Abstract
Significance: The conversion of protein-bound Tyr residues to 3-nitrotyrosine (3NY) can occur during nitrative stress and has been correlated to aging and many disease states. Proteomic analysis of this post-translational modification, using mass spectrometry-based techniques, is crucial for understanding its potential role in pathological and physiological processes. Recent Advances: To overcome some of the disadvantages inherent to well-established nitroproteomic methods using anti-3NY antibodies and gel-based separations, methods involving multidimensional chromatography, precursor ion scanning, and/or chemical derivatization have emerged for both identification and quantitation of protein nitration sites. A few of these methods have successfully detected endogenous 3NY modifications from biological samples. Critical Issues: While model systems often show promising results, identification of endogenous 3NY modifications remains largely elusive. The frequently low abundance of nitrated proteins in vivo, even under inflammatory conditions, is especially challenging, and sample loss due to derivatization and cleaning may become significant. Future Directions: Continued efforts to avoid interference from non-nitrated peptides without sacrificing recovery of nitrated peptides are needed. Quantitative methods are emerging and are crucial for identifying endogenous modifications that may have significant biological impacts. Antioxid. Redox Signal. 19, 1247–1256.
Introduction
Nitrative stress leads to 3-nitrotyrosine formation
Under conditions of nitrative stress, particularly when nitric oxide combines with superoxide to form peroxynitrite, protein Tyr residues can undergo nitration to 3-nitrotyrosine (3NY), and increases in 3NY have been correlated to aging and numerous disease states, including cardiovascular disease, neurodegeneration, and autoimmune diseases (26). Tyr nitration can alter the protein structure and function and may play a role in redox signaling (2, 11, 30).
To understand the biological impact of Tyr nitration, protein targets must be identified, most often by mass spectrometry (MS)-based proteomics. Because many reviews of nitroproteomic methods exist (2, 7, 9, 16, 24, 45, 47), including several dedicated to MS techniques (4, 28, 35, 36), the present review will focus on recent improvements in the field.
Some considerations for the development of nitroproteomic methods
The field of nitroproteomics has long been dominated by two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), followed by Western blotting with antibodies against 3NY and analysis of in-gel-digested spots by MS. This remains the most sensitive technique available, routinely indicating endogenous nitration. However, it is laborious and suffers from problems with specificity, background signals, and reproducibility, due in part to variations among antibody batches and suppliers (2, 16, 46). Antibodies may be either too specific, depending on the antigen used to raise them, or not specific enough, as cross-reactivity can occur, for instance, with the Trp derivative 5-hydroxy-6-nitrotryptophan (25). Methods for enrichment by immunoprecipitation can suffer from some of the same limitations. Furthermore, 2D-PAGE can be biased against proteins on the extremes of size, isoelectric point, and hydrophobicity. Finally, unambiguous identification of nitrated proteins and residues is difficult because a single gel spot may contain multiple proteins, and tandem mass spectrometry (MS/MS) data are often not obtained for the tryptic peptides containing 3NY (2, 16).
Multidimensional chromatography, either on-line or off-line, can partially automate the prefractionation process for 3NY proteomics. However, specifically offline prefractionation can be time-consuming and generate large amounts of samples for MS analysis, thus requiring significant instrument time. Due to the large dynamic range required for proteomic analysis, as well as concerns about column capacity and ion suppression of low-abundance species by an overwhelming matrix of high-abundance species, detection of low endogenous levels of 3NY [on the order of one to five residues per 10,000 Tyr residues in samples under inflammatory conditions (30)] depends upon sufficient simplification and/or purification of the sample.
To address these limitations, many methods for chemical derivatization of 3NY have been developed, mostly to enrich 3NY-containing proteins and/or peptides. These methods often involve several derivatization steps, almost always including the reduction of 3NY to 3-aminotyrosine (3AY) by sodium dithionite (SDT) (40), followed by reaction of the aromatic amine with a labeling reagent, along with one or more cleaning steps. Derivatization methods also present opportunities for relative and absolute quantitation using isotopic labeling and/or fluorogenic tagging. Some derivatization strategies have focused on simplification and avoidance of sample losses that inevitably accompany derivatization and cleaning.
The reduction of 3NY to 3AY can be a challenging step. The most popular method uses SDT (40), but concomitant 3AY and Tyr sulfation can interfere with MS analysis (12, 13). An alternative method using thiols and heme yields 3AY as a single product but is more sensitive to conditions and difficult to automate (3, 12, 13). An electrochemical 3NY reduction has not yet been widely applied in the field of nitroproteomics (2). 3AY may also form inadvertently during sample processing, as observed for the Arabidopsis thaliana proteome (21), and may even form in vivo (2). These phenomena should be accounted for in proteomic database searches.
Labeling the 3AY aromatic amine with high specificity is crucial, due to the very large excess of aliphatic amine groups (N-termini and Lys ɛ-amino groups) over low-abundance, endogenous 3NY sites in biological samples, approximated to be ca. 28,000-fold molar excess (1). Some groups have attempted to exploit the lower pKa of the aromatic amine, which is 4.75 in the free amino acid (40), relative to aliphatic amines (pKa ca. 8–10) to impart specificity in labeling. However, others observed that such pH-based selectivity is insufficient (1, 13, 48), and most methods have added steps to block reactive amines before the reduction of 3NY.
Some considerations for MS of nitrated peptides and proteins
When matrix-assisted laser desorption ionization (MALDI) is used for ionization, 3NY photodegradation can occur with a loss of one or two oxygen atoms or reduction of 3NY to 3AY (6, 19, 20, 28, 34). While some studies use this characteristic pattern to aid in 3NY identification, it raises limits of detection because the MS signal intensity is shared among three or four peaks. Electrospray ionization (ESI) avoids this problem but does have some disadvantages, such as a lower tolerance for sample contaminants and matrix components. Regardless of the ionization source, both 3NY- and 3AY-containing peptides can have low ionization efficiency and/or poor MS/MS fragmentation (1, 13, 17, 19, 28, 44). 3AY may give deviating isotopic envelopes for fragment ions, possibly due to chemical rearrangements, which can interfere with protein database searching and prevent proper identification (13). Derivatization of 3AY can overcome these problems (13, 29). Removal of the nitro group, an electron predator, enables the use of electron capture and electron transfer dissociation as complementary fragmentation methods to improve sequence coverage (29); see Refs. (15, 22) for further discussion.
Multidimensional Chromatography
Technical and methodological advances have benefited nitroproteomic studies (2, 4, 5, 41). More recent developments are discussed below.
