Abstract
Significance: S-nitrosylation (SNO) has been identified throughout the body as an important signaling modification both in physiology and a variety of diseases. SNO is a multifaceted post-translational modification, in that it can either act as a signaling molecule itself or as an intermediate to other modifications. Recent Advances and Critical Issues: Through extensive SNO research, we have made progress toward understanding the importance of single cysteine-SNO sites; however, we are just beginning to explore the importance of specific SNO within the context of other SNO sites and post-translational modifications. Additionally, compartmentalization and SNO occupancy may play an important role in the consequences of the SNO modification. Future Directions: In this review, we will consider the context of SNO signaling and discuss how the transient nature of SNO, its role as an oxidative intermediate, and the pattern of SNO, should be considered when determining the impact of SNO signaling. Antioxid. Redox Signal. 19, 1209–1219.
Introduction
S-nitrosylation (sno) is a labile oxidative post-translational modification that has been implicated in the regulation of protein function in physiology (10, 13, 46, 66, 71, 110) and disease (15, 20, 25, 40, 59, 84, 95). The beauty and challenge of SNO lay in its transient nature, its redox regulation, its role as an oxidative intermediate, and the subtleties of its formation and removal. While many studies have identified targets of SNO and correlated these protein changes with functional outcomes, the implications of the transient and dynamic nature of SNO itself have not been extensively studied.
SNO Formation and Removal
Formation of SNO adducts
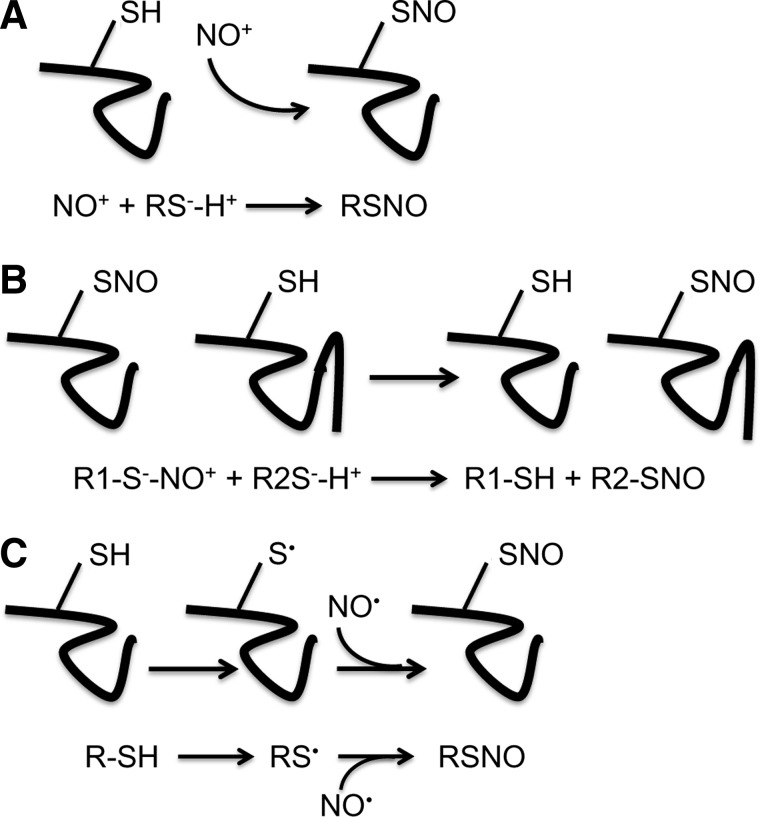
SNO is a reversible, nonenzymatic reaction between a nitric oxide (NO) moiety and the reduced thiol of a cysteine residue to form an S-nitrosothiol. SNO reactions are referred to as S-nitrosation or S-nitrosylation reactions, with the former referring to the chemically precise term for the formation of a thiol-SNO via addition of a nitroso group and the latter referring to the addition of a nitrosyl to a metal. However, SNO has been favored in common usage and will thus, be used in this review. The formation of SNO is achieved through one of three general pathways. In the first, NO becomes thiol reactive by the formation of a nitrosonium cation (NO+), which is then transferred to a thiolate (Fig. 1A). NO+ can be generated by several NO donors. In addition, a second mechanism of SNO formation is through transnitrosylation, whereby a NO+ is donated by either a small SNO-modified molecule or a proximal SNO-modified protein acting as a transnitrosylase (4, 49, 60, 79) (Fig. 1B). The existence of transnitrosylating proteins is clearly demonstrated by Kornberg et al., who show that through binding to Siah1, SNO-glyceraldehyde 3-phosphate dehydrogenase is transported to the nucleus where it then transnitrosylates sirtuin-1, histone deacetylase-2, and DNA-activated protein kinase (60). Finally, in the third SNO pathway, intramolecular electron transfer between redox-sensitive amino acids (120) or hydrogen transfer due to the presence of oxygen radicals (116) results in the formation of a reactive protein thiyl radical (RS•), which then acts as a nucleophile for a NO moiety, resulting in direct SNO formation (Fig. 1C). When first described