Abstract
Suppression of seed shattering was a key step during crop domestication that we have previously suggested to be convergent among independent cereal lineages. Positional, association, expression, and mutant complementation data all implicate a WRKY transcription factor, SpWRKY, in conferring shattering to a wild sorghum relative, Sorghum propinquum. We hypothesize that SpWRKY functions in a manner analogous to Medicago and Arabidopsis homologs that regulate cell wall biosynthesis genes, with low expression toward the end of floral development derepressing downstream cell wall biosynthesis genes to allow deposition of lignin that initiates the abscission zone in the seed–pedicel junction. The recent discovery of a YABBY locus that confers shattering within Sorghum bicolor and other cereals validated our prior hypothesis that some parallel domestication may have been convergent. Ironically, however, the shattering allele of SpWRKY appears to be recently evolved in S. propinquum and illustrates a case in which the genetic control of a trait in a wild relative fails to extrapolate even to closely related crops. Remarkably, the SpWRKY and YABBY loci lie only 300 kb apart and may have appeared to be a single genetic locus in some sorghum populations.
Keywords: cereal crop, seed dispersal, association mapping
Reduction of seed dispersal by “shattering” of the mature inflorescence is a critical step during the domestication of many cereal crops that dominate human calorie consumption. For example, in wild sorghums such as Sorghum propinquum and Sorghum halepense, shattering facilitates dispersal of sexual propagules, complementing dispersal of clonal propagules by subterranean rhizomes. Shattering is thought to have been strongly selected against during domestication because humans could more efficiently harvest grains that remain attached to the plant (1), and we have previously suggested that parallel domestications of divergent cereals may have involved mutations in some corresponding genes (2).
During plant development, the shattering of seeds involves the formation of a lignified abscission layer (or dehiscence zone) and is considered a process of programmed senescence (3). The pathway involving the formation of the abscission layer is well characterized in some eudicot species. SHATTERPROOF (SHP) genes SHP1 and SHP2 have been shown to specify valve margin cell identities in Arabidopsis (4), with their expression reinforced through negative regulation from FRUITFULL (FUL) in valve development (5) and REPLUMLESS (RPL) in the replum (6). Genes JAG, FIL, and YAB3 promote the expression of FUL and SHP but are negatively regulated by RPL (7). However, the abscission layer contributing to seed shattering in Arabidopsis is located at the valve–replum boundary and does not correspond to that of most cereal species, which is at the base of the pedicel. Therefore, it remains questionable whether orthologous genes are directly responsible for the seed dispersal mechanisms of dicots and cereals, respectively.
The genetic control of shattering varies among cereals, in many cases involving several genes. Two major genes that contribute to shattering in rice (Oryza sativa ssp.) have been identified: qSH1 and sh4, controlling 68% and 69% of the phenotypic variance in the studied crosses, respectively (8, 9). The fixation of sh4 occurred very early in rice domestication with the domesticated allele occurring in both subspecies indica and japonica, whereas qSH1 is much more recent, present only within temperate japonica genotypes (10, 11). In wheat, quantitative trait loci (QTLs) responsible for nonbrittle rachis are located in the homeologous regions of chromosome 3A (Br2), 3B (Br3), and 3D (Br1) (12, 13). Comparative mapping hinted that this part of the chromosomal regions might correspond to the orthologous region in barley, controlled by two tightly linked loci, Btr1 and Btr2, but do not appear to correspond to the region in other major cereals (12, 13). Indeed, although some shattering genes do appear to be in corresponding (orthologous) chromosomal locations among cereals (2), many do not; therefore, it is hypothesized that there are multiple pathways responsible for seed dispersal in the grasses (14). Steady progress in rice notwithstanding, more rice genes that control shattering are known (2, 3) but have not yet been identified; therefore, the above hypothesis remains to be tested.
Sorghum appears to be a favorable species to investigate the genetic basis of shattering, because only one genetic locus, Sh1, explains 100% of the phenotypic variance in a cross between an elite Sorghum bicolor breeding line (BTx623), and wild S. propinquum (2). The S. bicolor genome size is ∼730 Mb, has a high-quality reference sequence (15), and shows similar composition and high levels of synteny and microcolinearity with maize and rice (16) despite ∼50 My of divergence (17).
Recently, it was shown that a YABBY transcription factor near the Sh1 region contributes to seed shattering in crosses between the shattering species S. virgatum and a nonshattering breeding line Tx430 (18). There are three different mutations at the YABBY locus that resulted in either lower expression or produced a truncated transcript in the domesticated allele. The three different YABBY haplotypes suggested independent origins in different sorghum populations (18).
Here, we report that seed shattering of S. propinquum is conferred by a different locus than YABBY, specifically a WRKY transcription factor (SpWRKY). The expression of the WRKY gene appears to be modulated during floral and seed development and is implicated in the lignin deposition pathway. The three independent mutations in the YABBY locus validate our prior hypothesis that parallel domestications of divergent cereals may have involved mutations in some corresponding genes (2). In comparison, the shattering gene that we report here appears recently evolved in S. propinquum and illustrates a case in which the genetic control of a trait in a wild relative fails to extend to closely related crops. Remarkably, the SpWRKY and YABBY loci lie only 300 kb apart and may have appeared as a single genetic locus in some sorghum populations. We further note that both genes might regulate some of the same downstream genes involved in lignin deposition and thus play important roles in the shattering pathway, however, with different relative contributions in different species.
Results
Genetic Mapping of the Sh1 Locus and Genomic Alignments.
