized intensities for A–E demonstrate the difference in the variance σ2 of fluorescence intensity distributions corresponding to changes in chromatin concentration distributions. After DNA damage is induced (C–E), σ2 is lowered compared with control (F). (G) MSND measured for aphidicolin (n = 15), ICRF-193 (n = 9), and α-amanitin (n = 9) shows an increase in local displacements, whereas ATP depletion (n = 10) causes a reduction in local displacements. As a negative control we measured displacement in a sample fixed by formaldehyde (n = 5). Note the S-phase–specific behavior of cells treated with aphidicolin; cells in S phase (pink circles) react to aphidicolin perturbation, whereas cells not in S phase (purple circles) retain physiological behavior. All three drugs cause strong inhibition of local coherence: (H) ICRF-193, (I) α-amanitin, and (J) aphidicolin. For aphidicolin the effect is seen only in ∼35% of cells, which are shown by EdU stain to be in early-to-mid S phase, confirming the aphidicolin effect is S-phase specific (Fig. S6). (K) Cells not in S phase retained the local coherence in the chromatin dynamics upon treatment with aphidicolin. Fig. S7 shows high-resolution images of H–K. (Scale bar, 2 µm.)
The mechanochemical origin of chromatin movement is unclear. Lack of an effect of microtubule- and actin-targeting drugs showed that motion is generated within the nucleus and not by the action of cytoplasmic motors on the nuclear periphery (Fig. S5). As a start to investigating regulation and function of correlated motion of chromatin, we investigated the effect of drugs that target important nuclear enzymes: aphidicolin (DNA polymerase inhibitor), α-amanitin (RNA polymerase II inhibitor), and ICRF-193 (topoisomerase II inhibitor). In addition to inhibiting different nuclear enzymes with high specificity, these drugs all trigger DNA damage responses (35–37). We first determined their effects on chromatin architecture by measuring homogeneity in local density of H2B-GFP, from spatial variance σ2 of the fluorescence intensity (Fig. 4 C–F). As previously reported, DNA-damaging drugs caused chromatin to become more diffuse and homogeneous in staining, the opposite of the effect of ATP depletion (34, 38, 39). For aphidicolin the effect was seen in only ∼35% of cells. We suspected this is the fraction of cells that is undergoing DNA replication and tested it by incorporating EdU for 1 h to mark replicating cells (40), then adding aphidicolin, and then measuring chromatin morphology and EdU incorporation. All of the cells where chromatin became more homogeneous (lowered σ2) had been replicating (n = 23 of 67), confirming that aphidicolin’s effect on chromatin structure, and therefore presumably on dynamics, is S-phase specific (Fig. S6).
Analysis of H2B-GFP image sequences with DCS revealed that all three drugs caused strong inhibition of local coherence, and all three drugs increased local displacements (Fig. 4G); i.e., local motions were accelerated but became uncoupled from their neighbors (Fig. 4 H–K). MSND(t) fitted to Eq. 1 yielded A = 0.00324 ± 0.00012 µm2 and α= 0.52 ± 0.012 for aphidicolin, A = 0.00363 ± 0.00014 µm2 and α= 0.59 ± 0.02 for α-amanitin, and A = 0.00285 ± 0.00024 µm2 and α= 0.53 ± 0.02 for ICRF-193. We find similar behavior upon inducing direct DNA damage by treating the cells with neocarzinostatin (NCS), which causes DNA double-strand breaks directly, yielding A = 0.00338 ± 0.00016 µm2 and α= 0.59 ± 0.02 for MSND(t) fitted to Eq. 1 (Fig. S8). These data can be interpreted in two ways: Decreased condensation of chromatin, caused by the DNA damage response, may remove the physical links needed for coordination of motion over microns. Alternatively, decreased coordination may cause a decrease in condensation. In either case, our data revealed a unique aspect of how chromatin responds to DNA damage at a physical level. To decouple the contribution of the DNA damage response from loss of function of the drug target (DNA polymerase, RNA polymerase II, and topoisomerase II), we blocked ATR kinase and Chk1 by specific inhibitors VE-821 and Chir 124, respectively. These experiments revealed that ATR kinase activity was required for normal dynamics even in unperturbed cells; although blocking of ATR kinase and Chk1 did not influence the amplitude of local displacements, it did strongly inhibit the local coherence in chromatin dynamics (Fig. S9). This finding is consistent with our hypothesis that DNA damage and ATR activity influence coherence of chromatin dynamics, but made it difficult to separate direct vs. indirect effects of inhibiting nuclear enzymes.