Combined fractional diagonal chromatography
Ghesquière et al. (12) applied combined fractional diagonal chromatography (COFRADIC) to nitroproteomics. Protein digests are separated by reverse-phase (RP) high-performance liquid chromatography (HPLC) and fractions collected over 1-min intervals. After reduction of 3NY to 3AY with SDT, samples are again fractionated using the same RP-HPLC conditions, and peaks showing a hydrophilic shift are collected for MS identification. While this method requires tedious fraction collection, it successfully identified six in vivo nitration sites in four different proteins from serum in a mouse model of sepsis, specifically female C57BL6/J mice injected intravenously with pathogenic Salmonella enteritidis. Likewise, 3NY sites were identified from in vitro nitration models, such as bovine serum albumin (BSA) treated with tetranitromethane (TNM) and Jurkat cell lysate exposed to peroxynitrite.
The authors also observed sulfated 3AY as a side product of reduction of Jurkat cell samples by SDT, where ca. 84% of peptides were partially sulfated. Thus, it is important to include sulfation when searching MS data against proteomic databases in experiments that use SDT.
Larsen et al. (18) combined COFRADIC with off-line nano-HPLC-MALDI-MS with tandem time-of-flight detection in a study that was limited to BSA nitrated in vitro, for which this method identified five 3NY sites. Advantages include the small amount of sample required and the potential for automation.
Fluorescent Labels
Incorporation of fluorescent moieties into nitropeptides or nitroproteins can provide a powerful tool for detection, with high signal-to-noise ratios, as well as for absolute and/or relative quantitation and for visualization in gels, Western blots, or tissue samples.
6-substituted 2-phenylbenzoxazoles
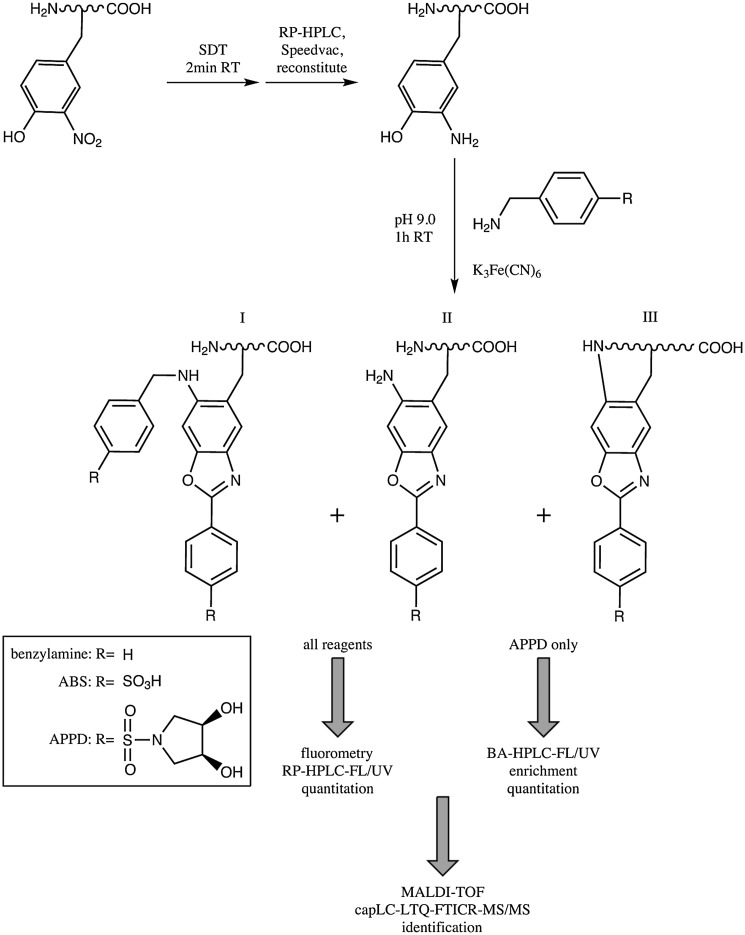
Several studies have demonstrated fluorogenic labeling of 3NY by reduction to 3AY with SDT followed by reaction with benzylamine (27, 38) or benzylamine derivatives, 4-(aminomethyl)benzenesulfonic acid (ABS) (37) or (3R,4S)-1-(4-(aminomethyl)phenylsulfonyl)pyrrolidine-3,4-diol (APPD) (8) (Fig. 1). A major advantage of this strategy is the selectivity for the o-aminophenol moiety of 3AY, which precludes the need to block other reactive amines in the peptide or protein. The products are highly fluorescent 6-substituted 2-phenylbenzoxazoles. For each reagent, three products have been characterized that differ in the substituent at the 6-position of the benzoxazole ring system but maintain similar fluorescent properties. Labeling can be followed by fluorescence detection and identification by MS. The method has also been applied for histochemical staining (39).
FIG. 1.
Fluorogenic derivatization of 3NY to 6-substituted 2-phenylbenzoxazoles. Adapted and modified from Refs. (8, 37, 38). 3NY, 3-nitrotyrosine; SDT, sodium dithionite; RT, room temperature; RP, reverse phase; HPLC, high-performance liquid chromatography; BA-HPLC, boronate-affinity high-performance liquid chromatography; FL, fluorescence detection; UV, ultraviolet absorbance detection; APPD, (3R,4S)-1-(4-(aminomethyl)phenylsulfonyl)pyrrolidine-3,4-diol; MALDI, matrix-assisted laser desorption ionization; TOF, time-of-flight; capLC, capillary liquid chromatography; LTQ, linear trap quadrupole (linear ion trap); FTICR, Fourier transform ion cyclotron resonance; MS/MS, tandem mass spectrometry.
For a synthetic nitropeptide, FSA(3NY)LER, tagged with ABS, the fluorescence quantum yield was determined as 0.77±0.08 (37), and limits of detection by fluorescence spectrometry were measured as 12 and 40 pmol for the nitropeptide alone and in the presence of 100 μg digested proteins from C2C12 murine myoblasts, a model background matrix, respectively. Each of the reagents is amenable to isotopic coding for MS-based relative quantitation (27, 38).
Notably, 3,4-dihydroxyphenylalanine and 5-hydroxytryptophan can also be labeled by this method, without reduction by SDT (37). The identity of the detected post-translational modification (PTM) can thus be deduced using controls not treated with SDT.
Affinity Labels
To avoid ion suppression of low-abundance nitropeptides, as well as to address chromatographic capacity limits, specific and efficient labeling of 3NY for affinity-based enrichment is a promising strategy for nitroproteomics. There has been great interest in developing antibody-free methods to overcome disadvantages of specificity, reproducibility, and throughput.
Boronate affinity
In the most recent development of a fluorogenic labeling strategy (27, 37, 38), Dremina et al. (8) describe a new benzylamine-derived reagent for derivatization and affinity enrichment of 3NY using boronate-affinity HPLC (BA-HPLC) (Fig. 1). The reagent APPD contains a benzylamine moiety for reaction with 3AY and a cis-diol moiety for pH-dependent coordination with boronic acid.