Sh1 was genetically mapped within a region flanked by two restriction fragment length polymorphism (RFLP) markers SOG0251 and SOG1273 (Fig. 1), separated by 0.42 cM (three recombinants out of 740 gametes). An S. propinquum bacterial artificial chromosome (BAC) library (19) was screened with overgo probes corresponding to the two flanking DNA markers, and hybridizing BACs were fingerprinted via restriction enzyme digestion. A physical contig made using FPC software (20) that spanned the two flanking markers included 10 overlapping S. propinquum BACs in a minimum tiling path. Sequence reads from the BACs were assembled into 30 contigs totaling 1.04 Mb (contig N50 = 63.9 kb). The S. propinquum BAC alignments correspond to a 1.13-Mb (11.33–12.47 Mb) region on S. bicolor chromosome 1. A total of 694 kb (61.3%) of this region can be aligned with the S. propinquum sequences using NUCMER (21), with average nucleotide identity of 94.5%. Most sequences missing in S. propinquum are low complexity repeats and transposable elements.
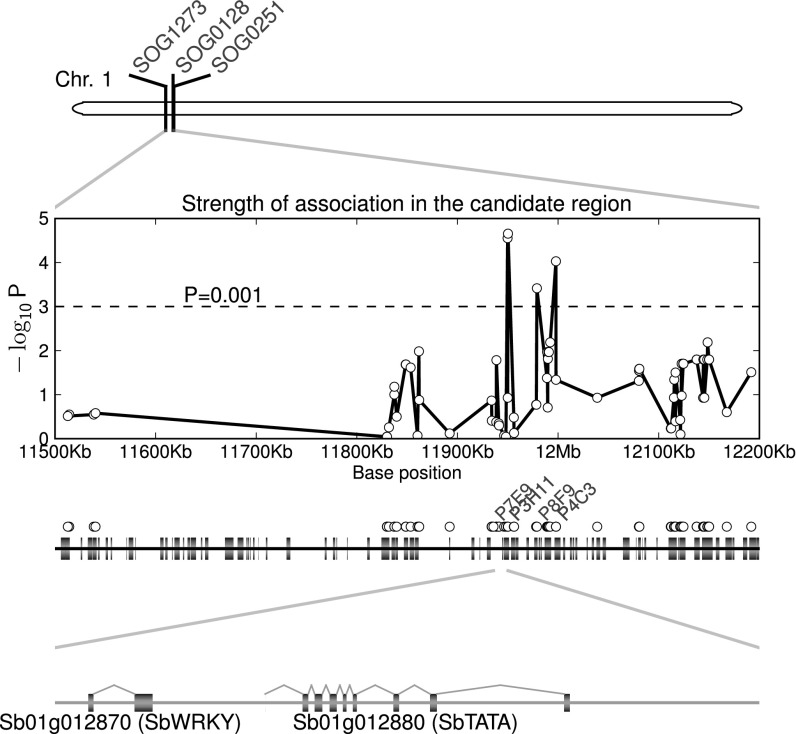
Fig. 1.
Fine mapping of the shattering trait from S. propinquum. Panels from top to bottom illustrate the markers we used to locate the candidate genes, RFLP markers flanking (SOG1273, SOG0251) or cosegregating (SOG0128) with the shattering trait. A set of amplicon-based SNP markers were used to test the strength of association of shattering in the delineated region (chr1: 11.5–12.2 Mb). Four SNPs (P7E9, P3H11, P8F9, and P4C3) were significantly associated with shattering (P < 0.001), and two candidate genes fall inside this region: Sb01g012870 (SbWRKY) and Sb01g012880 (SbTATA).
S. bicolor and S. propinquum coding regions were very similar at the gene level. Gene predictions revealed 80 S. propinquum gene models within the interval defined by RFLP markers flanking the locus, with a median size of 906 bp (Dataset S1). Among the 95 gene loci predicted on the BACs, 9 loci showed no protein sequence difference between S. proqinquum and the S. bicolor reference (of our nonshattering parental line). The median rate of synonymous substitutions per synonymous site (Ks) is 0.0215 for the orthologous genes situated in the shattering region. This median Ks value corresponds to ∼1.7 My of divergence between S. propinquum and S. bicolor, using a rate estimate of 6.5 × 10−9 synonymous substitutions per year (22).
Association Mapping.
We identified a total of 603 SNP sites within the coding regions of the 80 genes on the basis of sequence alignment between S. bicolor reference and S. propinquum BAC sequences. Polymorphic sites between S. bicolor and S. propinquum within the Sh1 region were sampled using a diversity panel of 11 shattering and 13 nonshattering accessions (Table S1). We obtained data for a total of 67 sites, initially targeting an average of one site per gene. Among the 67 sites tested (Dataset S2), 4 were significantly (P < 0.001) associated with the shattering trait (P7E9, P3H11, P8F9, and P4C3; Fig. 1). The four sites were also in linkage disequilibrium with one another. However, the intermediate sites between the two peaks were not significantly associated with the shattering trait, possibly reflecting mutations that are of more recent origin than those related to shattering and therefore are not informative with regard to shattering.
The highest peak in the association plot contains two SNP markers, P7E9 (P = 2.8 × 10−5) and P3H11 (P = 2.2 × 10−5), covering an ∼50-kb genomic region. Additional primers were designed to sample more sequence sites in the ∼50-kb genomic region, which extends from gene models Sb01g012870 to Sb01g012960 [gene identifiers from JGI annotation release Sbi1.4 (15)]. Based on the segregation pattern within this region (Dataset S2), we narrowed down the target locus to be contained between base position 11,941,320 and 11,956,003, because the level of association dropped to nonsignificance beyond these two flanking sites. This interval contains two candidate genes, encoding transcription factors Sb01g012870 (SbWRKY) and Sb01g012880 (SbTATA) (Fig. 1).
Expression Profile of Candidate Genes.