Conclusions
Our findings reveal a unique type of active movement in chromatin and raise the puzzling question of how microscopic forces couple over long distances to generate coherent movement. We suspect elastic coupling, which could generate coherent motion by at least two mechanisms: passive, where active motors in a small region push or pull on a larger region that responds passively, and self-organized, where forces serve to align, or activate prealigned, motors at a distance (41, 42). Self-organized motion of an active material is intriguing because it can give rise to complex behavior such as oscillations and traveling waves (43, 44). Chromatin-remodeling ATPases, which act on regions of high local curvature where DNA bends around nucleosomes (12), are interesting candidates for sensing force as well as generating it. Illuminating the physical nature of global chromatin dynamics is critical for understanding collective gene behavior and the underlying DNA biology.
The biological function of coherent chromatin motion is unclear; coherency might be functional or merely an epiphenomenon of dynamic organization. It is clearly an active, ATP-dependent, inherent attribute of global chromatin dynamics and therefore likely to impact on nuclear processes such as DNA replication, gene expression, and maintenance of genomic integrity. We find several different perturbations that cause similar loss of local coherence. The common factor is that all trigger DNA damage responses or, in the case of ATR inhibitors, detect and regulate damage responses. Loss of coherence in motion in DNA-damaged nuclei could be caused by global physical changes in chromatin (45), which might be needed to relax chromatin. We speculate that chromatin relaxation minimizes the effect of DNA damage by mechanically isolating the damage site through global loss of elastic coupling. More speculatively, coherent motion might serve to communicate information across the nucleus and thus help the nucleus mount an integrated response to DNA damage or physical deformation.
Materials and Methods
Cell Culture.
HeLa cells (CCL-2) were cultured according to the ATTC recommendations. The HeLa H2B-GFP line was a gift from James Orth (Harvard Medical School). Cells were cultured in a humidified, 5% CO2 (vol/vol) atmosphere at 37 °C in Gibco Dulbecco’s modified eagle medium (DMEM) supplemented with 10% FBS (vol/vol), 100 units/mL penicillin, and 100 mg/mL streptomycin (Invitrogen). Cells were plated 24 h before the experiment on 35-mm MatTek dishes with glass bottom no. 1.5 (MatTek). Before the experiment the medium was replaced by Gibco CO2-independent medium supplemented with l-glutamine (Invitrogen). Cells were then mounted on the microscope stage kept in a custom-built 37 °C microscope incubator enclosure with 5% CO2 (vol/vol) delivery during the entire experiment.
Chromosome Staining.
To stain chromosome territories we incorporated Cy3-dCTP (GE Healthcare) into the nuclei as described previously (33). Cells were scrape-loaded with 10 µM Cy3-dCTP in 10 mM PBS. Segregation of chromosome territories was obtained by growing the cells over 3–5 d.
Biochemical Perturbations.
To deplete ATP, 6 mM 2-deoxyglucose (DOG) and 1 µM trifluoromethoxy-carbonylcyanide phenylhydrazone (FCCP) dissolved in CO2-independent medium supplemented with l-glutamine were added to cells 2 h before imaging. For fixation experiments cells were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) at room temperature for 20 min and then washed three times in PBS every 5 min. Coverslips were mounted on glass microscope slides, using Prolong-Gold antifade reagent (Invitrogen). For cytoskeletal perturbations 10 µM latrunculin A, 10 µM blebbistatin, or 10 µM nocodazole, respectively, and for chromatin perturbations 0.5 µg/mL aphidicolin, 20 µg/mL α-amanitin or 2 µg/mL ICRF-193, 200 ng/mL neocarzinostatin, 10 µM VE-821, 1 µM Chir 124, respectively, in CO2-independent medium supplemented with l-glutamine were added to cells 30–120 min before imaging. All chemicals were from Sigma Aldrich, unless stated otherwise. VE-821 and Chir 124 were a gift from Jinhua Wang (Harvard Medical School).
EdU Stain.
