Significance
Animals form functional unities with communities of microbes. Often, these bacterial communities are highly specific to host species and resemble host phylogeny. But which factors determine community membership? Which host-factors are capable of selecting suitable bacteria by inhibiting colonization by potential foreign colonizers? In this study, we show that animals express a species-specific repertoire of antimicrobial peptides, which supports and maintains a species-specific bacterial community. Loss-of-function experiments showed that antimicrobial peptide composition is a predictor for bacterial colonization.
Keywords: host-microbe, Cnidaria, phylosymbiotic, core microbiota, holobiont
Abstract
Animals are colonized by coevolved bacterial communities, which contribute to the host’s health. This commensal microbiota is often highly specific to its host-species, inferring strong selective pressures on the associated microbes. Several factors, including diet, mucus composition, and the immune system have been proposed as putative determinants of host-associated bacterial communities. Here we report that species-specific antimicrobial peptides account for different bacterial communities associated with closely related species of the cnidarian Hydra. Gene family extensions for potent antimicrobial peptides, the arminins, were detected in four Hydra species, with each species possessing a unique composition and expression profile of arminins. For functional analysis, we inoculated arminin-deficient and control polyps with bacterial consortia characteristic for different Hydra species and compared their selective preferences by 454 pyrosequencing of the bacterial microbiota. In contrast to control polyps, arminin-deficient polyps displayed decreased potential to select for bacterial communities resembling their native microbiota. This finding indicates that species-specific antimicrobial peptides shape species-specific bacterial associations.
Epithelial surfaces of most animals are colonized by complex bacterial communities (1–4). This commensal microbiota has been shown to be beneficial for a broad range of host-physiological functions, including facilitation of nutrient supply (5–7), immune system maturation (8–10), gut development (11), and colonization resistance against pathogens (12). This finding is supported by observations of severe fitness disadvantages in germ-free animals (13) and evidence that dysregulation of host–bacterial homeostasis is involved in the occurrence of disorders, such as inflammatory bowel disease (14, 15). However, the processes that determine community membership in the microbiota are not fully understood, which has encouraged discussions as to what extent the microbiota is controlled by the host through top-down mechanisms, relative to microbiota-intrinsic or environmental-mediated factors (16, 17).
In 2007, Fraune and Bosch uncovered that two species of the cnidarian Hydra are colonized by remarkably different bacterial communities, despite being cultured under identical laboratory conditions for decades (1). These laboratory cultures were colonized by microbial communities similar to that of the same Hydra species freshly isolated from the wild, indicating strong host-mediated selective forces on the associated microbiota (1).
Compelling evidence for host-control over commensal bacteria also comes from reciprocal microbiota transplantations of zebrafish and mice into germ-free recipients (18). In that study, the authors demonstrated that the recipient host shapes the community structure of the transferred, foreign microbiota to resemble their native bacterial community (18). However, the study did not elucidate the factors responsible for host-mediated community control. Several host-factors are suggested to have influence on microbiota composition, ranging from oxygen conditions in the gut, nutrient intake, temperature, mucus barriers, and immunity (reviewed in ref. 17). All of these factors are likely to differ drastically between mouse and zebrafish.
Several studies have shown an active cross-talk between the host’s immune system and its associated microbiota. Commensal microbes are able to drive fundamental aspects of innate and adaptive immunity, such as T-cell maturation (9, 19), production of IgA, mucus secretion (20), and induction of innate immunity-effector molecules, such as antimicrobial peptides (AMPs) (21). Similarly, the host’s immune system appears to regulate the abundance and composition of the microbiota (22–26). Studies in mice have shown that the expression level of AMPs of the α-defensin family greatly affects community composition (25). In the cnidarian Hydra, sequentially expressed AMPs of the periculin family mediate the establishment of the bacterial microbiota during embryogenesis (27).
In Hydra, AMPs of the arminin peptide family are among the most highly expressed genes. Multiple arminins have been identified in Hydra magnipapillata, all of them being short, secreted peptides (28). The propeptide consists of a highly conserved, negatively charged N-terminal region and a rather nonconserved, highly cationic C-terminal part, which was predicted to be cleaved to generate the bacteriocidal fragment (28). Consistent with that prediction, the synthetically produced C-terminal fragment of Arminin 1a showed strong antibacterial activity against Escherichia coli, Bacillus megaterium, and several methicillin-resistant Staphylococcus aureus strains in concentrations equal or lower than 0.4 µM (28).
In the present study, we addressed the question whether species-specific AMPs shape species-specific bacterial communities. In particular, we investigated the effect of arminin deficiency in the cnidarian host Hydra. Arminin-deficient and control polyps were inoculated with native as well as foreign bacterial communities characteristic for the closely related species Hydra oligactis and Hydra viridissima. Whereas control polyps selected for bacterial communities resembling their native microbiota, this host-driven selection was significantly less pronounced in arminin-deficient polyps. These data provide strong evidence for a role of species-specific AMPs in selecting suitable bacterial partners, leading to host-species specific bacterial associations.
Results
Polyps of Hydra Are Associated with Species-Specific Bacterial Communities.
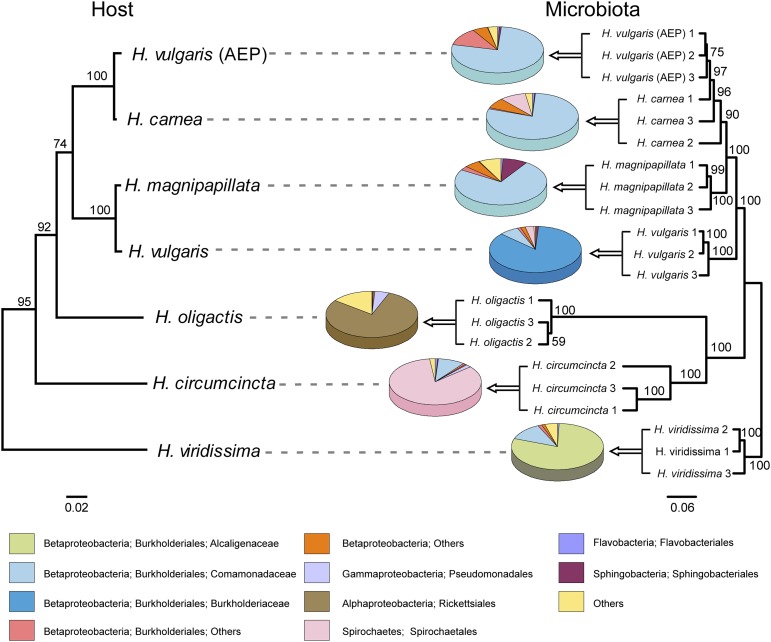
The associated bacterial communities of seven Hydra species were characterized by 454 pyrosequencing the variable regions 1 and 2 of the bacterial 16S rRNA gene, amplified from total DNA extracted from single Hydra polyps. All species were laboratory-reared under identical conditions including diet, medium, and temperature for more than three decades, and therefore share a highly similar physiology. Pyrosequencing resulted in 79,130 high-quality reads ranging from 1,310 to 10,130 reads per sample. For intersample comparisons, sequences were rarified to 1,300 reads per sample, grouped into operational taxonomic units (OTUs) at a ≥ 99% sequence identity threshold, and classified by the Ribosomal Database Project (RDP) classifier. The comparison of the hierarchical cluster tree and the phylogenetic tree of the Hydra species (29) reveals strong host-specificity of Hydra-associated bacterial communities, which reflects the phylogenetic relationship of their hosts (Fig. 1). Interestingly, the microbiota of Hydra vulgaris (AEP), the strain used to generate transgenic Hydra, clustered together with Hydra carnea. This finding is noteworthy because molecular phylogenetic analysis of Hydra revealed that H. vulgaris (AEP) is more closely related to H. carnea than to its eponym H. vulgaris (29). The microbiota of all seven Hydra species was dominated by Gram-negative bacteria. Betaproteobacteria of the family Comamonadaceae or Burkholderiaceae dominated in the closely related species H. magnipapillata, H. carnea, H. vulgaris (AEP), and H. vulgaris. The most basal species, H. viridissima, was colonized by host species-specific bacteria of the Alcaligenaceae family. This Hydra species harbors intracellular symbiotic algae of the genus Chlorella. Reads of amplified Chlorella chloroplast 16S rRNA genes were removed in silico. H. oligactis and Hydra circumcincta were characterized by the dominance of Alphaproteobacteria or Spirochaetes, respectively (Fig. 1).