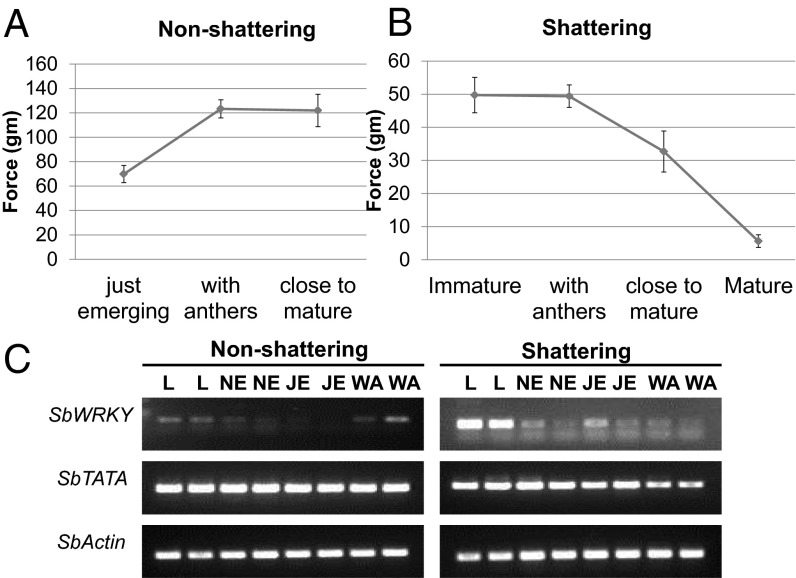
We used semiquantitative RT-PCR to assay expression of the two candidate genes at different floral developmental stages in shattering and nonshattering genotypes, confirming the shattering phenotype using the breaking tensile strength (BTS) method. For each stage, 10 individual florets were tested from two different panicles. In nonshattering S. bicolor, the BTS value actually increased from 69.8 g in the immature inflorescence to 123.1 g after anther dehiscence and remained stable in the mature inflorescence (122 g; Fig. 2A). The BTS value declined rapidly in shattering S. halepense, from 55.1 g in immature inflorescences just emerged from flag leaf to 7.5 g in mature inflorescences (Fig. 2B).
Fig. 2.
BTS of (A) nonshattering and (B) shattering inflorescences at different developmental stages. For each stage, 10 individual florets were tested from two different panicles. Bars represent ±SE (n = 2). (C) RT-PCR expression profile in nonshattering and shattering sorghum of SbWRKY, SbTATA, and SbActin as a control. L, leaf; NE, inflorescence still in flag leaf; JE, inflorescence just emerging from flag leaf; WA, after anther dehiscence.
Although SbTATA closely resembled the constitutively expressed actin control, the expression of SbWRKY was heavily modulated during the course of floral development (Fig. 2C). Both SbWRKY and SpWRKY were expressed strongly in leaves, but in floral tissues, the expression gradually decreased during inflorescence development (NE, inflorescence still in flag leaf; JE, inflorescence just emerging from flag leaf). However, near the anther dehiscence stage, SbWRKY had a strong pulse of expression in the nonshattering genotype, whereas the expression of SpWRKY continued to decrease. This clear difference of gene expression suggested a correlation of increased SbWRKY expression with nonshattering (i.e., SbWRKY is a negative regulator of shattering), also suggesting that the particular developmental stage of expression is critical for manifestation of the trait.
Mutant Complementation.
We transformed the two candidate genes SbWRKY and SbTATA from genetically dominant S. propinquum BAC YRL20H16 into recessive nonshattering sorghum cultivar Tx430, each construct including coding sequences and about 1 kb of 5′- and 3′- flanking sequences (Dataset S3). In transgenic sorghums validated to carry the S. propinquum allele, the SpWRKY construct induced discernible seed dropping in some transformants, whereas SbTATA transformants and controls did not.
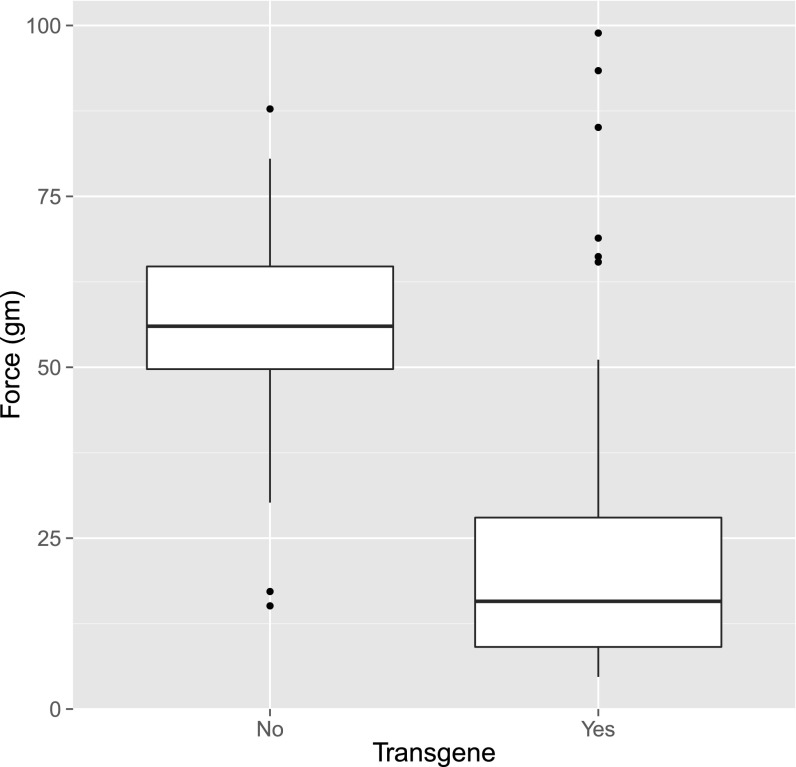
To quantify the effect of SpWRKY on seed shattering, nine plants containing different transgenic events were self-pollinated, and up to 24 progenies were evaluated for each event. The transgene was segregating in eight of the nine progeny groups (Fig. S1; group T3 lacked the transgene, possibly indicating that it had not been integrated into the nucleus in the original transgenic plant). Across 136 plants from the eight validated events, reduced BTS was highly correlated with the presence of the transgene (r = −0.641; P = 4.2 × 10−17), with correlation coefficients in the individual populations (events) ranging from −0.399 to −0.946. Segregants that lacked the transgene showed an average BTS of 57.8 g (SD = 13.99, n = 38), which was indistinguishable from that of the T3 population that lost the transgene (mean = 52.4, SD = 15.7, n = 17). Plants containing the SpWRKY transgene had an average BTS of 22.3 g (SD = 18.6, n = 105), which is a highly significant 61% reduction from the nonshattering plant (Fig. 3).