S-phase–specific staining was achieved using click chemistry as described previously (40). We incubated cells for 1 h with EdU before fixation. Alexa 555 azide (Invitrogen) was used as a fluorophore in the click reaction.
Microscopy and Image Acquisition.
Images were acquired with a Yokogawa CSU-X1 spinning-disk confocal head with an internal motorized high-speed emission filter wheel and Spectral Applied Research Borealis modification for increased light throughput and illumination homogeneity on a Nikon Ti inverted microscope equipped with a 100× Plan Apo NA 1.4 objective lens and the Perfect Focus System for continuous maintenance of focus. The microscope is mounted on a TMC vibration-isolation table. H2B-GFP fluorescence was excited with a 491-nm solid state laser [controlled with an acousto-optic tunable filter (AOTF)] and collected with a 405/491/561/642 multibandpass dichroic mirror (Semrock) and an ET525/50 emission filter (Chroma Technology). Images were acquired with a Hamamatsu ORCA-AG cooled CCD camera controlled with MetaMorph 7 (Molecular Devices) software. No binning was used; the pixel size for the 100× objective was 0.065 µm. The stream acquisitions were collected using an exposure time of 250 ms for H2B-GFP. For fixed samples we also used a Nikon TE-2000 microscope with a Yokogawa CSU10 spinning-disk confocal head equipped with a 100× Plan Apo NA 1.4 objective lens, the Perfect Focus System, a 491-nm solid state laser (controlled with an AOTF) and an Andor iXon 897 camera controlled by MetaMorph7 at binning 2, pixel size 0.107 µm, and exposure of 300 ms. Little bleaching was observed, as determined by a lack of decrease in average intensity during measurements. The streams of 16-bit images were saved as multitiff stacks. Images were converted to single-tiff images and analyzed in MatLab (The MathWorks) as described below. Cell viability after image acquisition was verified by checking that cells proceed into a proper cell division (Fig. S10).
DCS.
The calculation comprises three steps: (i) A time series of displacement fields for a time series of fluorescent images is computed using a standard PIV algorithm (46) for all image pairs separated by time lag Δt. (ii) The average spatial displacement autocorrelation function (SDACF) is calculated by averaging over all SDACFs calculated for every displacement field obtained in i to detect characteristic length scales. (iii) Steps i and ii are repeated for all accessible time lags Δt to obtain temporal evolution of SDACF(Δt), identifying characteristic timescales and length scales in the system. Optionally, a temporal autocorrelation of SDACF(Δt) is performed to characterize the temporal decorrelation in the system. A DCS method illustration is provided in Fig. S1.
DCS calculations i–iii were carried out using the Orchestra computation cluster at Harvard Medical School. Typically, DCS calculations for one nucleus and a stream of 25 s containing 100 frames lasted ∼5 h.
Specifically, the calculation of displacement fields was carried out using the MatPiv 1.60 package (46) (a GNU public license software, http://folk.uio.no/jks/matpiv/index2.html) for MatLab (The MathWorks) in combination with custom-made MatLab routines. We used 16 × 16 pixel interrogation windows with an overlap of 75% to provide high-resolution information. The calculations were also tested for interrogation windows of 32 × 32 pixels and 64 × 64 pixels, with overlap 0–75%, and the presence of correlated motion was confirmed here. The displacement fields were calculated for all time intervals accessible in the experiment, typically 250 ms to 25 s. The resulting displacement fields were filtered using filters “snrfilt”, “pkhfilt”, and “globfilt” from the MatPiv 1.60 package, evaluating signal-to-noise ratio, peak height of correlation functions, and global velocity distribution to remove spurious vectors.
We used custom-made MatLab code to calculate spatial displacement autocorrelation function cdx(Δr→,Δt)=<dx(r→,Δt), dx(r→ +Δr→,Δt)> for the x-component dx of the measured displacements d→(r→,Δt). There was no noticeable difference when calculated for the y-component. We calculate cdx(Δr,Δt) for every displacement field at all Δt and then fit the radially averaged cdx(Δr,Δt) to a power law with an exponential cutoff f(Δr) = A(Δr)ne−Δr/ξ, using custom-made MatLab code. Thus, we obtain scaling exponent n and correlation length ξ for every displacement field separately and then compute average n and ξ for given Δt. To evaluate the average extent of the spatial correlation in the displacement fields we calculate average spatial displacement autocorrelation function <cdx(Δr,Δt)>, for a Δt by averaging over all cdx(Δr,Δt) calculated for single DCS fields at given Δt.