Fig. 1.
Comparison of the phylogenetic tree of Hydra species and the cluster tree of the corresponding bacterial communities. (Left) Phylogenetic tree of Hydra species based on cytochrome oxidase genes [maximum likelihood, general time reversible (GTR+I)]. Bootstrap values are shown at the corresponding nodes. The branch-length indicator displays 0.02 substitutions per site. H. vulgaris (AEP) (EF059935), H. carnea (EF059940), H. magnipapillata (EF059934), H. vulgaris (EF059936), H. oligactis (EF059937), H. circumcincta (EF059938), H. viridis (EF059941). (Right) Jacknife environment cluster tree (weighted UniFrac metric, rarified to 1,300 sequences per sample) of 21 bacterial communities from seven different Hydra species. One-thousand replicates were calculated; nodes are marked with Jackknife support values. The branch-length indicator displays distance between samples in Unifrac units. Pie charts represent mean relative abundance of bacterial orders. The highly variable Burkholderiales order was separated into different families.
Hydra Species Maintain Their Specific Bacterial Associations in Cocultivation.
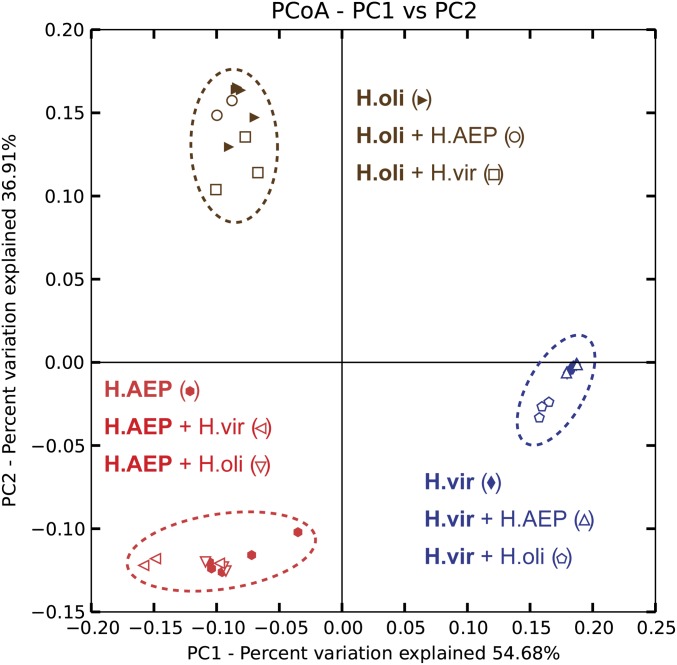
To demonstrate that the observed species-specificity of host–bacterial associations is not the result of long-term, separate cultivation and stochastic exposure to different bacteria, pairwise cocultivation of the morphologically distinguishable species H. vulgaris (AEP), H. oligactis, and H. viridissima were conducted. Following 5 wk of cocultivation, single polyps were subjected to 454 pyrosequencing of their microbiota. After removal of chimeric sequences and reads assigned to Chlorella sp. chloroplasts in H. viridissima, pyrosequencing resulted in 1,394–6,790 reads per sample, which were subsequently rarified to 1,350 reads per sample. As shown in Fig. 2, all three Hydra species maintain their specific bacterial profiles even under cocultivating conditions, leading to three species-specific clusters characteristic for H. vulgaris (AEP), H. oligactis, and H. viridissima. These results clearly validate the host as a major determinant for bacterial community composition.
Fig. 2.
Host-species–specific bacterial communities remain stable in pairwise coculture. Pairwise cocultivations were conducted between the morphological distinguishable species H. vulgaris (AEP), H. oligactis, and H. viridissima. Bacterial communities were clustered using principle coordinate analysis of the weighted Unifrac distance matrix. The percent variation explained by the principle coordinates is indicated at the axes. Reads were rarified to 1,350 reads per sample. H.oli, Hydra oligactis; H.AEP, Hydra vulgaris (AEP); H.vir, Hydra viridissima; +, cocultured. For cocultivation samples, the sequenced sample in written bold. Cocultivations were conducted in biological triplicates (n = 3); n = 5 for noncocultivated polyps (H.oli, H.AEP, H.vir). Certain samples cluster strongly together such that single symbols may be overlaid.
AMPs of the Arminin Family Show Species-Specific Composition in Different Species of Hydra.
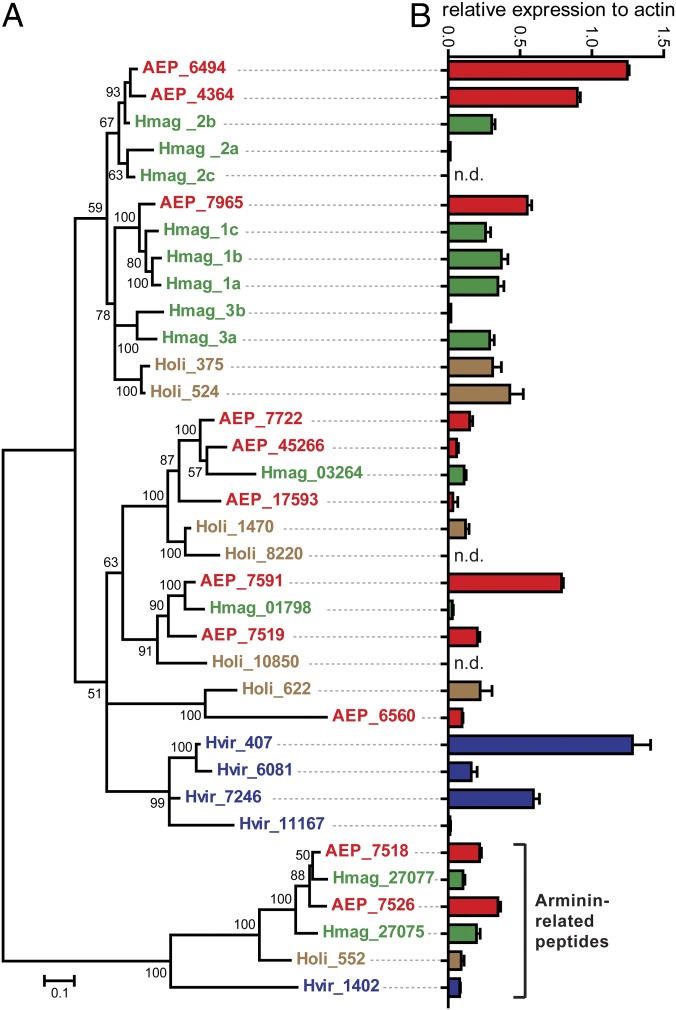
After assessing the host as one determining factor for the microbiota composition, candidate host factors capable of influencing the bacterial communities in a species-specific manner were examined. Because Hydra lacks an adaptive immune system, the focus was set on its innate immunity. Tissue homogenates of Hydra possess strong bacteriocidal activity (30), most likely a result of the expression of potent antimicrobial peptides (27, 28, 31). Because arminins (28) are among the most highly expressed AMPs in Hydra, the available transcriptomes of H. vulgaris (AEP), H. oligactis, and H. viridissima were screened for orthologs of arminins by comparing the conserved N-terminal region of the previously published H. magnipapillata genes (28) through a BLAST search. Compared with 10 genes in H. magnipapillata, nine orthologs were found in H. vulgaris (AEP), six in H. oligactis, and four in H. viridissima. In addition, a unique cluster of arminin-related peptides was discovered. Phylogenetic analysis of all identified orthologs, using the arminin-related genes as an outgroup, revealed that H. viridissima (Fig. 3, blue) expresses a unique cluster of four arminin genes (Fig. 3A). No species-specific clusters were identified for H. magnipapillata (Fig. 3, green), H. vulgaris (AEP) (Fig. 3, red), and H. oligactis (Fig. 3, brown). The expression level of the distinct arminins was determined by comparing microarray signal intensities to the expression level of the highly expressed housekeeping gene β-actin of the corresponding Hydra species (Fig. 3B). The expression of some arminins exceeds that of β-actin, indicating the biological relevance of this gene family in Hydra. The expression profile of arminin paralogs differed between Hydra species [e.g., the highest expressed paralogs of H. vulgaris (AEP) are grouped in a different cluster than the dominant arminins in H. oligactis (Fig. 3)]. The closer related species H. vulgaris (AEP) and H. magnipapillata show a more similar profile of arminin expression, albeit H. vulgaris (AEP) generally has higher expression levels. Taking these data together, this analysis indicates that each Hydra species is equipped with a unique repertoire of AMPs.