Fig. 3.
Transgenic progeny shattering strength estimated with breaking tensile strength for the group of individuals that lack the transgene (n = 38) and individuals with the transgene (n = 105). The box of each group depicts lower quartile, median, and upper quartile. Dots, if any, indicate which observations that are considered outliers.
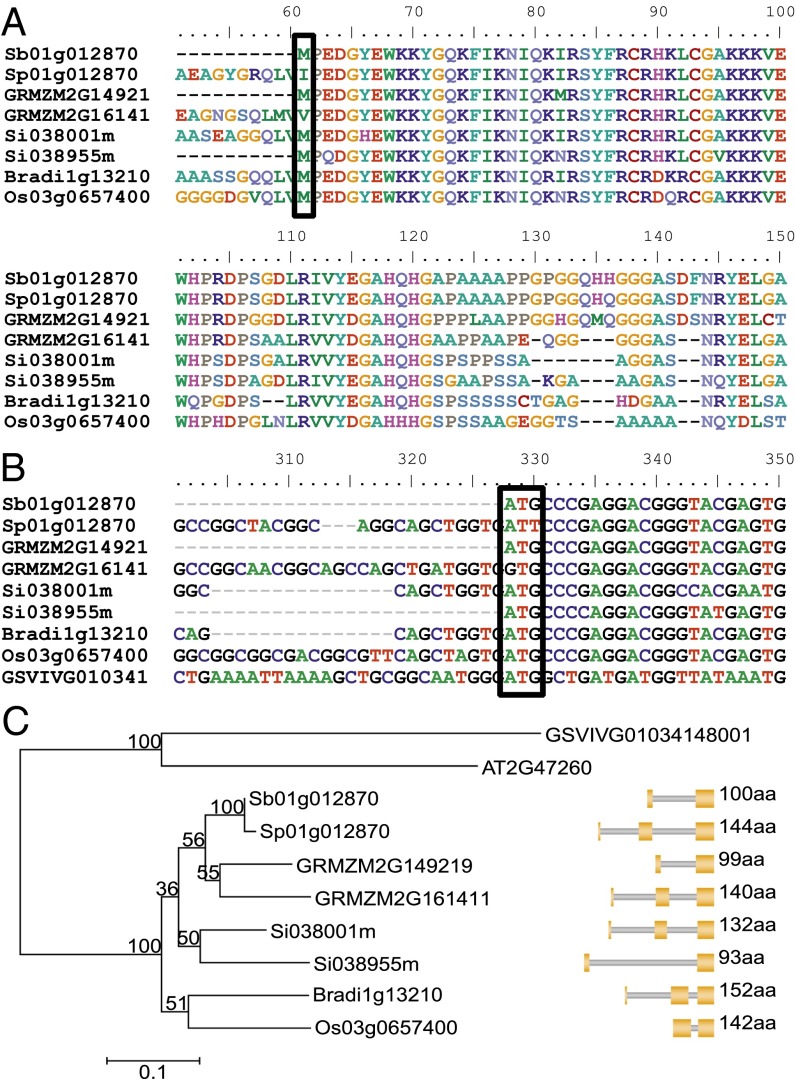
The dominant SpWRKY allele conferring shattering in S. propinquum and the recessive SbWRKY in nonshattering S. bicolor BTx623 encode proteins that differ at two amino acid positions in the aligned portions. SpWRKY proteins cannot start at the S. bicolor start, because of the ATG to ATT mutation in SpWRKY that results in a methionine (M) to isoleucine (I) substitution in the protein sequence (column 61 in Fig. 4A) and renders the SpWRKY protein 44 amino acid residues longer than SbWRKY. The second amino acid difference is a substitution of histidine (H) to glutamine (Q) (column 136 in Fig. 4A). There are no amino acid differences in the two WRKY domains, particularly in the conserved (WKKYGQK) sequence thought to be directly involved in DNA binding (23).
Fig. 4.
Sequence alignments of SbWRKY and related homologs. (A) Partial protein alignments of SbWRKY orthologs from five grasses. The WRKY domain is located between columns 62 and 115. S. propinquum and S. bicolor alleles differ at start codon position, resulting in a shorter S. bicolor protein. There is only one copy each in sorghum, rice, and Brachypodium, but two copies in maize and Setaria. (B) Partial DNA alignments of the orthologs of SbWRKY from five grasses and grape. The columns highlighted in the solid box marks the aligned positions for start codons of the short proteins. (C) Neighbor-joining tree among selected SpWRKY homologs. The number next to the branch nodes are bootstrap values with 500 samples. Exon structure for individual gene homolog is illustrated with coding exons in orange blocks and protein length.
Discussion
Evolution of SbWRKY Orthologs in Two Sorghum Species and Other Major Grass Species.
SpWRKY appears to be a gain-of-function mutation in S. propinquum after its divergence from a common ancestor shared with S. bicolor. A parsimony model based on multiple sequence alignments favors the loss of the SbWRKY start codon by the ATG to ATT (methionine to isoleucine) mutation in S. propinquum after its divergence from a common lineage shared with S. bicolor (Fig. 4B). Transcriptome sequences from several wild S. bicolor races [S. bicolor ssp verticilliflorum race virgatum (unnamed), drummondii (IS 21691), arundinaceum (IS 14216), and aethiopicum (IS 27587)] and a genomic sequence from S. timorense (outgroup to the S. bicolor–S. propinquum divergence) all show ATG (S. bicolor–like) instead of ATT (S. propinquum–like) in the start codon position (Fig. S2).