MSND was calculated from the displacements d→(r→,Δt) obtained in step i for every Δt over the entire image sequence, MSND(Δt)=<|d→(r→,Δt)|2>, and fitted using custom-made MatLab code to f(Δt) = A + BΔtα.
Supplementary Material
Acknowledgments
A.Z. thanks Erich Sackmann for helpful discussions and critical reading of the manuscript and Nikon Imaging Center at Harvard Medical School for help with light microscopy. A.Z. also thanks Paul Wiseman and his group for helpful discussions about image correlation. The calculations in this paper were run on the Orchestra cluster supported by the Harvard Medical School Research Information Technology Group. This research was supported by the National Institutes of Health (Grant R01-GM39565). A.Z. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG 2040-10).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220313110/-/DCSupplemental.
References
- 1. Alberts B (2002) Molecular Biology of The Cell (Garland Science, New York)
- 2.Lengauer C, et al. Chromosomal bar codes produced by multicolor fluorescence in situ hybridization with multiple YAC clones and whole chromosome painting probes. Hum Mol Genet. 1993;2(5):505–512. doi: 10.1093/hmg/2.5.505. [DOI] [PubMed] [Google Scholar]
- 3.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2(4):292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 4.Cremer T, Küpper K, Dietzel S, Fakan S. Higher order chromatin architecture in the cell nucleus: On the way from structure to function. Biol Cell. 2004;96(8):555–567. doi: 10.1016/j.biolcel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubner MR, Spector DL. Chromatin Dynamics. Annu Rev Biophys. 2010;39:471–489. doi: 10.1146/annurev.biophys.093008.131348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cremer T, et al. Chromosome territories—a functional nuclear landscape. Curr Opin Cell Biol. 2006;18(3):307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8(2):104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 9.Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell. 2008;132(6):929–934. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4(5):e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meaburn KJ, Gudla PR, Khan S, Lockett SJ, Misteli T. Disease-specific gene repositioning in breast cancer. J Cell Biol. 2009;187(6):801–812. doi: 10.1083/jcb.200909127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Racki LR, Narlikar GJ. ATP-dependent chromatin remodeling enzymes: Two heads are not better, just different. Curr Opin Genet Dev. 2008;18(2):137–144. doi: 10.1016/j.gde.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408(6814):877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 14.Abney JR, Cutler B, Fillbach ML, Axelrod D, Scalettar BA. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J Cell Biol. 1997;137(7):1459–1468. doi: 10.1083/jcb.137.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404(6778):604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 16.Kimura H, Cook PR. Kinetics of core histones in living human cells: Little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153(7):1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiesmeijer K, Krouwels IM, Tanke HJ, Dirks RW. Chromatin movement visualized with photoactivable GFP-labeled histone H4. Differentiation. 2008;76(1):83–90. doi: 10.1111/j.1432-0436.2007.00234.x. [DOI] [PubMed] [Google Scholar]
- 18.Mora-Bermúdez F, Gerlich D, Ellenberg J. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat Cell Biol. 2007;9(7):822–831. doi: 10.1038/ncb1606. [DOI] [PubMed] [Google Scholar]
- 19.Levi V, Ruan QQ, Plutz M, Belmont AS, Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys J. 2005;89(6):4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuang CH, et al. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16(8):825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 21.Weber SC, Spakowitz AJ, Theriot JA. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc Natl Acad Sci USA. 2012;109(19):7338–7343. doi: 10.1073/pnas.1119505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belmont AS, Straight AF. In vivo visualization of chromosomes using lac operator-repressor binding. Trends Cell Biol. 1998;8(3):121–124. doi: 10.1016/s0962-8924(97)01211-7. [DOI] [PubMed] [Google Scholar]
- 23.Carmo-Fonseca M, Platani M, Swedlow JR. Macromolecular mobility inside the cell nucleus. Trends Cell Biol. 2002;12(11):491–495. doi: 10.1016/s0962-8924(02)02387-5. [DOI] [PubMed] [Google Scholar]