by Stamler et al. (104, 106), direct SNO was attributed to protein interaction with NO+ (69); however, later evidence suggested that SNO can be achieved through the action of the nitroxyl anion (NO−) as well in the presence of activated protein thiyls (37). Additionally, reactive nitrogen species, such as peroxynitrite (ONOO−), can act as nitrosylating reagents by serving as sources of NO+. It is important to consider, however, that in the absence of a transnitrosylation mechanism, the ability of NO or NO donors to directly SNO proteins is limited by NO diffusion and is thus restricted to proteins localized near sources of NO.
FIG. 1.
Formation of SNO adducts. (A) A nitrosonium cation (NO+) acts as an activated NO moiety to form a SNO bond on a thiolate (RS−H+). (B) A NO+ is donated from a protein-SNO to a reduced protein through a transnitrosylation reaction. (C) A cysteine thiol is activated to a thiyl radical (S•), which can then react with a NO radical (NO•) to form a protein-SNO. NO, nitric oxide; SNO, S-nitrosylation.
Sources of NO
NO is endogenously produced by nitric oxide synthase (NOS) or as the product of enzymatic nitrite reduction. There are three isoforms of NOS: neuronal NOS (NOS1), inducible NOS (NOS2), and endothelial NOS (eNOS; NOS3). The localized expression of the different NOS isoforms plays an important role in dictating the cellular SNO pattern (24, 52, 112), as the in vivo half-life of NO is less than 1 s (21). Both NOS1 and NOS3 are expressed physiologically in a spatially localized manner, while NOS2 expression is increased upon cellular stresses, such as inflammation (42, 89) and hypoxia (41, 75). The ability of NO to modify proteins is also regulated by the presence of reactive oxygen species (ROS), such as superoxide, which may limit NO bioavailability by forming ONOO− (27), changing the ability of a protein cysteine to be SNO or promoting further oxidation of cysteines in a concentration-dependent manner (2).
Removal of SNO adducts
Denitrosylation is achieved through both enzymatic and nonenzymatic mechanisms. The major mechansism of SNO reduction is a transnitrosylation reaction, in which a thiol-containing agent acts as a receiver for the NO moiety of the SNO adduct. Ascorbate, which has been utilized experimentally as a specific reducing agent for SNO, removes SNO by acting as a nucleophile for a transnitrosylation reaction resulting in a free thiol and an O-nitrosoascorbate (29). Similarly, it appears that the enzyme, thioredoxin, reduces SNO through a series of transnitrosylation reactions (11). Additionally, reduced glutathione (GSH), which is an abundant cellular antioxidant, participates in transnitrosylation, resulting in the formation of S-nitrosoglutathione (GSNO). GSNO, the first transitrosylating agent to be characterized (99), can either act as a NO donor to another protein or be reduced to GSH through an enzymatic reaction with S-nitroso glutathione reductase (10, 70). SNO adducts also can be reversed by photolysis under ultraviolet light (33) or through treatment with inorganic copper or mercury, thus providing specific tools for studying SNO. It is important to note that the stability of SNO is limited and thus, must be measured by indirect means, such as the biotin switch method, in conditions of relative darkness. While complicating the measurement of SNO, this inherent instability is favorable for dynamic signaling.