The WRKY gene family is large in plants, e.g., 113 members in rice (24), but we were able to infer the direct ortholog(s) of SpWRKY in related grass genomes based on genomic colinearity (25). SpWRKY is orthologous to two maize proteins encoded by GRMZM2G149219 and GRMZM2G161411, two Setaria proteins Si038955m and Si038001m, rice OsWRKY60 (Os03g0657400), and Brachypodium protein Bradi1g13210 (Fig. 4A). It is more difficult to discern the direct orthologs(s) among the 21 similar proteins in grape and 19 in Arabidopsis because of the lesser colinearity across longer evolutionary divergence. The two maize gene copies were derived from the maize lineage genome duplication (26). The two Setaria genes are tandem copies adjacent to one another in the ancestral location, thus retaining genomic colinearity to the SpWRKY locus.
Mutation in the Start Codon in the Shattering Allele Results in a Longer Protein.
The earlier start codon of SpWRKY than SbWRKY has been confirmed empirically, with the S. propinquum transcript being 132 bp longer than the S. bicolor transcript. Long (∼140 amino acids) and short WRKY proteins (∼100 amino acids) in sorghum also exist in other grass genomes, with the short proteins often lacking an ∼40 amino acid N terminus. These extra ∼40 amino acids in the long proteins are much more variable in the grasses than the (highly conserved) WRKY domain (Fig. 4A) or the 3′-terminal exon, contrary to general conservation patterns across plant genes. The main difference among the gene homologs is whether they have one or two additional exons in the 5′ end, which adds up to either two or three exons in total (Fig. 4C). Long SpWRKY proteins often contain three exons, with the only exception being Os03g0657400, which might have fused the first two exons. Based on the nucleotide alignments of the identified orthologs, all other orthologous genes in the related grass species have a G in the third nucleotide position of the start codon, with only S. propinquum showing a T. In addition, every grass ortholog in the comparison translates this specific codon to methionine with the exception of SpWRKY and the maize ortholog GRMZM2G161411, the latter with a TTG codon that translates to valine (V).
All grasses that we compared have at least one copy of the long WRKY protein, whereas species with two gene copies (maize and Setaria) also contain one short WRKY protein. The single orthologs in rice and Brachypodium are long. There are two copies in both maize and Setaria: one short and one long. The duplication into two copies in maize and Setaria occurred more recently and independently in the respective lineages after their divergence from other grasses (Fig. 4C). The QTL intervals affecting shattering on maize chromosomes 1 and 5 (2), respectively, harbor GRMZM2G149219 (a short protein with 99 amino acids) and GRMZM2G161411 (a long protein with 140 amino acids).
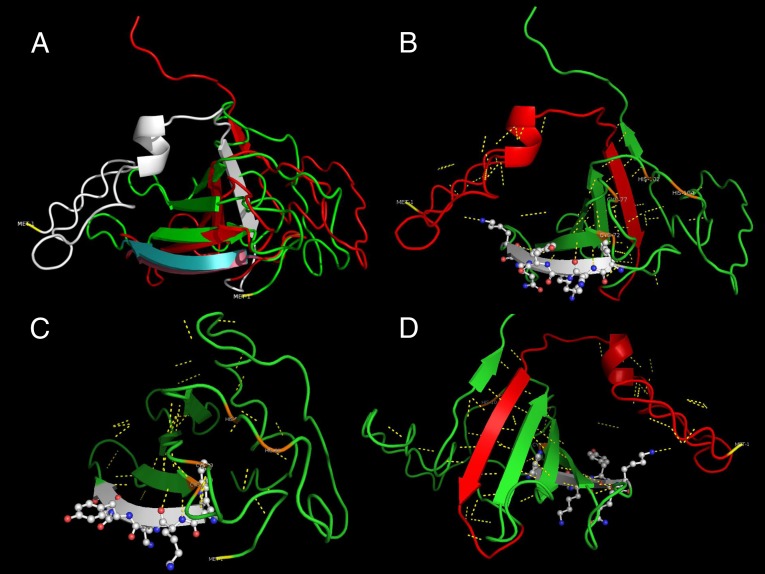
The different lengths of SbWRKY and SpWRKY proteins may impact gene function. The WRKY domain starts right after the first amino acid in the short SbWRKY protein (Fig. 4A). A sequence similarity search against GenBank using only the 44 N-terminal amino acids did not reveal any significant hits at E < 0.01. 3D structural prediction of the extended sequence in the long proteins using I-TASSER (27) suggested that part of the extended sequence folds into a β-sheet with two polar interactions with the WRKY domain, which may impact its ability to bind DNA (Fig. 5). We failed to detect significant signal peptide regions in the missing N-terminal sequence.
Fig. 5.
3D structure comparison and functional prediction of SbWRKY and SpWRKY peptides. (A) Superimposition of SbWRKY (green) and SpWRKY (red) transcription factors aligned at the first four amino acid residues of WRKY domains. Structure of the SbWRKY domain (cyan) departs very rapidly from that of SpWRKY domain (purple). The truncated 44 amino acid residues of SpWRKY are shown in white. (B) Conserved WRKY DNA binding domain is shown on second β-plated sheet (white). The first β-sheet in the truncated section in SbWRKY (red) intercalates between the fifth and sixth β-sheets forming a ribcage supported by polar interactions (yellow interrupted lines). The model also predicts a zinc-finger domain among Cys-72, Cys-77, His-58, and His-104 residues (orange). (C) Model of the truncated SbWRKY. Lack of the first β-sheet disrupts the integrity. The predicted zinc-finger domain maintains it functional structure among the four residues (orange). (D) Closer view of the β-sheets after 180° rotation of the SpWRKY model. There are 17 polar interactions, of which 15 maintain β-sheet orientation. The remaining two interactions are with the WRKY domain and might play a central role in DNA binding.