- 24.Bronstein I, et al. Transient anomalous diffusion of telomeres in the nucleus of mammalian cells. Phys Rev Lett. 2009;103(1):018102. doi: 10.1103/PhysRevLett.103.018102. [DOI] [PubMed] [Google Scholar]
- 25. Stixova L, et al. (2011) Heterogeneity in the kinetics of nuclear proteins and trajectories of substructures associated with heterochromatin. Epigenetics Chromatin 4:5. [DOI] [PMC free article] [PubMed]
- 26.Misteli T. Protein dynamics: Implications for nuclear architecture and gene expression. Science. 2001;291(5505):843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- 27.Edelmann P, Bornfleth H, Zink D, Cremer T, Cremer C. Morphology and dynamics of chromosome territories in living cells. Biochim Biophys Acta Rev Cancer. 2001;1551:M29–M40. doi: 10.1016/s0304-419x(01)00023-3. [DOI] [PubMed] [Google Scholar]
- 28.Zink D, et al. Structure and dynamics of human interphase chromosome territories in vivo. Hum Genet. 1998;102(2):241–251. doi: 10.1007/s004390050686. [DOI] [PubMed] [Google Scholar]
- 29.Bornfleth H, Edelmann P, Zink D, Cremer T, Cremer C. Quantitative motion analysis of subchromosomal foci in living cells using four-dimensional microscopy. Biophys J. 1999;77(5):2871–2886. doi: 10.1016/S0006-3495(99)77119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albiez H, et al. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006;14(7):707–733. doi: 10.1007/s10577-006-1086-x. [DOI] [PubMed] [Google Scholar]
- 31.Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol. 2003;160(5):685–697. doi: 10.1083/jcb.200211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hihara S, et al. Local nucleosome dynamics facilitate chromatin accessibility in living mammalian cells. Cell Rep. 2012;2(6):1645–1656. doi: 10.1016/j.celrep.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 33.Manders EMM, Kimura H, Cook PR. Direct imaging of DNA in living cells reveals the dynamics of chromosome formation. J Cell Biol. 1999;144(5):813–821. doi: 10.1083/jcb.144.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llères D, James J, Swift S, Norman DG, Lamond AI. Quantitative analysis of chromatin compaction in living cells using FLIM-FRET. J Cell Biol. 2009;187(4):481–496. doi: 10.1083/jcb.200907029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee KB, Wang D, Lippard SJ, Sharp PA. Transcription-coupled and DNA damage-dependent ubiquitination of RNA polymerase II in vitro. Proc Natl Acad Sci USA. 2002;99(7):4239–4244. doi: 10.1073/pnas.072068399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park I, Avraham HK. Cell cycle-dependent DNA damage signaling induced by ICRF-193 involves ATM, ATR, CHK2, and BRCA1. Exp Cell Res. 2006;312(11):1996–2008. doi: 10.1016/j.yexcr.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 37.Shechter D, Costanzo V, Gautier J. Regulation of DNA replication by ATR: Signaling in response to DNA intermediates. DNA Repair. 2004;3(8–9):901–908. doi: 10.1016/j.dnarep.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 38.Ziv Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8(8):870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 39.Kruhlak MJ, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172(6):823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105(7):2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guérin T, Prost J, Martin P, Joanny JF. Coordination and collective properties of molecular motors: Theory. Curr Opin Cell Biol. 2010;22(1):14–20. doi: 10.1016/j.ceb.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 42.Holzbaur ELF, Goldman YE. Coordination of molecular motors: From in vitro assays to intracellular dynamics. Curr Opin Cell Biol. 2010;22(1):4–13. doi: 10.1016/j.ceb.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Julicher F, Prost J. Spontaneous oscillations of collective molecular motors. Phys Rev Lett. 1997;78:4510–4513. [Google Scholar]
- 44.Hilfinger A, Chattopadhyay AK, Jülicher F. Nonlinear dynamics of cilia and flagella. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;79(5 Pt 1):051918. doi: 10.1103/PhysRevE.79.051918. [DOI] [PubMed] [Google Scholar]
- 45.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10(4):243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sveen JK (2004) An Introduction to MatPIV v. 1.6.1, eprint series “Mechanics and Applied Mathematics” (Department of Mathematics, University of Oslo, Oslo)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.