Both fluorescence spectrometry and BA-HPLC data (with fluorescence detection) can be used for quantitation of 3NY by this method, with limits of detection calculated as 19 nmol and 13 pmol, respectively, for a digest of rabbit phosphorylase b nitrated in vitro by peroxynitrite, alone or in a matrix of digested C2C12 murine myoblast proteins. Side product(s) can increase background and detection limits, but enrichment by BA-HPLC attenuates this problem.
Biotin
The high-affinity interaction of biotin with avidin and streptavidin makes it an attractive labeling moiety. As reviewed elsewhere (2, 24), several groups have developed methods for biotinylation followed by enrichment on immobilized avidin or streptavidin.
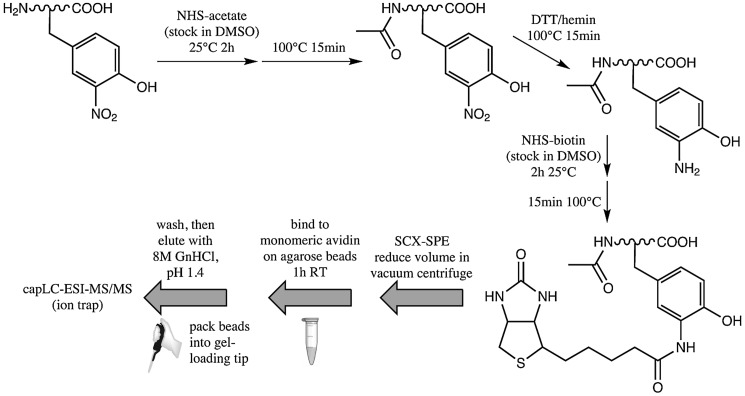
The method of Abello et al. (1), summarized in Figure 2, reduces the need for cleaning steps and pH changes, thus minimizing sample losses. Aliphatic amines are acetylated by acetic acid N-hydroxysuccinimide ester (NHS-acetate), 3NY is reduced to 3AY using the dithiothreitol/hemin system, and a large excess of biotinyl N-hydroxysuccinimide ester (NHS-biotin) completes the derivatization. Following both N-hydroxysuccinimide (NHS)-based reactions, heating in a bath of boiling water degrades the remaining reagent and reverses any undesirable O-acetylation or O-biotinylation. The single cleaning step required, a strong cation exchange solid-phase extraction (SPE) used to remove the remaining biotin reagent, has poor recovery. After cleaning, the biotinylated peptides are enriched on a monomeric avidin resin, washed, and eluted by guanidinium hydrochloride under acidic conditions.
FIG. 2.
Method for biotinylation of 3NY for affinity-based enrichment. This particular method minimizes sample cleaning steps. Adapted and modified from Ref. (1). NHS, N-hydroxysuccinimide; NHS-acetate, acetic acid NHS ester; DMSO, dimethyl sulfoxide; DTT, dithiothreitol; NHS-biotin, biotinyl NHS ester; ESI, electrospray ionization; SCX, strong cation exchange; SPE, solid-phase extraction; GnHCl, guanidinium hydrochloride.
The peptide angiotensin II (Ang II) was nitrated in vitro by peroxynitrite and spiked into a tryptic digest of BSA as a model system. Although the peptide was successfully labeled and recovered, the SPE cleaning step is a major source of sample loss. Therefore, the authors propose future work to perform all derivatization reactions on intact proteins, such that size-based separation methods (e.g., gel filtration or dialysis) can be used instead.
Metal chelation
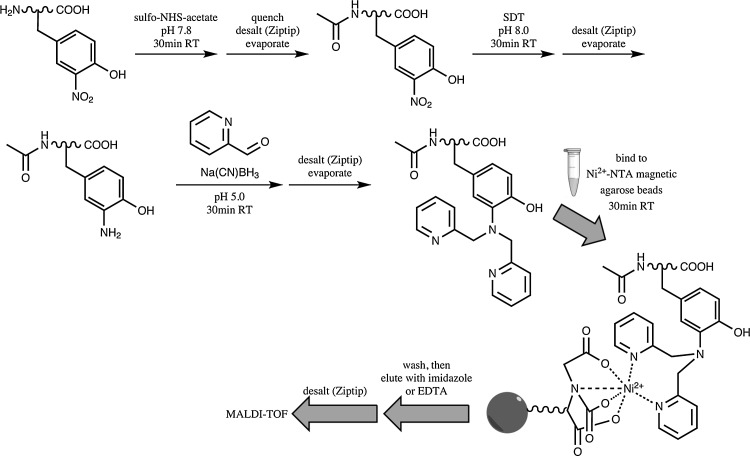
A new adaptation of immobilized metal affinity chromatography (IMAC) has been developed by Lee et al. (19) for the enrichment of 3NY-containing peptides (Fig. 3). Primary amines are blocked by acetylation with sulfo-NHS-acetate before SDT reduction of 3NY to 3AY, after which the latter undergoes Schiff base formation and reductive amination with a large excess of pyridine-2-carboxaldehyde and sodium cyanoborohydride to give peptides that are bispyridinylated at the original nitration sites, with small amounts of monopyridinylation. Desalting steps are required following each of these reactions. The labeled peptides now chelate nickel (II) attached to magnetic agarose beads. After binding and washing, peptides are released with imidazole or ethylenediaminetetraacetic acid, desalted, and identified by MALDI-MS with time-of-flight detection.
FIG. 3.
Immobilized metal affinity chromatography method for enrichment of 3NY-containing peptides, by bispyridinylation of 3NY. Adapted and modified from Ref. (19). NTA, nitrilotriacetic acid; EDTA, ethylenediaminetetraacetic acid.
This approach was applied to synthetic nitrated Ang II and to BSA nitrated by TNM. The authors demonstrate detection of as little as 100 fmol of nitropeptide spiked into a matrix of 10 μg HeLa cell lysate. As absolute signal intensity values are not presented with the MS data, it is unclear whether significant peptide losses occur during derivatization and cleaning steps. Also, while the mass shifts are generally in accord with expected values for each reaction step, all of the observed m/z values for Ang II and its derivatives appear higher than the corresponding theoretical values, with mass errors of 580–1080 ppm, a phenomenon also seen in the data of Kim et al. (17) (described below).