Fig. 3.
The arminin family of antimicrobial peptides. (A) Phylogenetic analysis of the arminin AMP family from four different Hydra species. The tree was built by Bayesian interference of phylogeny. A total of 3 million generations were calculated using the general time reversible model and the invgamma rate variation and four chains with a burn-in of 25%. Posterior probabilities are shown at the corresponding nodes. Genes are colored according to species. Numbers indicate contig numbers in the species transcriptome. AEP, H. vulgaris (AEP); Hmag, H. magnipapillata; Holi, H. oligactis; Hvir, H. viridissima. (B) Relative expression of each arminin, compared with the expression of β-actin in the corresponding species. Expression data were retrieved from microarray data, conducted in three biological replicates. Bar charts represent mean + SD. Certain paralogues had no corresponding microarray probes (n.d., not detected).
The expression of arminins in H. vulgaris (AEP) was analyzed in more detail. As shown in Fig. S1, most arminins are expressed exclusively in endodermal epithelial cells, thus likely being secreted to the gastric cavity, a compartment resembling the mammalian intestine. Paralog 6560 was shown to be expressed in the ectodermal epithelium. None of the paralogs are expressed in the tentacles or in the hypostome region, restricting the localization to the body column.
Silencing of Arminin Decreases the Antibacterial Activity of Hydra Tissue.
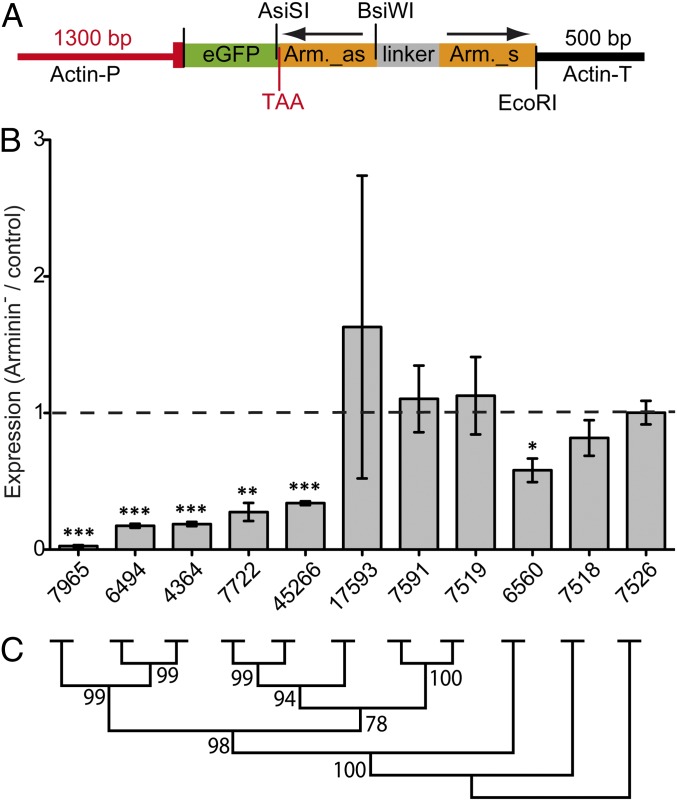
To broadly interfere with the host’s expression of endogenous arminins, transgenic H. vulgaris (AEP) polyps expressing a hairpin cassette containing the arminin7965 antisense and sense sequences fused to the reporter gene egfp (Fig. 4A) were generated. By several rounds of asexual proliferation, two stable lines were established originating from a mosaic founder polyp: the control line, which contained no remaining eGFP+ cells, and the arminin-deficient (Arminin−) line, expressing the hairpin-transgene in the complete endodermal epithelial cell lineage. By using this control line, the effect of transgenic interference could be analyzed without the need to account for different genomic backgrounds. Neither control nor Arminin− polyps displayed any obvious morphological phenotypes (Fig. S2).
Fig. 4.
Successful knockdown of arminin family members in H. vulgaris (AEP). (A) Arminin–hairpin construct for generation of transgenic Hydra (as, antisense; s, sense; TAA, stop codon; P, promoter; T, terminator). (B) Relative expression of arminins quantified by qRT-PCR with specific primers. cDNA of control polyps served as reference (dashed line). cDNA amounts were equilibrated by EF1α, the graphic shows means + SEM (n = 3). Statistic was carried out using two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001. (C) Neighbor-joining tree of arminins from H. vulgaris (AEP). Bootstrap values are shown at the corresponding nodes (1,000 replicates).
Arminin-deficient polyps show a 97% decrease of the endogenous arminin7965 transcript as determined by quantitative real-time PCR (qRT-PCR) (Fig. 4B). Because the 5′ sequences of paraloguous arminins shows high sequence identity (Fig. S3A), the hairpin-mediated RNAi targeted several arminins, leading to a significant decrease in expression of the genes 7965, 6494, 4364, 7722, and 45266. This decrease in expression is correlated with the level of sequence identity to gene 7965, the sequence used to generate the hairpin transgene (Fig. 4C).
The decreased expression of several arminins in Arminin− polyps drastically changed the AMP composition of the transgenic H. vulgaris (AEP) polyps, with the highest expressed paralog switching from 6494 to 7591 (Fig. S3 B and C). Total arminin expression was reduced by ∼50%. To assess the impact on protein level, peptide extracts of Arminin− polyps and control polyps were tested against E. coli DH5α, which was previously reported to be sensitive to arminin (28). The minimal inhibitory concentration (MIC) of Arminin− extracts was doubled compared with control extracts (Table S1). In other words, the bacteriocidal activity of Hydra tissue extract was reduced by 50% in Arminin− polyps. This effect was also clearly visible in radial diffusion assays against E. coli DH5α (Table S1).
Species-Specific Arminins Select for Coevolved Bacterial Colonizers.