Likely Mechanism of SpWRKY in the Shattering Pathway.
The WRKY superfamily of plant transcriptional factors has been implicated in a variety of physiological and developmental processes unique to plants, including leaf senescence (28, 29), trichome initiation (30), and embryo morphogenesis (31). The WRKY domain functions through direct interactions with the W-box domain in the promoter region of downstream gene targets (23). Overexpression of gene homologs results in ectopic lignin deposition in Medicago (32, 33), tobacco (34), and rice (35). Disruption of a WRKY gene in Medicago and Arabidopsis caused secondary cell wall thickening associated with the deposition of lignin, xylan, and cellulose, implying that the gene is a negative regulator (33).
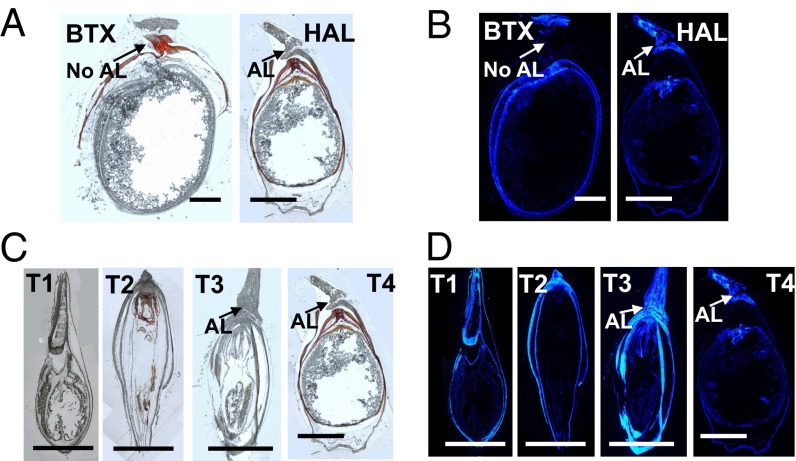
Lignin deposition at the seed–stalk interface is closely correlated with shattering of the sorghum inflorescence. Lignin staining of nonshattering sorghum revealed no deposition of lignin in the seed pedicel, in contrast to the fluorescence seen in the pericarp (Fig. 6B). In shattering sorghum (HAL; S. halepense) there is heavy lignin deposition in the stalk–glume junction, stopping abruptly as the glume starts. Across four stages of seed maturity (Fig. 6 C and D), lignin deposition is tightly correlated with the emergence of the abscission layer (AL) at later stages of seed development.
Fig. 6.
UV lignin stains of the seeds from BTX (S. bicolor, nonshattering) and HAL (S. halepense, shattering), showing parts of the seed structure, pedicel, glume, and abscission layer (AL). T4 mature stage of seeds from BTX and HAL, (A) unstained and (B) lignin stained. Various development stages (T1 through T4, in progressive maturity level: T1, early histo-differentiation; T2, embryonic cell expansion; T3, endosperm filling; T4, preabscision desiccation) of seeds from HAL, (C) unstained and (D) lignin stained. (Scale bar, 1 mm.)
We hypothesize that SpWRKY functions in a manner analogous to Medicago and Arabidopsis homologs shown to repress cell wall biosynthesis genes (33). The low expression of SpWRKY toward the end of floral development may derepress its downstream cell wall biosynthesis genes, allowing deposition of lignin hence initiating the abscission zone in the seed–pedicel junction. The general pattern of expression of SbWRKY is similar to SpWRKY through the first stages of floral development, with a spike of expression of during anther dehiscence (Fig. 2C; WA, without anthers). The high expression of SbWRKY in the final floral development stage may keep cell wall biosynthesis genes repressed in S. bicolor, also repressing the shattering phenotype. This difference in the expression profile between SbWRKY and SpWRKY alleles may be related to or partially caused by the shift in position of the start codon. However, the concomitant changes in both sequences and their expression patterns may necessitate further experiments to determine the exact molecular mechanism that differentiates SbWRKY and SpWRKY with regard to shattering.
Despite conventional wisdom that shattering of sorghum is genetically dominant (2), correlation of transgene copy numbers with the level of shattering indicates that the effect of SbWRKY is to some degree dependent on dosage. In the segregating populations derived from self-pollinated sorghums containing different transformation events, we also estimated SpWRKY copy number based on the intensity of amplified bands. There is a very strong negative correlation between BTS and the inferred copy number. Plants with no SP allele had a median BTS value of 56 g. Plants that contain the transgene had median values of 38.7 g (∼31% reduction) for those with very faint SpWRKY bands, 24.4 g (∼56% reduction) for faint, 13.9 g (∼75% reduction) for medium, and 12.8 g (∼77% reduction) for strong (Fig. S3).
Relationship to the YABBY Protein and the Genetics of Shattering Trait in Sorghum Species.
The YABBY protein (Sb01g013270; Chr1: 12.29 Mb) reported to account for shattering in an intraspecific sorghum population (18) is outside of the region to which we delineated Sh1 by genetic recombination but physically is a remarkably close ∼300 kb to SpWRKY (Sb01g012870; Chr1: 11.94 Mb). The proximity of these two genes, each implicated in the shattering phenotype, challenges the proposition of a single controlling locus as previously assumed. Indeed, in crosses segregating for functional polymorphisms at both loci, their close genetic linkage may have interfered with being recognized as separate loci.
We showed that the SpWRKY allele evolved in S. propinquum after its divergence from S. bicolor, partially explaining how there got to be (at least) two shattering genes in the Sorghum genus. Both WRKY and YABBY genes contain DNA-binding domains known to regulate the transcription of genes involved in cell wall biosynthesis. Similar to the role of the WRKY gene in ectopic lignin deposition (33), the fruit from the Arabidopsis double mutant fil yab3 [YAB3 is a homolog of YABBY (18)] lacks lignin in cells normally found in the valve margin (7).