Fluorine affinity
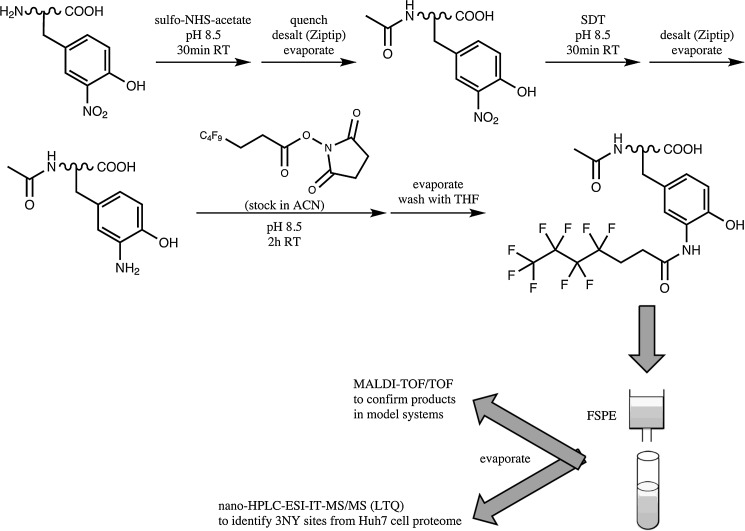
Some of the same researchers have also developed a strategy to derivatize 3NY with highly fluorinated moieties for enrichment by fluorinated SPE (FSPE) (17) (Fig. 4). After acetylation of aliphatic amines with sulfo-NHS-acetate and reduction of 3NY to 3AY with SDT, peptides are tagged with N-succinimidyl 3-(perfluorobutyl)propionate, with cleaning steps after each reaction. Labeled peptides are enriched using FSPE, and the eluates are analyzed by MS.
FIG. 4.
Method for fluorination of 3NY for purification by FSPE. Adapted and modified from Ref. (17). ACN, acetonitrile; THF, tetrahydrofuran; TOF/TOF, tandem time-of-flight; FSPE, fluorinated solid-phase extraction; IT, ion trap.
Using this strategy for synthetic nitrated Ang II mixed with its non-nitrated counterpart, successful derivatization and enrichment are demonstrated by MALDI-MS. Interestingly, the absolute MS intensity for the tagged peptide is much greater after FSPE than before, and, while not discussed in the article, this may be an indication of ion suppression in the crude sample that is overcome by the affinity purification step.
However, as in the IMAC study (19), there are systematic deviations of observed m/z values from the theoretical values, especially for nitrated Ang II and its derivatives, which show large mass errors of+580–1038 ppm. The mass shifts after each reaction are generally as expected, except for Δm/z=46.1 between native and nitrated Ang II (the expected value is 45.0). Inadequate instrument calibration is one possible explanation for this combination of phenomena, but the article does not discuss it. Also, there are some additional peaks in the final product spectrum that are not explained. Among them is a small peak near 1100 m/z, which could represent 3AY-Ang II (1104 m/z), but the absence of annotation for this peak precludes a definitive determination. At the characteristically low ionization efficiency of the 3AY-peptide, even a small peak can represent a significant amount of peptide that was not tagged.
The decrease in absolute intensity of the MALDI peak for the non-nitrated, acetylated peptide following subsequent steps probably results from sample losses due to cleaning, or perhaps due to reaction conditions (e.g., possible side products or peptide degradation). These losses can become very important when looking for low-abundant PTMs, like 3NY.
The method was able to tag, enrich, and detect 100 pmol of nitrated Ang II in a background of 10 μg of a BSA tryptic digest. More notably, 28 endogenously nitrated peptides from 28 proteins were identified in the Huh7 human hepatoma cell line, and the 3NY sites were confirmed by MS/MS with manual validation. Some of the peptide spectra show XCorr scores [cross-correlation values generated by the SEQUEST algorithm (10)] that are lower than ideal, possibly indicating a poor fit to the theoretical spectra for the assigned peptide sequences. The most intense peaks in the MS/MS spectra are assigned, but only one spectrum shown has backbone cleavages on both sides of the tagged Tyr, which is stronger evidence for nitration site assignment. Not all of the Lys residues are acetylated, even though the text lists this modification as a requirement for acceptance of the identification. In contrast, all of the N-termini are acetylated in the identified peptides. For two of the identifications, the nitration site is located in a specific functional domain.
Solid-Phase Chemistry
Several methods use covalent reactions between the peptide of interest and a solid-phase reagent, followed by chemical cleavage to recover the peptide with a vestigial tagging moiety that can be identified during MS analysis. Recent studies have endeavored to improve upon the earlier methods of Zhang et al. (48) and Nikov et al. (23).
Chemoprecipitation using solid-phase active ester reagent
To simplify the derivatization procedure and avoid sample losses incurred by cleaning steps, Prokai-Tatrai et al. (29) developed a chemoprecipitation technique in which amine groups are blocked by reductive methylation, 3NY is reduced to 3AY, and the latter reacts with a solid-phase active ester reagent (SPAER) on glass beads, followed by washing and gentle acid-catalyzed hydrolysis to remove the peptides, which retain a 4-formylbenzoylamido group (Fig. 5). By solution methods, the dimethylation step requires clean-up by SPE, but a one-pot method using solid-phase reagents is also reported. Importantly, dimethylation prevents nonselective labeling of amine groups while retaining their ability to ionize for facile detection in positive-mode HPLC-ESI-MS/MS.
FIG. 5.
SPAER method for enrichment of 3NY by chemoprecipitation. This represents the solution-phase method, but a solid-phase method was also tested. Adapted and modified from Ref. (29). Incorporation of isotopic labels for relative quantitation is further described in Ref. (14). SPAER, solid-phase active ester reagent; TFA, trifluoroacetic acid.
In addition to identifying tagged derivatives of three synthetic nitropeptides in a BSA tryptic digest matrix (>200-fold molar excess of unmodified over nitrated peptides), 32 unique tagged peptides were identified from 12 different proteins in a tryptic digest of human plasma samples that were nitrated in vitro by TNM, some of which were previously found nitrated in aging and disease states. Eight contaminating, unmodified peptides from plasma proteins were also identified. This is attributed to nonspecific adsorption to the beads, which the authors propose to address by more thorough washing with organic solvents.
Building upon this work, Guo et al. (14) developed a method for relative quantitation by using isotopically labeled formaldehyde for reductive methylation, which did not alter the chromatographic properties of the products. This method was successfully applied to two model systems: synthetic nitropeptides spiked into a matrix containing the tryptic digest of human serum albumin and the model protein ubiquitin nitrated in vitro and spiked into a matrix of human plasma (14).
Quantitation
An important strategy for quantitation of 3NY consists of using isotopically coded reagents for differential labeling of samples from two (or more) experimental conditions and mixing these samples before MS analysis, thus allowing for quantitative comparisons of ion peak areas. While this is largely applied to relative quantitation, there are ways to achieve absolute quantitation by including an isotopically labeled internal standard. Isobaric tagging strategies have also emerged, which maintain a single m/z value for precursor ions but yield specific reporter ions after fragmentation, with the latter used for quantitation. These include tandem mass tags (TMT) (42) and isobaric tags for relative and absolute quantitation (iTRAQ) (32).