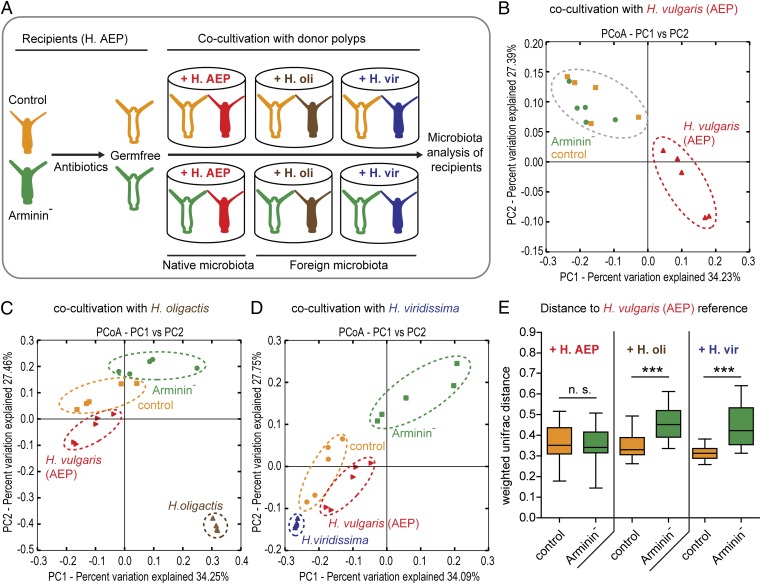
The expression profile of arminins is specific to different Hydra species (Fig. 3) and contributes a major portion to the antibacterial activity of the host’s tissue (Table S1). Thus, a logical question is whether host-species–specific expression of arminins contributes to the observed species-specific bacterial associations (Fig. 1). To answer this question, germ-free control and Arminin− polyps were generated by antibiotic treatment (Fig. S4). These polyps were separated in single wells and cocultivated with either H. vulgaris (AEP), H. oligactis, or H. viridissima for 5 wk (Fig. 5A). Following that period, the recipients as well as the donor polyps were subjected to 454 sequencing of the bacterial microbiota. Five biological replicates (= single polyps) were conducted for each treatment. H. viridissima chloroplast sequences were removed in silico. Resulting high-quality reads ranged from 1,329 to 6,755 per sample. Sequences were rarified to 1,300 sequences per sample, grouped into OTUs at a ≥ 97% sequence-identity threshold and classified by the RDP classifier.
Fig. 5.
Species-specific arminins select for species-specific bacteria. (A) Schematic representation of the experimental design. Germ-free control polyps as well as germ-free Arminin− polyps [= transgenic H. vulgaris (AEP)] were cocultivated with either H. vulgaris (AEP), H. oligactis (H.oli), or H. viridissima (H.vir) polyps for 5 wk. (B–D) Bacterial communities were clustered using PCoA of the weighted Unifrac distance matrix. All three PCoA plots are based on the same distance matrix containing all 45 samples, and therefore all samples are presented relative to each other. The percent variation explained by the principle coordinates is indicated at the axes. (B) When inoculated with native bacteria retrieved from H. vulgaris (AEP), control and Arminin− polyps show no differences in bacterial recolonization. (C and D) When cocultivated with H. oligactis (C) or H. viridissima (D), control and Arminin− polyps cluster separately, with recolonized control polyps clustering proximate to H. vulgaris (AEP) reference polyps. (E) Comparison of the weighted Unifrac distances to the native H. vulgaris (AEP) microbiota. Control polyps infected with a H. oligactis or H. viridissima microbiota show significantly lower Unifrac distances to their bona fide wild-type status than Arminin− polyps. Statistics were carried out using two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001.
When inoculated with native, H. vulgaris (AEP)-specific microbiota, recipient control and Arminin− H. vulgaris (AEP) polyps displayed no differences in their bacterial recolonization, as indicated by their clustering in a principle coordinate analysis (PCoA) based on the weighted Unifrac metric (ANOSIM R = 0.15, P = 0.141) (Fig. 5B and Table S2). In contrast, inoculation with foreign bacterial communities provided by H. oligactis cocultivation led to differential recolonization in control and Arminin− polyps. As indicated by PCoA, microbial profiles of recolonized control and Arminin− polyps clustered separately (ANOSIM R = 0.62, P = 0.007), with recolonized control polyps clustering in close proximity to H. vulgaris (AEP) polyps (Fig. 5C and Table S2). Similar but even more drastic differential recolonization between control and Arminin− polyps was observed when recipients were inoculated with the microbiota from H. viridissima (ANOSIM R = 0.82, P = 0.008) (Fig. 5D and Table S2). In addition, we quantified the recipient’s approximation to the native microbiota by comparing weighted Unifrac distances between recolonized control polyps, Arminin− polyps, and H. vulgaris (AEP) polyps. As shown in Fig. 5E, the microbiota of control polyps recolonized by H. oligactis or H. viridissima bacterial communities significantly better resembled the native microbiota of H. vulgaris (AEP) than the reestablished communities in Arminin− polyps, indicating a loss of selective preferences because of disturbed arminin expression. Interindividual microbiota variation and bacterial species diversity were not significantly different between recolonized control and Arminin− polyps (Fig. S5 A–D).
To test the influence on community clustering of other β-diversity measures incorporating phylogeny (i.e., Unifrac) versus those that are OTU-based (Bray–Curtis, Jensen–Shannon), we additionally included the Bray–Curtis, Jensen–Shannon, and unweighted Unifrac distances (Fig. S6 and Table S2). Although the Bray–Curtis and Jensen–Shannon metrics strongly support the presence of the described clusters seen with the weighted Unifrac metric (Fig. 5 B–D), the unweighted Unifrac metric reveals little or none of the previously described clustering (Fig. S6 and Table S2). These results indicate that the observed patterns are characterized by differences in both taxon abundance and phylogeny.
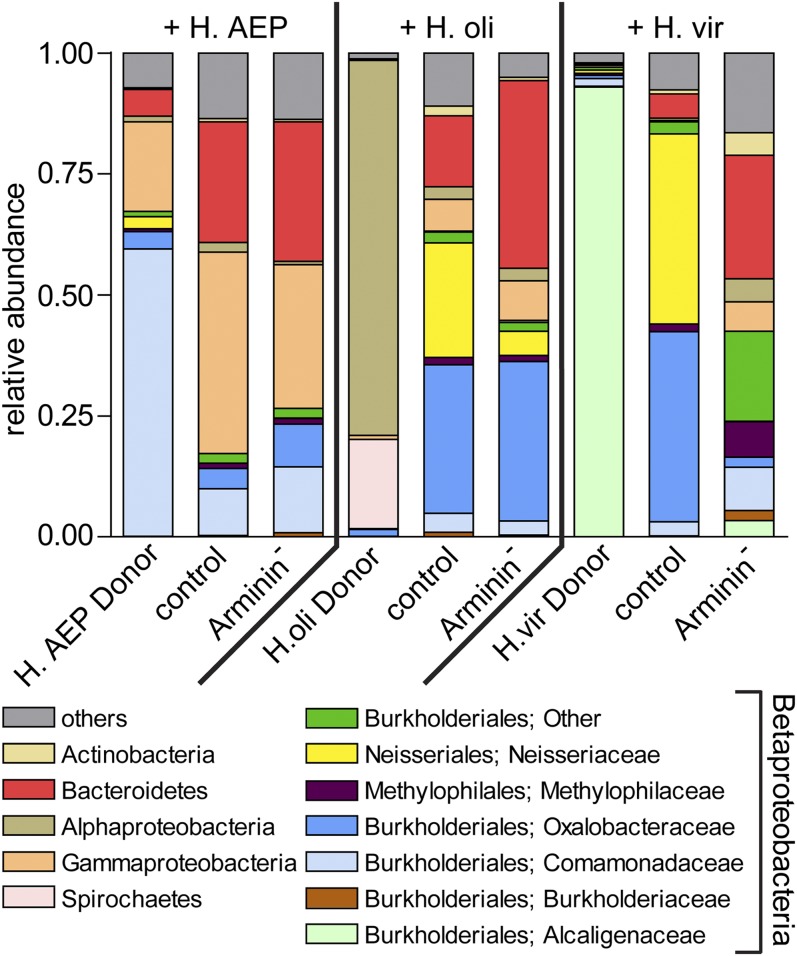
Fig. 6 summarizes the bacterial communities of donor and recipient polyps. When recolonized by H. vulgaris (AEP)-specific bacteria, both control and Arminin− polyps tended toward bacterial types abundant in the donor microbiota. Bacteroidetes, Comamonadaceae, and Gammaproteobacteria were transferred by cocultivation and established associations with the recipient polyps (Fig. 6). The outcome of the recolonization process differed, however, when polyps were inoculated with bacterial communities provided by H. oligactis or H. viridissima. Dominant bacterial taxa of H. oligactis or H. viridissima were not transmitted to recipient polyps. Instead, recipient polyps enriched bacteria from the donor’s rare bacterial associates (Fig. 6 and Fig. S7A). After inoculation with H. oligactis-specific bacteria, Betaproteobacteria (Neisseriales, Methylophilales, and Burkholderiales) constituted 64% of the microbiota of recolonized control polyps, compared with 45% in recolonized Arminin− polyps. Arminin− polyps showed an increased colonization by species belonging to Bacteroidetes (two-tailed t test, P = 0.0099) and a significantly lower colonization with those belonging to Neisseriales (two-tailed t test, P = 0.048), compared with recolonized control polyps (Fig. 6). When recolonized by H. viridissima-specific bacteria, Betaproteobacteria accounted for 87% of the microbiota of recolonized control polyps in contrast to 43% in recolonized Arminin− polyps. In detail, Arminin− polyps displayed a significantly increased colonization by Bacteroidetes (two-tailed t test, P < 0.001) and a decreased prevalence of Neisseriales (two-tailed t test, P = 0.021) and Oxalobacteraceae (two-tailed t test, P = 0.034). Neisseriales accounted for 39% of the microbiota in recolonized control polyps and were absent in Arminin− polyps (Fig. 6).