It is possible that both WRKY and YABBY might regulate some of the same downstream genes involved in lignin deposition and thus each play important roles in the shattering pathway; however, there would be different relative contributions in the two species of sorghum studied. Our transgenic introduction of the SpWRKY allele into a background (RTx430) containing the nonshattering YABBY allele restored the shattering phenotype, suggesting another possibility of epistatic interaction between YABBY and WRKY. It remains a subject for further investigation what the modes of interaction between the two loci are, if any. It would be interesting, for example, to see if the expressions of the two genes overlap spatially and/or temporally. Thurber et al. suggested that the time of formation and degradation of abscission layers may vary between different shattering rice species, partially explaining the fact that there are separate loci controlling the trait (3).
In contrast to the hypothesis of convergent pathways for crop domestication (2, 18), the existence of two different sorghum shattering genes WRKY and YABBY supports an alternative hypothesis of multiple genetic pathways for crop domestication traits. There is a growing list of examples of major crop domestication phenotypes having diverse genetic architecture. Another related example is the seed threshability trait of the common wheat. In A- and D-genome diploid progenitors, two separate nonhomologous genes (sog and Tg) arose by independent mutations, but both are known to contribute to the tenacity of the wheat glume (36).
Implications for Sorghum Domestication.
The finding of recent and independent mutations at orthologs of the WRKY locus, both among S. bicolor genotypes (in our association study) and among divergent plant species, seems to suggest that the S. propinquum WRKY shattering locus is unstable and perhaps would have been less likely to reach high frequency during domestication than the S. bicolor YABBY locus. Given that SpWKRY is a gain-of-function mutation and thus far found only in S. propinquum, it likely represents the evolution of the genetic architecture of shattering in the wild species and played little or no role in the domestication of sorghum.
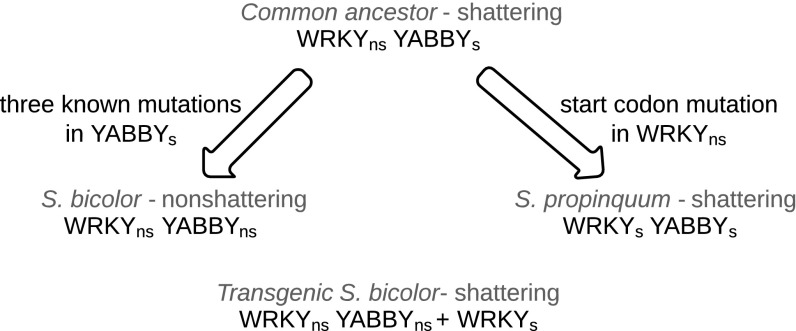
Nevertheless, the discovery of SpWRKY adds to our understanding of the genetics of shattering and the evolution of domestication-related traits. Seed shattering is often under opposing selection in crops and their wild relative; whereas humans tend to select nonshattering crops, seed dispersal in wild species may tend to favor maintaining or increasing the level of shattering. The seed shattering phenotype was probably the ancestral condition, whereas loss of shattering is a derived trait in the domesticate. This implies that the ancestral sorghum that predated the divergence of S. bicolor and S. propinquum contained the shattering allele of YABBY but the nonshattering allele of WRKY (Fig. 7). After the two sorghum species diverged, several nonshattering alleles of YABBY emerged in the S. bicolor population and were subsequently selected by humans (18), whereas the evolution of SpWRKY in S. propinquum perhaps reinforced or further increased the level of shattering (Fig. 7).
Fig. 7.
Inferred evolutionary history of two shattering loci in two Sorghum species. Subscripts: s, shattering allele; ns, nonshattering allele.
Our finding that the shattering phenotype is controlled by a different locus in the wild relative than in the domesticated relative is not an isolated case. All US weedy rice individuals share the same nonshattering mutation at the sh4 locus, which has been associated with loss of shattering during rice domestication, yet must have reacquired the shattering phenotype through unidentified regulatory genes other than sh4 (3). Similarly, we showed by transformation that the SpWRKY gene in the wild sorghum is fully capable of restoring the shattering phenotype even in the presence of homozygosity for the nonshattering allele of the domestication locus YABBY.
Additionally, the discovery of recent and independent duplications of maize and Setaria orthologs each followed by length mutations, together with our analysis of other sorghum species, suggests a tendency to recurring mutation at the locus. We currently only possess one valid S. propinquum accession (a second accession listed in the US Department of Agriculture National Plant Germplasm System has been substantially introgressed with S. bicolor chromatin, based on microsatellite genotyping); thus, we presently lack data about intraspecific S. propinquum sequence variation at the locus. Variation in both S. bicolor and S. propinquum would reconcile our association results with the fact that the specific allele that we cloned evolved in S. propinquum.
Materials and Methods
A diversity panel of sorghum varieties were collected from Cornell University and the US Department of Agriculture–Agricultural Research Service germplasm collection, selected to represent a wide range of geographical locations. The shattering phenotype for each accession was carefully validated by measuring BTS (measured in grams) using a digital force gauge (DPS-4; IMADA). Resequencing used BigDye terminator chemistry, and the chromatograms were examined using SEQUENCHER software (version 4.1; GENECODES). The association analyses were performed using TASSEL software using a general linear model. Candidate genes in the high association region were isolated from the BAC YRL20H16, cloned in pZP211, and transformed into nonshattering RTx430. Detailed methods and associated references are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank B. Richardson for assistance with light microscopy and the late K. F. Schertz for key contributions to the early stages of this work. Financial support was provided by US Department of Agriculture Grant 01-35301-10595 (to A.H.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305213110/-/DCSupplemental.