Isobaric tags for relative and absolute quantitation
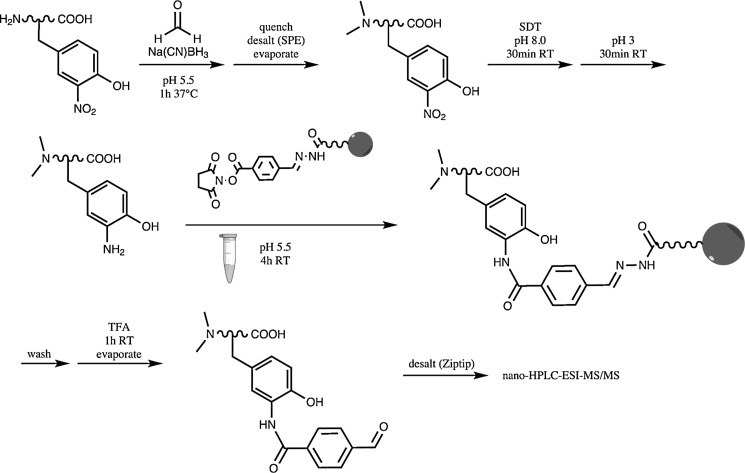
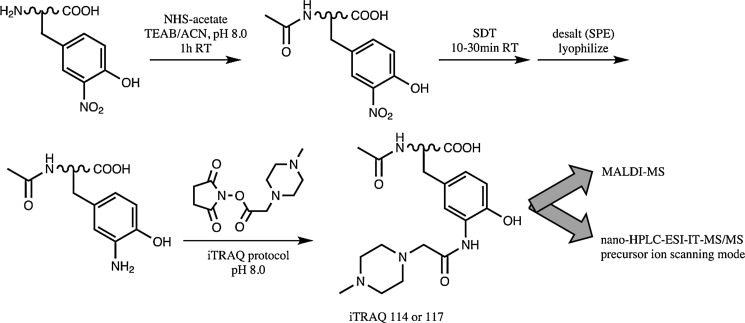
The iTRAQ method was developed based on labeling primary amines for profiling whole proteomes (32). By acetylating primary amines with NHS-acetate, then reducing 3NY to 3AY with SDT, and finally labeling 3AY with iTRAQ reagents, with some cleaning procedures required, Chiappetta et al. (6) have adapted the iTRAQ methodology for identification and quantitation of 3NY sites (Fig. 6). Nano-HPLC-ESI-MS/MS was performed in the precursor ion scanning mode for specific reporter ions, which enabled the use of the mass spectrometer's duty cycle for analyzing only peptides of interest and thus enhanced the limit of detection without the need for affinity purification/enrichment.
FIG. 6.
Relative quantitation of 3NY with an adaptation of the iTRAQ method. Adapted and modified from Ref. (6). TEAB, tetraethylammonium bicarbonate; iTRAQ, isobaric tag for relative and absolute quantitation.
This method was applied to BSA nitrated in vitro by TNM, alone or in the presence of 30 mg of Escherichia coli protein extract, and to bovine milk proteins nitrated in vitro with TNM. For the latter, nine 3NY sites were identified, which were found in both high- and low-abundance proteins, with accurate relative quantitation for seven of them.
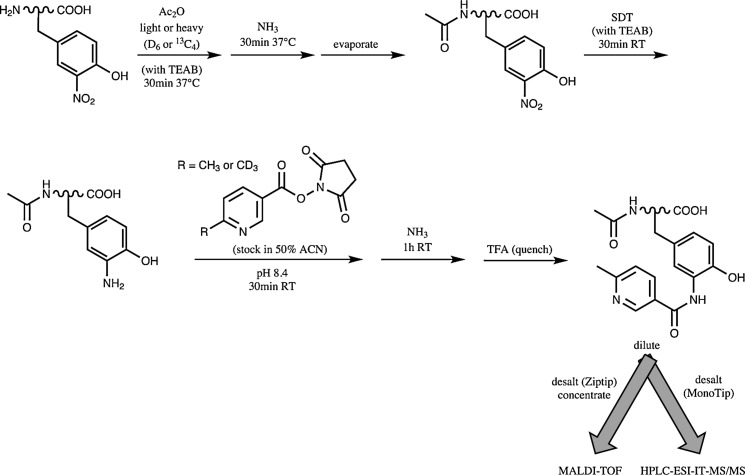
Two-dimensional isotopic coding
Building on previous work (43), Tsumoto et al. (44) developed a method to increase the ionization efficiency of tryptic nitropeptides, prevent their photodecomposition during MALDI experiments, and incorporate isotopic labels (Fig. 7). Due to the two-dimensional nature of this isotopic labeling strategy, with the first dimension applied to all peptides and the second dimension applied only to 3NY, both non-nitrated and nitrated peptides are quantitatively compared between two samples in a single experiment. This feature was used to quantify the degree of nitration achieved by in vitro methods for specific Tyr residues. Although not explicitly discussed, it could also be very useful to control for protein expression levels, an important concern for comparative proteomic profiling of PTMs. Any 3NY labeling strategy that includes a step to block aliphatic amines could likely be adapted to take advantage of this feature.
FIG. 7.
Method for isotopic coding of 3NY for quantitation. Adapted and modified from Ref. (44). Ac2O, acetic anhydride.
Following acetylation of aliphatic amines with light or heavy acetic anhydride, 3NY is reduced to 3AY by SDT for subsequent tagging with 1-(6-methyl[D0/D3]nicotinoyloxy)succinimide, with appropriate cleaning procedures, followed by MS analysis.
Two model systems were used, synthetic nitrated Ang II and BSA nitrated in vitro with peroxynitrite. Ionization efficiency of tagged peptides was greatly improved compared to their 3NY- or 3AY-containing counterparts, recovering to about the same level as the acetylated, non-nitrated peptide. Since no absolute intensities are shown for the mass spectra, it is not possible to compare samples for peptide losses during derivatization and cleaning steps.