Fig. 6.
Bar charts representing the microbiota of donor and recipient polyps. Mean relative abundances (n = 5) of bacterial classes. The highly abundant Betaproteobacteria were split into different families.
To evaluate the origin of single OTUs in more detail, we analyzed if the bacteria successfully recolonizing the recipient polyps are indeed present in the donor species (Fig. S7). The heat map (Fig. S7A) illustrates 48 representative OTUs by showing the abundance of each OTU in the different treatments. It is apparent that many bacteria, which recolonize the tissue of control and Arminin− polyps, are also present in the corresponding donor species, albeit often in low abundances in the case of the foreign donor species H. oligactis and H. viridissima. For example, in the Bacteroidetes group, OTU 1367 and 1621 are the major recolonizers in all three experiments and are also detectable in all three donor species. It is further evident that the polyps that were recolonized by foreign bacteria harbor a different set of Betaproteobacteria compared with the polyps which got recolonized by its native bacteria. That is, polyps recolonized with H. vulgaris (AEP) harbor mainly Comamonadaceae (OTU 761 and 778) but polyps recolonized with foreign microbiota harbor mainly Oxalobacteraceae (OTU 899, 1251, and 1525). All of these OTUs are detectable in the corresponding donor species.
In addition, recolonizing OTUs can be identified, which are present in the donor species but absent in H. vulgaris (AEP), like the OTUs 1679 (Bacteroidetes), 1071 (Alphaproteobacteria), or 1215 (Spirochaetes).
Taken together, these data elucidate that the bacteria, which recolonize the different polyps, are indeed originating mainly from the corresponding donor species and not from the original microbiota returning. In case of the recolonization with foreign microbiota, a large fraction of successful bacteria seem to originate from the donor’s rare microbiota. Additionally, we found recolonizing OTUs of unknown origin (Fig. S7B). Because these OTUs were found only in the case of recolonization by foreign microbiota provided by H. oligactis or H. viridissima and not in recolonization by H. vulgaris (AEP)-specific bacteria, we argue that these colonizers might be undetected members of the very rare microbiota of the corresponding donor species.
Because antimicrobial activity was reduced to ∼50% in Arminin− polyps, the absolute abundance of bacteria was quantified for recipient polyps after recolonization. Bacterial load did not differ significantly between treatments. All recipient polyps were colonized by a similar amount of bacteria, regardless of the origin of inoculated microbiota or antibacterial status of the tissue (Fig. S5E), indicating that all niches offered by the host are colonized by bacteria.
Discussion
Mutualistic associations between animals and microbes can evolve by distinct selective forces. Because association with a beneficial microbiota increases host fitness (4), selective pressures should act on host-mechanisms (e.g., immune effectors), ensuring suitable bacterial colonization. Additionally, transmitted bacteria are selected for being beneficial to the host, because the increase in host fitness ensures the future availability of the habitat (16). These interlinked dependencies between the host and its associated microbes (i.e., the holobiont) led to the hypothesis of the “hologenome theory of evolution,” considering the holobiont as a unit of natural selection (32).
Species-Specific AMPs Select for Suitable Bacterial Partners.
Our analysis of associated bacterial communities of seven species of the cnidarian Hydra revealed highly species-specific bacterial associations (Fig. 1), which reflect phylogenetic relationships of the hosts. For this strong correlation between host phylogeny and microbial community composition, the term “phylosymbiotic microbiota” was recently introduced (33). Our observation is consistent with similar findings in insects (34, 35) and mammals, including a detailed survey of the gut microbiota in hominid species (3, 36). The observed associations in Hydra are extremely stable, as the analyzed species are cultivated under identical environmental conditions for up to 30 y. As shown by Ubeda et al., long-term separate cultivation imposes the risk of resulting in culture-specific shifts in the microbial community by stochastic events (37). These legacy effects of the associations were excluded by exposing the host species to identical potential colonizers by cocultivation (Fig. 2). Taken together, these data strongly indicate a role of the host tissue in mediating host–bacterial associations.
Several studies provide compelling evidence that experimental manipulation of AMPs affects the resident microbiota by changing their composition (25, 27, 38, 39) or behavior (40). However, direct evidence for species-specific AMPs acting as determinants for host-specific bacterial communities was lacking. The present data indicate that arminin-deficient Hydra polyps have a decreased ability to select suitable bacterial partners from a pool of foreign potential colonizers, as they are colonized differently than control polyps, which select for bacterial types partially resembling their native microbiota (Figs. 5 C–E and 6). The microbiota of recolonized recipient polyps failed to fully resemble that of their native microbiota, perhaps because of either native bacteria being absent in the donor microbiota or failing to be transmitted horizontally. Because only a single timepoint (5 wk postrecolonization) was analyzed, this situation might represent a transient state and change in later timepoints. However, the weighted Unifrac metric indicated similarity to the bona fide H. vulgaris (AEP) microbiota, which was significantly less pronounced in Arminin− polyps (Fig. 5E). Because related bacterial species are more likely performing similar ecological functions, the host might select for certain bacterial divisions by expressing AMPs to which these taxa are less susceptible.
As total bacterial abundance did not differ significantly between treatments, all available niches seemed to be occupied by transferred bacteria, regardless of the AMP deficiency or origin of the transferred bacteria (Fig. S5E). This finding is in line with studies analyzing the effect of changed expression levels of α-defensin AMPs in mice (25), which indicated that AMPs are host-derived regulators of microbial diversity rather than unselective bacteriocides. Similar to mammalian α-defensins (41), arminins seem to be constitutively expressed because they are not differentially regulated by myeloid differentiation primary response gene 88 deficiency or the absence of bacteria (42). This result suggests a broader role for AMPs in mediating the host–microbe interface (41).
Arminin Peptides Are Taxonomically Restricted to Hydra.
Analyzing the composition of the arminin peptide family in four different Hydra species, we found species-specific clusters of arminins which varied greatly in their expression profiles, likely causing distinct antimicrobial activity in different Hydra species. Moreover, all peptides of the arminin family are specific for the genus Hydra and are not present in the genomes of other animal taxa. This specificity may reflect habitat-specific adaptations, supporting the view that secretion of taxonomically restricted AMPs reflects habitat-specific adaptations to facilitate the control of habitat-specific bacterial colonizers (43).