Specificity of SNO Target Sites
Much attention has been given to the importance of compartmentalization and localization of NO sources in determining the location of SNO, but only a few studies have considered how different cellular compartments might biochemically promote or inhibit SNO formation (31, 60, 118). For example, the redox environment of the endoplasmic reticulum is relatively more oxidizing compared with the nucleus or the mitochondria (98). On one hand, a more oxidizing environment might favor prolonged SNO by inhibiting denitrosylation or inhibiting the reduction of oxidized denitrosylases. On the other hand, an oxidizing environment might favor the progression of a SNO adduct to a disulfide or a higher order oxidative post-translational modification. The availability of intraorganellular free glutathione may also affect the concentration of protein-bound SNO by either serving as a substrate for the formation of SNO-glutathione or acting as a transnitrosylase. GSNO is a powerful transnitrosylating agent and thus has the potential to alter the SNO of proteins that are remote from sources of NO. This potential may play an important role in the ability of the SNO signal to propagate to different compartments. Additionally, compartment-specific targeting of other transnitrosylases or denitrosylases may affect localized SNO levels. Therefore, it is important to consider protein-SNO within individual organelles and interpret SNO signals from whole cell lysates in the context of organellular subsets.
In addition to the cellular compartment, the reactivity of the protein thiol group contributes to the formation of SNO. While most reduced thiols are active at pH 8.5, some particularly reactive cysteines can be modified at as low as pH 6.5. This reactivity is determined by the charge of the surrounding secondary structure, which stabilizes the cysteine sulfur in the more reactive thiolate anion (S−) form and stabilizes the transition state of the SNO reaction (26). Therefore, for a given protein, there may be cysteines that are preferentially SNO. Indeed, this appears to be the case. In a previous study, we treated whole heart homogenates with GSNO and detected only a small subset of SNO cysteine residues, rather than SNO of all available cysteine residues, for many protein targets (58). For example, heat shock protein-71 has four available cysteine residues, but only one SNO cysteine was detected. Similarly, NADH-ubiquinone oxidoreductase has 18 available cysteine residues, but only six SNO cysteines were detected. Whole heart homogenates were treated with GSNO in the presence of sodium dodecyl sulfate to facilitate access to all available cysteine residues.
To help explain SNO specificity, the existence of a SNO motif has been explored (39, 58, 82, 117). An acid–base motif within eight angstroms of a cysteine is thought to act as a preferential SNO motif (74). However, the specific features that determine preferential SNO sites are still largely undetermined and may depend on a variety of factors, including secondary structure, the charge of surrounding amino acids, accessibility of the cysteine, and coordinating post-translational modifications. In particular, a distinction between cysteines that are SNO by direct oxidation and those that participate in transitrosation reactions has not been made, and this may lend insight into why SNO patterns differ among proteins. This may also explain why it has been difficult to identify a consensus SNO motif.
Important Properties of SNO
Reactivity
While it is well documented that SNO adducts are labile, the importance of this characteristic is perhaps under appreciated. Unlike irreversible cysteine modifications, such as sulfinic (SO2H) and sulfonic acid (SO3H), SNO is readily reversed and can serve as a temporary modulator of function. Unlike glutathiolation, another reversible oxidative post-translational modification, SNO can be nonenzymatically reduced, allowing for localized changes in pH, increased levels of GSH or even free proximal cysteines to reduce protein SNO (88, 103). SNO shares many characteristics of sulfenic acid (SOH) modifications; however, because of the reactivity of sulfenic acid adducts compared to SNO, sulfenic acid more readily acts as an intermediate to further oxidation (92), while SNO is able to directly modify protein function without further oxidation in addition to serving as an intermediate.
The reactivity of SNO allows for nitrosylated proteins to serve as transnitrosylases, enhancing the SNO signal beyond the diffusion limits of NO and allowing for modification in specific compartments that typically lack NOS, such as the nucleus (60, 112). SNO can also shield cysteines from further oxidation, such as is seen in cardioprotective ischemic preconditioning (58, 59), or serve as a platform for further oxidation, such as in atherosclerosis (2, 107). The diversity of regulatory mechanisms for SNO might account for the ability of SNO to regulate a variety of cellular conditions. Indeed, studies have implicated regulation by SNO in sex-specific protein changes (13, 81, 110), aging (95, 97), cardiovascular disease (5, 36, 58, 59, 85, 107, 109), and renal function (113), to name a few.