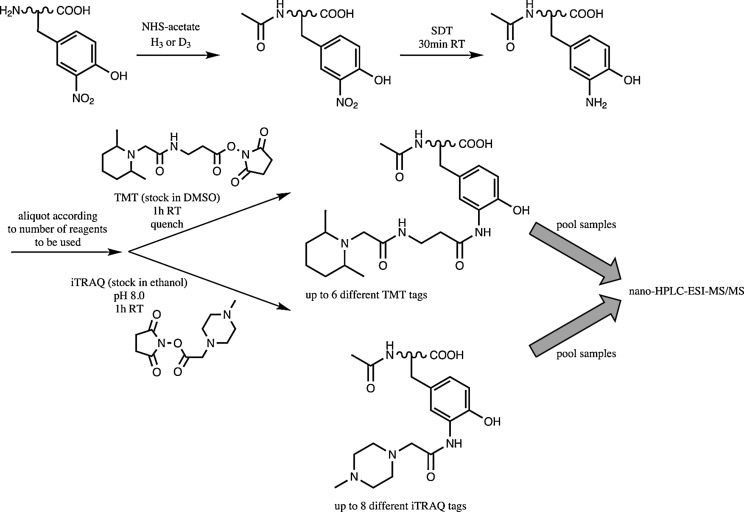
Combined precursor isotopic labeling and isobaric tagging
Robinson and Evans (31) developed the method of combined precursor isotopic labeling and isobaric tagging (cPILOT) to multiplex as many as 16 samples, using the previously reported isobaric tagging methods TMT (42) and iTRAQ (32) (Fig. 8). This allows for simultaneous comparison of numerous biological samples, which is more cost efficient. After acetylating amines with light or heavy NHS-acetate to give duplexed samples, SDT reduces 3NY to 3AY, and sets of either TMT or iTRAQ reagents complete the derivatization, with up to 16-plex experiments possible in theory and no cleaning steps required until immediately before MS.
FIG. 8.
Multiplexed method for relative quantitation of 3NY in up to 16 samples. Adapted and modified from (31). TMT, tandem mass tags.
Using BSA nitrated in vitro by peroxynitrite as a model, a 12-plex experiment (duplexed acetylation combined with 6-plex TMT) showed overlap in the precursor ion isolation (i.e., insufficient resolution), causing inaccurate quantitation (31). For investigation of unknown, in vivo nitration sites in the mouse spleen proteome, scaling back to 4-plex still resulted in some overlap, but the relative quantitation was accurate nevertheless. Notably, five endogenous nitration sites were identified from five unique proteins. Possible improvements include enrichment strategies before labeling to increase proteomic identifications and an increase in the mass shift for the first dimension of isotopic labeling to overcome the problem of precursor ion overlap.
Conclusions
Despite many successful proof-of-concept studies with model systems, detection of endogenous 3NY remains a significant challenge. At present, most endogenous 3NY assignments are still based on 2D-PAGE/Western blot and immunoprecipitation methods (2). Of the recent studies covered in this review, only three (12, 17, 31) successfully identified in vivo nitration sites, for a total of 39 sites in 37 different proteins. Other successful studies (33, 49) did not derivatize 3NY, but used prefractionation and advanced HPLC-MS equipment, most notably capillary columns that were 65 cm in length, compared to 10–15 cm for most other studies.
This difficulty may be due to a combination of very low levels of endogenous 3NY and sample losses during derivatization and cleaning steps. Many studies (1, 6, 8, 14, 17, 19, 44) rely on the disappearance of mass spectral peaks for the starting materials to demonstrate a complete conversion for each reaction. However, this is not a truly quantitative technique, especially when MALDI is used, and furthermore this data cannot account for sample losses (e.g., due to adsorption on plastic or glassware) or undetected side products.
Successful nitroproteomics requires selective separation and/or purification of nitropeptides from the matrix of nonmodified peptides to reduce ion suppression and remain within the limits of column capacity for HPLC-MS/MS. This must be done with a minimum of sample losses, especially considering the low abundance of 3NY in vivo and the small amounts of biological material that are often available from precious specimens. Indeed, several of the most successful methods to date have either avoided 3NY derivatization completely (33, 49) or kept it to a minimum (12).
Abbreviations Used
- 2D-PAGE
two-dimensional polyacrylamide gel electrophoresis
- 3AY
3-aminotyrosine
- 3NY
3-nitrotyrosine
- ABS
4-(aminomethyl)benzenesulfonic acid
- Ac2O
acetic anhydride
- ACN
acetonitrile
- Ang II
angiotensin II
- APPD
(3R,4S)-1-(4-(aminomethyl)phenylsulfonyl)pyrrolidine-3,4-diol
- BA-HPLC
boronate-affinity high-performance liquid chromatography
- BSA
bovine serum albumin
- capLC
capillary liquid chromatography
- COFRADIC
combined fractional diagonal chromatography
- cPILOT
combined precursor isotopic labeling and isobaric tagging
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- ESI
electrospray ionization
- FL
fluorescence detection
- FSPE
fluorinated solid-phase extraction
- FTICR
Fourier transform ion cyclotron resonance
- GnHCl
guanidinium hydrochloride
- HPLC
high-performance liquid chromatography
- IMAC
immobilized metal affinity chromatography
- IT
ion trap
- iTRAQ
isobaric tag for relative and absolute quantitation
- LTQ
linear trap quadrupole (linear ion trap)
- MALDI
matrix-assisted laser desorption ionization
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- NHS
N-hydroxysuccinimide
- NHS-acetate
acetic acid N-hydroxysuccinimide ester
- NHS-biotin
biotinyl N-hydroxysuccinimide ester
- NTA
nitrilotriacetic acid
- PTM
post-translational modification
- RP
reverse phase
- RT
room temperature
- SCX
strong cation exchange
- SDT
sodium dithionite
- SPAER
solid-phase active ester reagent
- SPE
solid-phase extraction
- TEAB
tetraethylammonium bicarbonate
- TFA
trifluoroacetic acid
- THF
tetrahydrofuran
- TMT
tandem mass tags
- TNM
tetranitromethane
- TOF
time-of-flight
- TOF/TOF
tandem time-of-flight
- UV
ultraviolet absorbance detection
Acknowledgments
We gratefully acknowledge support by NIH (AG12993, AG23551, and AG25350), the Madison and Lila Self Graduate Fellowship program at The University of Kansas, Amgen, and the Department of Pharmaceutical Chemistry at The University of Kansas.