These findings are in line with studies investigating the evolution of insect immunity. The genomic equipment of AMPs varies greatly within insects, with many AMPs being specific only to a few, closely related species (44). Similar to arminins in Hydra, AMP variety with independent gene expansions within the genus Drosophila indicates that these peptides belong to a fast evolving group of molecules (44, 45).
Host–Bacterial Interactions and Their Role in Speciation.
In 1927, the microbiologist Ivan E. Wallin hypothesized in his book, Symbionticism and the Origin of Species (46), that the acquisition of bacterial endosymbionts favors the origin of new species. Recently, Brucker and Bordenstein summarized three general observations which link bacterial colonization and speciation (47). First, microbial colonizers are universally present in eukaryotic hosts. Second, host–microbe associations are very specific and third, host immune genes are rapidly evolving in response to microbial colonizers.
Strong evidence for the validity of the hypothesis of microbe-assisted speciation comes from studies in aphids. In a comparison between two different strains of the pea aphid (Acyrthosiphon pisum), difference in host-plant preference was caused by the symbiosis with an endosymbiotic bacterium, which enabled the aphid to use white clover (Trifolium repens) as a food source (48). Plant preference could easily lead to reproductive isolation, thus favoring speciation. Furthermore, host-associated bacteria have been shown to cause hybrid lethality in wasps of the genus Nasonia (33) and positive assortative mating in Drosophila (49). Inoculation with a single bacterial colonizer, Lactobacillus plantarum, caused significant sexual isolation, an effect that was reversible by the administration of antibiotics. Sharon et al. state bacterial modulation of sex pheromones as a mechanism behind the observed mating preferences (49). Thus, bacterial symbionts can contribute to reproductive isolation and thus accelerate speciation. However, how do animals change their bacterial partners? One significant driver of bacterial community composition is diet (2, 49). In addition, host factors, such as differential phagocytic activity (26) or antimicrobial peptides, have been shown to drastically influence the microbiota (25, 27).
AMPs appear to evolve fast as a result of positive selection (50), and changes in antimicrobial activity are also likely to occur in animals via means of gene duplication and diversification. The data in this study indicate that changes in antimicrobial activity drastically influence the composition of the microbiota. Given the high impact of beneficial microbes on host physiology, development, and fitness (4), changing the microbiota might allow adaptation to different niches faster than evolution of novel metabolic pathways in the host genome. As shown in Fig. 1, different bacterial communities were observed in different species of the cnidarian Hydra. Because these Hydra species inhabit different habitats (51), it is tempting to speculate that differences in the microbiota contribute to diversification of their host species.
Conclusion
Hydra species are characterized by coevolved, species-specific bacterial communities. These host–bacterial associations proved to be highly stable in different environmental conditions. Our data point to a critical influence of AMP composition on selective preferences during the establishment of host–bacterial interactions.
Materials and Methods
Animals.
Experiments were carried out using Hydra vulgaris (AEP) (29), Hydra oligactis (strain 10/02), Hydra viridissima (strain A99), Hydra magnipapillata (stain 105), Hydra carnea (strain Darmstadt), Hydra vulgaris (strain Basel), and Hydra circumcincta (strain M7). All laboratory-cultured strains are avaiable at the University of Kiel. All animals were cultured under constant, identical environmental conditions including culture medium, food (first-instar larvae of Artemia salina, fed three times per week) and temperature according to standard procedures (52). For cocultivation, single polyps of two species were cultured in single wells of 12-well plates (Greiner Bio One) for 5 wk with regular feeding. For isolation of DNA, polyps were separated according to morphology or GFP expression. For all experiments, adult polyps without buds or gonads were used.
Generation of Transgenic H. vulgaris (AEP) Polyps.
Hairpin-mediated silencing of target genes in Hydra can be achieved as previously described (42). For generation of H. vulgaris (AEP) transgenics, a cassette consisting of a 318-bp-long fragment (containing the complete coding sequence) of arminin7965 and its corresponding antisense sequence, separated by a spacer of 300 bp, was cloned in the LigAF1 vector (53) behind the eGFP. The resulting vector was injected into H. vulgaris (AEP) embryos as previously described (53). Founder polyps showed stable eGFP expression in a group of endodermal cells and were expanded further by clonal propagation. By selecting for eGFP-expression, mass cultures of both, polyps with no transgenic cells (control) and polyps with full endodermal expression of eGFP (Arminin−), were generated.
Microscopic Analysis of Transgenic H. vulgaris (AEP) Polyps.
Polyps were relaxed in 2% (wt/vol) urethane before fixation in 3.5% (vol/vol) glutaraldehyde in 0.05 mol/L cacodylate buffer, pH 7.4, for 18 h at 4 °C. After washing with 0.075 mol/L cacodylate buffer for 30 min, animals were postfixed with 1% (vol/vol) OsO4 in 0.075 mol/L cacodylate buffer for 2 h at 4 °C. After additional washing for 30 min, the tissue was dehydrated in ethanol and embedded in Agar 100 resin (Agar Scientific). Semithin sections were stained according to Richardson with a solution containing 0.5% Methylene blue, 0.5% borax, and 0.5% Azur II in ddH2O at 60 °C for 1–2 min. Semithin sections were analyzed on a Zeiss Axioscope fluorescence microscope with an Axiocam (Zeiss) digital camera.
Generation of Germ-Free Hydra.
A mixed culture of control and Arminin− polyps was incubated for 1 wk in an antibiotic solution containing 50 µg/mL each of ampicillin, rifampicin, streptomycin, and neomycin, with daily exchange of the solution as previously described (42). Following 3 d of recovery in autoclaved Hydra medium, control and Arminin− polyps were separated by screening for GFP and the absence of bacteria was verified by plating homogenized polyps on R2A-Agar (ROTH) plates. After incubation at 18 °C for 3 d, the CFU were counted. Absence of CFU indicated successful antibiotic treatment.
For culture-independent analysis, total DNA was extracted from single polyps using the DNeasy Blood & Tissue Kit (Qiagen). The 16S rRNA genes were amplified using the universal primers Eub-27F and Eub-1492R (54) in a 30-cycle PCR. Sterility was verified by the absence of a PCR-product.
DNA Extraction and Sequencing of 16S rRNA Genes.
For total DNA extraction, single polyps were subjected to the DNeasy Blood & Tissue Kit (Qiagen) after being washed three times with sterile filtered culture medium. Extraction was performed following the manufacturer’s protocol, except that DNA was eluted in 50 µL. For sequencing of the bacterial 16S rRNA genes, the variable regions 1 and 2 (V1V2) were amplified using the universal forward primer V2_B_Pyro_27F (5′- CTATGCGCCTTGCCAGCCCGCTCAGTCAGAGTTTGATCCTGGCTCAG-3′), which consists of the 454 FLX Amplicon primer B (underlined), a two base linker (italics), and the universal 16S primer 27F (bold), and the barcoded reverse primer V2_A_338R (5′- CGTATCGCCTCCCTCGCGCCATCAGNNNNNNNNNNCATGCTGCCTCCCGTAGGAGT-3′), which contains the 454 FLX Amplicon primer A (underlined), a sample specific 10-mer barcode (Ns), a two-base linker (italics), and the universal 16S primer 338R (bold). Twenty-five microliter PCR reactions were performed using the Phusion Hot-Start II DNA polymerase (Finnzymes) following the manufacturer’s instructions. PCR conditions consisted of an initial denaturation step (98 °C, 30 s) followed by 30 cycles of denaturation (98 °C, 9 s), annealing (55 °C, 30 s), and elongation (72 °C, 20 s). PCR was terminated by a final elongation of 72 °C for 10 min. All reactions were performed in duplicates, which were combined after PCR. PCR products were extracted from agarose-gels with the Qiagen MinElute Gel Extraction Kit and quantified with the Quant-iT dsDNA BR Assay Kit on a NanoDrop 3300 Fluorometer. Equimolar amounts of purified PCR product were pooled and further purified using Ampure Beads (Agencourt). A sample of each library was run on an Agilent Bioanalyzer before emulsion PCR and sequencing as recommended by Roche. Amplicon libraries were subsequently sequenced on a 454 GS-FLX using Titanium sequencing chemistry.