References
- 1.Kislev ME, Weiss E, Hartmann A. Impetus for sowing and the beginning of agriculture: Ground collecting of wild cereals. Proc Natl Acad Sci USA. 2004;101(9):2692–2695. doi: 10.1073/pnas.0308739101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paterson AH, et al. Convergent domestication of cereal crops by independent mutations at corresponding genetic Loci. Science. 1995;269(5231):1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- 3.Thurber CS, Hepler PK, Caicedo AL. Timing is everything: Early degradation of abscission layer is associated with increased seed shattering in U.S. weedy rice. BMC Plant Biol. 2011;11:14. doi: 10.1186/1471-2229-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liljegren SJ, et al. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404(6779):766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- 5.Ferrándiz C, Liljegren SJ, Yanofsky MF. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science. 2000;289(5478):436–438. doi: 10.1126/science.289.5478.436. [DOI] [PubMed] [Google Scholar]
- 6.Roeder AH, Ferrándiz C, Yanofsky MF. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr Biol. 2003;13(18):1630–1635. doi: 10.1016/j.cub.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 7.Dinneny JR, Weigel D, Yanofsky MF. A genetic framework for fruit patterning in Arabidopsis thaliana. Development. 2005;132(21):4687–4696. doi: 10.1242/dev.02062. [DOI] [PubMed] [Google Scholar]
- 8.Konishi S, et al. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312(5778):1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- 9.Li C, Zhou A, Sang T. Rice domestication by reducing shattering. Science. 2006;311(5769):1936–1939. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- 10.Konishi S, Ebana K, Izawa T. Inference of the japonica rice domestication process from the distribution of six functional nucleotide polymorphisms of domestication-related genes in various landraces and modern cultivars. Plant Cell Physiol. 2008;49(9):1283–1293. doi: 10.1093/pcp/pcn118. [DOI] [PubMed] [Google Scholar]
- 11.Zhang LB, et al. Selection on grain shattering genes and rates of rice domestication. New Phytol. 2009;184(3):708–720. doi: 10.1111/j.1469-8137.2009.02984.x. [DOI] [PubMed] [Google Scholar]
- 12.Nalam VJ, Vales MI, Watson CJ, Johnson EB, Riera-Lizarazu O. Map-based analysis of genetic loci on chromosome 2D that affect glume tenacity and threshability, components of the free-threshing habit in common wheat (Triticum aestivum L.) Theor Appl Genet. 2007;116(1):135–145. doi: 10.1007/s00122-007-0653-7. [DOI] [PubMed] [Google Scholar]
- 13.Nalam VJ, Vales MI, Watson CJ, Kianian SF, Riera-Lizarazu O. Map-based analysis of genes affecting the brittle rachis character in tetraploid wheat (Triticum turgidum L.) Theor Appl Genet. 2006;112(2):373–381. doi: 10.1007/s00122-005-0140-y. [DOI] [PubMed] [Google Scholar]
- 14.Li W, Gill BS. Multiple genetic pathways for seed shattering in the grasses. Funct Integr Genomics. 2006;6(4):300–309. doi: 10.1007/s10142-005-0015-y. [DOI] [PubMed] [Google Scholar]
- 15.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457(7229):551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 16.Bowers JE, et al. Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses. Proc Natl Acad Sci USA. 2005;102(37):13206–13211. doi: 10.1073/pnas.0502365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vicentini A, Barber JC, Aliscioni SS, Giussani LM, Kellogg EA. The age of the grasses and clusters of origins of C4 photosynthesis. Global Change Biol. 2008;14(12):2963–2977. [Google Scholar]
- 18.Lin Z, et al. Parallel domestication of the Shattering1 genes in cereals. Nat Genet. 2012;44(6):720–724. doi: 10.1038/ng.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y-R, et al. A Sorghum propinquum BAC library, suitable for cloning genes associated with loss-of-function mutations during crop domestication. Mol Breed. 1999;5(6):511–520. [Google Scholar]
- 20.Soderlund C, Longden I, Mott R. FPC: A system for building contigs from restriction fingerprinted clones. Comput Appl Biosci. 1997;13(5):523–535. doi: 10.1093/bioinformatics/13.5.523. [DOI] [PubMed] [Google Scholar]
- 21.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaut BS, Morton BR, McCaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA. 1996;93(19):10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5(5):199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 24.Gao G, et al. DRTF: A database of rice transcription factors. Bioinformatics. 2006;22(10):1286–1287. doi: 10.1093/bioinformatics/btl107. [DOI] [PubMed] [Google Scholar]
- 25.Tang H, et al. Synteny and collinearity in plant genomes. Science. 2008;320(5875):486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 26.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 27.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 2001;28(2):123–133. doi: 10.1046/j.1365-313x.2001.01131.x. [DOI] [PubMed] [Google Scholar]
- 29.Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16(9):1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14(6):1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagacé M, Matton DP. Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta. 2004;219(1):185–189. doi: 10.1007/s00425-004-1253-2. [DOI] [PubMed] [Google Scholar]
- 32.Naoumkina MA, He X, Dixon RA. Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol. 2008;8:132. doi: 10.1186/1471-2229-8-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, et al. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc Natl Acad Sci USA. 2010;107(51):22338–22343. doi: 10.1073/pnas.1016436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guillaumie S, et al. The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol Biol. 2010;72(1-2):215–234. doi: 10.1007/s11103-009-9563-1. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, et al. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol. 2007;65(6):799–815. doi: 10.1007/s11103-007-9244-x. [DOI] [PubMed] [Google Scholar]
- 36.Sood S, Kuraparthy V, Bai G, Gill BS. The major threshability genes soft glume (sog) and tenacious glume (Tg), of diploid and polyploid wheat, trace their origin to independent mutations at non-orthologous loci. Theor Appl Genet. 2009;119(2):341–351. doi: 10.1007/s00122-009-1043-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.