References
- 1.Abello N. Barroso B. Kerstjens HAM. Postma DS. Bischoff R. Chemical labeling and enrichment of nitrotyrosine-containing peptides. Talanta. 2010;80:1503–1512. doi: 10.1016/j.talanta.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Abello N. Kerstjens HAM. Postma DS. Bischoff R. Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J Proteome Res. 2009;8:3222–3238. doi: 10.1021/pr900039c. [DOI] [PubMed] [Google Scholar]
- 3.Balabanli B. Kamisaki Y. Martin E. Murad F. Requirements for heme and thiols for the nonenzymatic modification of nitrotyrosine. Proc Natl Acad Sci U S A. 1999;96:13136–13141. doi: 10.1073/pnas.96.23.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bigelow DJ. Qian WJ. Quantitative proteome mapping of nitrotyrosines. Methods Enzymol. 2008;440:191–205. doi: 10.1016/S0076-6879(07)00811-7. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff R. Schlüter H. Amino acids: chemistry, functionality and selected non-enzymatic post-translational modifications. J Proteomics. 2012;75:2275–2296. doi: 10.1016/j.jprot.2012.01.041. [DOI] [PubMed] [Google Scholar]
- 6.Chiappetta G. Corbo C. Palmese A. Marino G. Amoresano A. Quantitative identification of protein nitration sites. Proteomics. 2009;9:1524–1537. doi: 10.1002/pmic.200800493. [DOI] [PubMed] [Google Scholar]
- 7.Deeb R. Nuriel T. Gross SS. Untargeted discovery of nitric oxide-modified proteins. In: Ignarro LJ, editor. Nitric Oxide: Biology and Pathobiology. San Diego, CA: Academic Press; 2010. pp. 327–389. [Google Scholar]
- 8.Dremina ES. Li X. Galeva NA. Sharov VS. Stobaugh JF. Schöneich C. A methodology for simultaneous fluorogenic derivatization and boronate affinity enrichment of 3-nitrotyrosine-containing peptides. Anal Biochem. 2011;418:184–196. doi: 10.1016/j.ab.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan MW. A review of approaches to the analysis of 3-nitrotyrosine. Amino Acids. 2003;25:351–361. doi: 10.1007/s00726-003-0022-z. [DOI] [PubMed] [Google Scholar]
- 10.Eng JK. McCormack AL. Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 11.Feeney MB. Schöneich C. Tyrosine modifications in aging. Antioxid Redox Signal. 2012;17:1571–1579. doi: 10.1089/ars.2012.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghesquière B. Colaert N. Helsens K. Dejager L. Vanhaute C. Verleysen K. Kas K. Timmerman E. Goethals M. Libert C. Vandekerckhove J. Gevaert K. In vitro and in vivo protein-bound tyrosine nitration characterized by diagonal chromatography. Mol Cell Proteomics. 2009;8:2642–2652. doi: 10.1074/mcp.M900259-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghesquière B. Goethals M. Van Damme J. Staes A. Timmerman E. Vandekerckhove J. Gevaert K. Improved tandem mass spectrometric characterization of 3-nitrotyrosine sites in peptides. Rapid Commun Mass Spectrom. 2006;20:2885–2893. doi: 10.1002/rcm.2676. [DOI] [PubMed] [Google Scholar]
- 14.Guo J. Prokai-Tatrai K. Prokai L. Relative quantitation of protein nitration by liquid chromatography–mass spectrometry using isotope-coded dimethyl labeling and chemoprecipitation. J Chromatogr A. 2012;1232:266–275. doi: 10.1016/j.chroma.2011.12.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones AW. Mikhailov VA. Iniesta J. Cooper HJ. Electron capture dissociation mass spectrometry of tyrosine nitrated peptides. J Am Soc Mass Spectrom. 2010;21:268–277. doi: 10.1016/j.jasms.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 16.Kanski J. Schöneich C. Protein nitration in biological aging: proteomic and tandem mass spectrometric characterization of nitrated sites. Methods Enzymol. 2005;396:160–171. doi: 10.1016/S0076-6879(05)96016-3. [DOI] [PubMed] [Google Scholar]
- 17.Kim JK. Lee JR. Kang JW. Lee SJ. Shin GC. Yeo W-S. Kim K-H. Park HS. Kim KP. Selective enrichment and mass spectrometric identification of nitrated peptides using fluorinated carbon tags. Anal Chem. 2011;83:157–163. doi: 10.1021/ac102080d. [DOI] [PubMed] [Google Scholar]
- 18.Larsen T. Bache N. Gramsbergen J. Roepstorff P. Identification of nitrotyrosine containing peptides using combined fractional diagonal chromatography (COFRADIC) and off-line nano-LC-MALDI. J Am Soc Mass Spectrom. 2011;22:989–996. doi: 10.1007/s13361-011-0095-y. [DOI] [PubMed] [Google Scholar]
- 19.Lee JR. Lee SJ. Kim TW. Kim JK. Park HS. Kim D-E. Kim KP. Yeo W-S. Chemical approach for specific enrichment and mass analysis of nitrated peptides. Anal Chem. 2009;81:6620–6626. doi: 10.1021/ac9005099. [DOI] [PubMed] [Google Scholar]
- 20.Lee SJ. Lee JR. Kim YH. Park YS. Park SI. Park HS. Kim KP. Investigation of tyrosine nitration and nitrosylation of angiotensin II and bovine serum albumin with electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:2797–2804. doi: 10.1002/rcm.3145. [DOI] [PubMed] [Google Scholar]
- 21.Lozano-Juste J. Colom-Moreno R. León J. In vivo protein tyrosine nitration in Arabidopsis thaliana. J Exp Bot. 2011;62:3501–3517. doi: 10.1093/jxb/err042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikhailov VA. Iniesta J. Cooper HJ. Top-down mass analysis of protein tyrosine nitration: comparison of electron capture dissociation with “slow-heating” tandem mass spectrometry methods. Anal Chem. 2010;82:7283–7292. doi: 10.1021/ac101177r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikov G. Bhat V. Wishnok JS. Tannenbaum SR. Analysis of nitrated proteins by nitrotyrosine-specific affinity probes and mass spectrometry. Anal Biochem. 2003;320:214–222. doi: 10.1016/s0003-2697(03)00359-2. [DOI] [PubMed] [Google Scholar]
- 24.Nuriel T. Deeb RS. Hajjar DP. Gross SS. Protein 3-nitrotyrosine in complex biological samples: quantification by high-pressure liquid chromatography/electrochemical detection and emergence of proteomic approaches for unbiased identification of modification sites. Methods Enzymol. 2008;441:1–17. doi: 10.1016/S0076-6879(08)01201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nuriel T. Hansler A. Gross SS. Protein nitrotryptophan: formation, significance and identification. J Proteomics. 2011;74:2300–2312. doi: 10.1016/j.jprot.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pacher P. Beckman JS. Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pennington JP. Schöneich C. Stobaugh J. Selective fluorogenic derivatization with isotopic coding of catechols and 2-amino phenols with benzylamine: a chemical basis for the relative determination of 3-hydroxy-tyrosine and 3-nitro-tyrosine peptides. Chromatographia. 2007;66:649–659. [Google Scholar]