16S rRNA Gene Sequence Analysis.
The 16S rRNA gene amplicon sequence analysis was conducted using the Qiime 1.5.0 package (55). Using the sequence fasta-file, a quality file and a mapping file which assigned the 10-nt barcodes to the corresponding sample as input, the sequences were analyzed using the following parameters: length between 300 and 400 bp, no ambiguous bases, and no mismatch to the primer sequence. Chimeric sequences were identified using Chimera Slayer (56). Identified chimeric sequences were checked manually. Putative chimeric sequences, which were present in at least two independent samples, were retained. Sequences were rarified to the lowest number of sequences, grouped into OTUs at a ≥ 97% or ≥ 99% sequence identity threshold and classified by the RDP classifier. H. vulgaris (AEP) donor polyps displayed a fraction of the spirochaet bacterium Turneriella parva. Electron microscopic pictures provide evidence that this bacterium infects the mesogloea of Hydra (57). However, this infection was never observed before in five independent lines of Hydra vulgaris (AEP) (42, 58), nor was it transmittable in cocultivation experiments (Fig. 6) and was therefore considered as an exceptional condition. To concentrate on the transmittable, epibiotic microbiota, T. parva sequences were removed in silico from the H. vulgaris (AEP) donor samples to resemble the wild-type H. vulgaris (AEP) microbiota. Similarly, Chloroplast sequences were removed from H. viridissima samples. The 454 data are deposited at Metagenomics RAST (ID nos. 3512, 3514, and 3526).
Statistical Analyses of Bacterial Communities.
To avoid potential biases in α- and β-diversity estimates caused by the sensitivity of these metrics to sampling effort, a random subset of sequences was generated to normalize the read distribution. Bacterial community analyses including the weighted Unifrac, unweighted Unifrac, and Bray–Curtis distances, statistical analysis of clustering (using the statistical methods ADONIS and ANOSIM), and constrained analysis of principal coordinates were carried out using Qiime 1.5.0 package (55). Bacterial community analyses including the Jensen–Shannon distance were carried out using the VEGAN R package (R Development Core Team 2011).
Phylogenetic Analysis.
For the calculation of the arminin tree, a nucleotide alignment of the coding sequences was used. The nucleotide alignment was build with the TranslatorX (59) program, which aligns protein-coding nucleotide sequences based on their corresponding amino acid translations. As an outgroup, the arminin-like peptides of the four different species were selected. Bayesian posterior probabilities were calculated using MrBayes v3.1.2 (60). A total of 3 million generations were calculated using the general time-reversible model and four chains with a burn-in of 25% and the invgamma rate variation. The tree was visualized using Mega 5 (61).
Sequence alignment for the cytochrome oxidase genes was generated using Clustal_X (62) and subsequently imported into the Mega 5 sequence analysis software package. A model-test was used to estimate the best-fit substitution models for phylogenetic analyses. For the maximum-likelihood analyses, genes were tested using the General Time Reversible (GTR + I) model. A bootstrap test with 1,000 replicates for maximum likelihood and random seed was conducted.
In Situ Hybridzation.
Gene-expression localization of arminins was analyzed by whole-mount in situ hybridization as previously described (63). Primers for probe generation were positioned in nonconserved regions and their specificity was proofed by sequencing the amplified fragment.
Arminin Expression Quantification by qRT-PCR.
Total RNA was isolated from 15 polyps using the TRIZOL-plus protocol (Invitrogen) and cDNA was generated using the first-strand cDNA Synthesis Kit (Fermentas) according to the manufacturer’s protocol. qRT-PCR was conducted in biological triplicates (n = 3), using the GoTaq qPCR Master Mix (Promega) and a 7300 real-time PCR system (ABI). Template amounts were equilibrated for the Hydra EF1α (elongation factor 1α) gene (EF1α_F 5′-GCAGTACTGGTGAGTTTGAAG-3′ and EF1α_R 5′-CTTCGCTGTATGGTGGTTCAG -3′). Fold-changes were normalized to control polyps.
Quantification of Bacterial 16S Genes by qRT-PCR.
Total bacterial quantification was performed with the original DNAs used for 454 sequencing. Template amounts were equilibrated for the Hydra actin gene (hyActinF 5′-GAATCAGCTGGTATCCATGAAAC-3′ and hyActinR 5′-AACATTGTCGTACCACCTGATAG-3′). Bacterial DNA was quantified with universal bacteria primers Eub341_F and Eub534_R (64). The fold-change was calculated using the formula fold-change = 2−ΔΔCt with Ct being the PCR threshold cycle. Fold-changes were normalized to control polyps to one replicate of control + H. vulgaris AEP.
Peptide Extraction from Hydra Tissue.
For peptide extraction, ∼1,000 control or Arminin− polyps were homogenated in 100 mL of 1 M HCl, 5% (vol/vol) formic acid, 1% (vol/vol) trifluoro acetic acid (TFA), and 1% (wt/vol) NaCl at 4 °C overnight, as previously described (65). After centrifugation at 30,000 × g for 1 h, the supernatants were applied to tC18 6cm3 (500 mg) SepPak Vac cartridges (Milford) for solid-phase extraction. Bound material was eluted with 84% acetonitrile in 0.1% TFA (vol/vol). The eluates were lyophilized and redissolved in 0.01% TFA (vol/vol). The protein concentration of the resulting elutions was determined using the Micro BCA Protein Assay Kit (Pierce) according to manufacturer’s instructions.
Test for Antimicrobial Activity of Hydra Tissue Extracts.
For radial diffusion assay, E. coli (DH5α) cells were seeded on R2A Agar forming a uniform layer of cells. Fifty micrograms of extracted proteins from tissue of control or Arminin− polyps were pipetted on circular filter plates placed on the agar. After incubation at 37 °C for 16 h, bacterial growth inhibition zones were clearly visible.
MIC assay was performed using 96-well microtiter plates. The plates were precoated with sterile-filtered 0.1% BSA for at least 30 min. BSA was removed and the wells were filled with a twofold dilution series of the extracted peptides, starting with 50 µg/mL in 90 µL 10 mM sodiumphosphate buffer (NaP, pH 7.4) supplemented with 10% LB-media. Finally, each well was inoculated with 100 CFU of E. coli DH5α, reaching a final volume of 100 µL solution per well. The microtiter plates were incubated overnight at 37 °C in a moisture chamber and MIC was determined by the absence of a bacterial cell pellet. Experiments were carried out with three biological replicates.
Supplementary Material
Acknowledgments
We thank Doris Willoweit-Ohl, Jörg Wittlieb, Friederike Anton-Erxleben, and Antje Thomas for their expert technical assistance; Heinke Buhtz for her technical support in the 454 sequencing process; and René Augustin and Cleo Pietschke for critically reading the manuscript. This work was funded by the Deutsche Forschungsgemeinschaft through Grants BO 848/17-1 and FR 3041/2-1, and grants from the Deutsche Forschungsgemeinschaft Cluster of Excellence programs “The Future Ocean” and “Inflammation at Interfaces.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.M.-N. is a guest editor invited by the Editorial Board.
Data deposition: The 454 data are deposited in the Metagenomics RAST (MG-RAST) database, metagenomics.anl.gov (Project IDs: 3512, 3514, and 3526). The arminin sequences have been deposited in the GenBank database, http://www.ncbi.nlm.nih.gov/genbank/ (accession nos. KC701494–KC701520).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304960110/-/DCSupplemental.