- 28.Petersson A-S. Steen H. Kalume DE. Caidahl K. Roepstorff P. Investigation of tyrosine nitration in proteins by mass spectrometry. J Mass Spectrom. 2001;36:616–625. doi: 10.1002/jms.161. [DOI] [PubMed] [Google Scholar]
- 29.Prokai-Tatrai K. Guo J. Prokai L. Selective chemoprecipitation and subsequent release of tagged species for the analysis of nitropeptides by liquid chromatography–tandem mass spectrometry. Mol Cell Proteomics. 2011;10:M110.002923. doi: 10.1074/mcp.M110.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci U S A. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson RAS. Evans AR. Enhanced sample multiplexing for nitrotyrosine-modified proteins using combined precursor isotopic labeling and isobaric tagging. Anal Chem. 2012;84:4677–4686. doi: 10.1021/ac202000v. [DOI] [PubMed] [Google Scholar]
- 32.Ross PL. Huang YN. Marchese JN. Williamson B. Parker K. Hattan S. Khainovski N. Pillai S. Dey S. Daniels S. Purkayastha S. Juhasz P. Martin S. Bartlet-Jones M. He F. Jacobson A. Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Sacksteder CA. Qian W-J. Knyushko TV. Wang H. Chin MH. Lacan G. Melega WP. Camp DG. Smith RD. Smith DJ. Squier TC. Bigelow DJ. Endogenously nitrated proteins in mouse brain: links to neurodegenerative disease. Biochemistry. 2006;45:8009–8022. doi: 10.1021/bi060474w. [DOI] [PubMed] [Google Scholar]
- 34.Sarver A. Scheffler NK. Shetlar MD. Gibson BW. Analysis of peptides and proteins containing nitrotyrosine by matrix-assisted laser desorption/ionization mass spectrometry. J Am Soc Mass Spectrom. 2001;12:439–448. doi: 10.1016/S1044-0305(01)00213-6. [DOI] [PubMed] [Google Scholar]
- 35.Scaloni A. Mass spectrometry approaches for the molecular characterization of oxidatively/nitrosatively modified proteins. In: Dalle-Donne I, editor; Scaloni A, editor; Butterfield DA, editor. Redox Proteomics: From Protein Modifications to Cellular Dysfunction and Diseases. Hoboken, NJ: Wiley-Interscience; 2006. pp. 59–99. [Google Scholar]
- 36.Schöneich C. Sharov VS. Mass spectrometry of protein modifications by reactive oxygen and nitrogen species. Free Radic Biol Med. 2006;41:1507–1520. doi: 10.1016/j.freeradbiomed.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Sharov V. Dremina E. Galeva N. Gerstenecker G. Li X. Dobrowsky R. Stobaugh J. Schöneich C. Fluorogenic tagging of peptide and protein 3-nitrotyrosine with 4-(aminomethyl)benzenesulfonic acid for quantitative analysis of protein tyrosine nitration. Chromatographia. 2010;71:37–53. doi: 10.1365/s10337-009-1409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharov VS. Dremina ES. Pennington J. Killmer J. Asmus C. Thorson M. Hong SJ. Li X. Stobaugh JF. Schöneich C. Selective fluorogenic derivatization of 3-nitrotyrosine and 3,4-dihydroxyphenylalanine in peptides: a method designed for quantitative proteomic analysis. Methods Enzymol. 2008;441:19–32. doi: 10.1016/S0076-6879(08)01202-0. [DOI] [PubMed] [Google Scholar]
- 39.Sharov VS. Pal R. Dremina ES. Michaelis EK. Schöneich C. Fluorogenic tagging of protein 3-nitrotyrosine with 4-(aminomethyl)benzene sulfonate in tissues: a useful alternative to immunohistochemistry for fluorescence microscopy imaging of protein nitration. Free Radic Biol Med. 2012;53:1877–1885. doi: 10.1016/j.freeradbiomed.2012.08.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokolovsky M. Riordan JF. Vallee BL. Conversion of 3-nitrotyrosine to 3-aminotyrosine in peptides and proteins. Biochem Biophys Res Commun. 1967;27:20–25. doi: 10.1016/s0006-291x(67)80033-0. [DOI] [PubMed] [Google Scholar]
- 41.Spickett C. Pitt A. Protein oxidation: role in signalling and detection by mass spectrometry. Amino Acids. 2012;42:5–21. doi: 10.1007/s00726-010-0585-4. [DOI] [PubMed] [Google Scholar]
- 42.Thompson A. Schäfer J. Kuhn K. Kienle S. Schwarz J. Schmidt G. Neumann T. Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 43.Tsumoto H. Murata C. Miyata N. Kohda K. Taguchi R. Efficient identification and quantification of proteins using isotope-coded 1-(6-methylnicotinoyloxy)succinimides by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:3815–3824. doi: 10.1002/rcm.3279. [DOI] [PubMed] [Google Scholar]
- 44.Tsumoto H. Taguchi R. Kohda K. Efficient identification and quantification of peptides containing nitrotyrosine by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry after derivatization. Chem Pharm Bull (Tokyo) 2010;58:488–494. doi: 10.1248/cpb.58.488. [DOI] [PubMed] [Google Scholar]
- 45.Tyther R. McDonagh B. Sheehan D. Proteomics in investigation of protein nitration in kidney disease: technical challenges and perspectives from the spontaneously hypertensive rat. Mass Spectrom Rev. 2011;30:121–141. doi: 10.1002/mas.20270. [DOI] [PubMed] [Google Scholar]
- 46.Wisastra R. Poelstra K. Bischoff R. Maarsingh H. Haisma HJ. Dekker FJ. Antibody-free detection of protein tyrosine nitration in tissue sections. Chembiochem. 2011;12:2016–2020. doi: 10.1002/cbic.201100148. [DOI] [PubMed] [Google Scholar]
- 47.Yeo WS. Lee SJ. Lee JR. Kim KP. Nitrosative protein tyrosine modifications: biochemistry and functional significance. BMB Rep. 2008;41:194–203. doi: 10.5483/bmbrep.2008.41.3.194. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Q. Qian WJ. Knyushko TV. Clauss TR. Purvine SO. Moore RJ. Sacksteder CA. Chin MH. Smith DJ. Camp DG., 2nd Bigelow DJ. Smith RD. A method for selective enrichment and analysis of nitrotyrosine-containing peptides in complex proteome samples. J Proteome Res. 2007;6:2257–68. doi: 10.1021/pr0606934. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X. Monroe ME. Chen B. Chin MH. Heibeck TH. Schepmoes AA. Yang F. Petritis BO. Camp DG. Pounds JG. Jacobs JM. Smith DJ. Bigelow DJ. Smith RD. Qian W-J. Endogenous 3,4-dihydroxyphenylalanine and dopaquinone modifications on protein tyrosine: links to mitochondrially derived oxidative stress via hydroxyl radical. Mol Cell Proteomics. 2010;9:1199–1208. doi: 10.1074/mcp.M900321-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]