References
- 1.Fraune S, Bosch TC. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci USA. 2007;104(32):13146–13151. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6(10):776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110(9):3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douglas AE, Minto LB, Wilkinson TL. Quantifying nutrient production by the microbial symbionts in an aphid. J Exp Biol. 2001;204(Pt 2):349–358. doi: 10.1242/jeb.204.2.349. [DOI] [PubMed] [Google Scholar]
- 6.Sandström J, Telang A, Moran NA. Nutritional enhancement of host plants by aphids—A comparison of three aphid species on grasses. J Insect Physiol. 2000;46(1):33–40. doi: 10.1016/s0022-1910(99)00098-0. [DOI] [PubMed] [Google Scholar]
- 7.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobber R, Hertogh-Huijbregts A, Rozing J, Bottomly K, Nagelkerken L. The involvement of the intestinal microflora in the expansion of CD4+ T cells with a naive phenotype in the periphery. Dev Immunol. 1992;2(2):141–150. doi: 10.1155/1992/57057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Weiss BL, Maltz M, Aksoy S. Obligate symbionts activate immune system development in the tsetse fly. J Immunol. 2012;188(7):3395–3403. doi: 10.4049/jimmunol.1103691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101(13):4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stecher B, Hardt WD. The role of microbiota in infectious disease. Trends Microbiol. 2008;16(3):107–114. doi: 10.1016/j.tim.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Shin SC, et al. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334(6056):670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 14.French N, Pettersson S. Microbe-host interactions in the alimentary tract: The gateway to understanding inflammatory bowel disease. Gut. 2000;47(2):162–163. doi: 10.1136/gut.47.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ott SJ, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53(5):685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Bevins CL, Salzman NH. The potter’s wheel: The host’s role in sculpting its microbiota. Cell Mol Life Sci. 2011;68(22):3675–3685. doi: 10.1007/s00018-011-0830-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127(2):423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149(7):1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersson J, et al. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300(2):G327–G333. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313(5790):1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen L, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455(7216):1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rehman A, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60(10):1354–1362. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- 24.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332(6032):974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11(1):76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol. 2009;11(2):483–493. doi: 10.1111/j.1462-2920.2008.01788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraune S, et al. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci USA. 2010;107(42):18067–18072. doi: 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Augustin R, et al. Activity of the novel peptide arminin against multiresistant human pathogens shows the considerable potential of phylogenetically ancient organisms as drug sources. Antimicrob Agents Chemother. 2009;53(12):5245–5250. doi: 10.1128/AAC.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemmrich G, Anokhin B, Zacharias H, Bosch TC. Molecular phylogenetics in Hydra, a classical model in evolutionary developmental biology. Mol Phylogenet Evol. 2007;44(1):281–290. doi: 10.1016/j.ympev.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 30.Kasahara S, Bosch TC. Enhanced antibacterial activity in Hydra polyps lacking nerve cells. Dev Comp Immunol. 2003;27(2):79–85. doi: 10.1016/s0145-305x(02)00073-3. [DOI] [PubMed] [Google Scholar]
- 31.Bosch TC, et al. Uncovering the evolutionary history of innate immunity: The simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. 2009;33(4):559–569. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Rosenberg E, Sharon G, Zilber-Rosenberg I. The hologenome theory of evolution contains Lamarckian aspects within a Darwinian framework. Environ Microbiol. 2009;11(12):2959–2962. doi: 10.1111/j.1462-2920.2009.01995.x. [DOI] [PubMed] [Google Scholar]
- 33.Brucker RM, Bordenstein SR. The hologenomic basis of speciation: Gut bacteria cause hybrid lethality in the genus Nasonia. Science. 2012 doi: 10.1126/science.1240659. [DOI] [PubMed] [Google Scholar]
- 34.Hongoh Y, et al. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl Environ Microbiol. 2005;71(11):6590–6599. doi: 10.1128/AEM.71.11.6590-6599.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brucker RM, Bordenstein SR. The roles of host evolutionary relationships (genus: Nasonia) and development in structuring microbial communities. Evolution. 2012;66(2):349–362. doi: 10.1111/j.1558-5646.2011.01454.x. [DOI] [PubMed] [Google Scholar]
- 36.Ochman H, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8(11):e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ubeda C, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209(8):1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105(52):20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryu JH, et al. The homeobox gene Caudal regulates constitutive local expression of antimicrobial peptide genes in Drosophila epithelia. Mol Cell Biol. 2004;24(1):172–185. doi: 10.1128/MCB.24.1.172-185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Login FH, et al. Antimicrobial peptides keep insect endosymbionts under control. Science. 2011;334(6054):362–365. doi: 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- 41.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9(5):356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 42.Franzenburg S, et al. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc Natl Acad Sci USA. 2012;109(47):19374–19379. doi: 10.1073/pnas.1213110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG. More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25(9):404–413. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 44.Lazzaro BP. Natural selection on the Drosophila antimicrobial immune system. Curr Opin Microbiol. 2008;11(3):284–289. doi: 10.1016/j.mib.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sackton TB, et al. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39(12):1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- 46.Wallin IE. Symbionticism and the Origin of Species. Baltimore, MD: Williams and Wilkins; 1927. [Google Scholar]
- 47.Brucker RM, Bordenstein SR. Speciation by symbiosis. Trends Ecol Evol. 2012;27(8):443–451. doi: 10.1016/j.tree.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 48.Tsuchida T, Koga R, Fukatsu T. Host plant specialization governed by facultative symbiont. Science. 2004;303(5666):1989. doi: 10.1126/science.1094611. [DOI] [PubMed] [Google Scholar]
- 49.Sharon G, et al. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci USA. 2010;107(46):20051–20056. doi: 10.1073/pnas.1009906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tennessen JA. Molecular evolution of animal antimicrobial peptides: Widespread moderate positive selection. J Evol Biol. 2005;18(6):1387–1394. doi: 10.1111/j.1420-9101.2005.00925.x. [DOI] [PubMed] [Google Scholar]
- 51.Bosch TC, Krylow SM, Bode HR, Steele RE. Thermotolerance and synthesis of heat shock proteins: these responses are present in Hydra attenuata but absent in Hydra oligactis. Proc Natl Acad Sci USA. 1988;85(21):7927–7931. doi: 10.1073/pnas.85.21.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lenhoff HM, Brown RD. Mass culture of hydra: An improved method and its application to other aquatic invertebrates. Lab Anim. 1970;4(1):139–154. doi: 10.1258/002367770781036463. [DOI] [PubMed] [Google Scholar]
- 53.Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci USA. 2006;103(16):6208–6211. doi: 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haas BJ, et al. Human Microbiome Consortium Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21(3):494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stagni A, Lucchi ML. Is fresh water Hydra vulgaris attenuata a spirochaetales’ reservoir host? Experientia. 1969;25(6):662–663. doi: 10.1007/BF01896582. [DOI] [PubMed] [Google Scholar]
- 58.Franzenburg S, et al. Bacterial colonization of Hydra hatchlings follows a robust temporal pattern. ISME J. 2013;7(4):781–790. doi: 10.1038/ismej.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abascal F, Zardoya R, Telford MJ. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38(Web Server issue):W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 61.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Augustin R, et al. Dickkopf related genes are components of the positional value gradient in Hydra. Dev Biol. 2006;296(1):62–70. doi: 10.1016/j.ydbio.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 64.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59(3):695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Augustin R, Siebert S, Bosch TC. Identification of a kazal-type serine protease inhibitor with potent anti-staphylococcal activity as part of Hydra’s innate immune system. Dev Comp Immunol. 2009;33(7):830–837. doi: 10.1016/j.dci.2009.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.