Mechanism of SNO-induced protein changes
SNO is thought to exert its effects both through direct alteration of protein function and through thiol protection. In the former, SNO adducts result in steric changes that alter protein conformation and activate or inhibit the target protein (88, 100). For example, SNO can alter enzyme activity by targeting the active site, such as was first described in tissue-type plasminogen activator (105). In the latter, SNO adducts prevent the formation of irreversible oxidation by occupying the cysteine thiol then quickly reverse to restore normal function when the ROS returns to baseline (59). However, under specific redox conditions, SNO may also act as an intermediate to further post-translational modifications that can alter protein activity, degradation or protein–protein interactions (7, 47, 95). Thus, the exact mechanism of SNO-induced functional changes is largely contextual, depending on the redox environment, cellular compartment, and protein chemistry.
It is important to note that while SNO can be a protective modification in many cases, formation of SNO adducts can also lead to pathological states. In the brain, SNO has been correlated with the progression of Alzheimer's Disease due to its role in protein misfolding. In one instance, SNO preferentially modifies apolipoproteins-E2 and E3 at specific cysteine residues, affecting protein folding and low-density lipoprotein metabolism (1). Although many cases of Alzheimer's Disease are linked to genetic variations in apolipoprotein-E4, these SNO proteins may explain onset of the disease in nonfamilial or sporadic Alzheimer's Disease (1). Similarly, detrimental SNO events have been found in other organs, such as the heart, where increased SNO of tropomyosin has been linked to heart failure through inhibition of cardiomyocyte contractility (15), and the liver, where increased SNO is linked to decreased aldehyde dehydrogenase activity after ethanol exposure (80). Thus, SNO signaling can be beneficial or detrimental depending on the level of SNO, the duration of the signal, and the specific SNO target.
SNO occupancy
Although SNO has been shown to influence cell function by altering protein activity, localization, or stability, or by shielding critical thiols from oxidation, the percentage of cysteine-SNO that is required to elicit these effects is not known. In the absence of compartmentalization, it is reasonable to assume that the effectiveness of SNO in altering protein activity or shielding cysteines from oxidation is directly related to the percentage of the total protein that is SNO. In other words, if 1 out of 50 proteins contains a specific SNO modification, will this be functionally relevant? We have recently developed a method for testing this using differential labeling in an effort to understand the occupancy of a given cysteine (57). Using GSNO treatment, we found that SNO occupancy ranged from 23% for cysteine and glycine-rich protein 3 (Cys168) to nearly 80% for fatty acid-binding protein 4 (Cys2). These SNO occupancy levels were derived from total protein homogenate; however, the effects of compartmentalization must also be considered. If a protein is localized to a specific organelle when SNO, then the percentage of protein within this organelle that is SNO is more relevant than the percentage of total cellular protein.
Methods and Challenges of Measuring SNO
Tools for modulating SNO levels
For many reasons, studying the functional consequences of SNO presents a challenge. NO donors have been used extensively for driving SNO formation, but there are several complicating issues in using NO donors, including variation in the NO moiety released, cell permeability, and spatio-temporal specificity of NO release (54, 73, 76). In addition, NO donors may drive the formation of glutathione adducts or ROS rather than SNO. The properties of some common and emerging NO donors are described in Table 1. Recently, organelle-targeted NO donors have been used to study SNO in the mitochondria (19, 93), improving the spatial regulation of SNO formation. It is likely that more targeted approaches, such as this will improve the understanding of SNO compartmentalization and help avoid some experimental artifacts. Tools for the disruption of SNO also suffer from nonspecificity. For example, NOS inhibitors prevent both SNO and NO signaling through guanylylcyclase and thus, have multiple effects that complicate interpretation of the SNO effects. Similarly, the specificity of SNO-reducing agents, such as ascorbate, is controversial (28, 29, 34, 50, 64). Cysteine to serine or alanine protein mutants are also valuable tools to prevent SNO of specific cysteines. While this approach is more targeted than pharmacological approaches, the effects of other post-translational modifications at this site or changes in protein function due to the mutation itself may be overlooked, necessitating the need for careful experimental design. Unfortunately, there are no known mutations that mimic the action of a SNO adduct, although a cysteine to aspartate mutation has been used to mimic oxidation (90). Future work focused on overcoming experimental hurdles in the study of SNO should be encouraged.
Table 1.
Common Nitric Oxide Donors
| Dissociation time | Major NO species | NO byproducts | Special considerations | |
|---|---|---|---|---|
| Angeli's salt | 2.3 min | HNO | NO2− | Also promotes glutathiolation |
| DEA NONOate | 2 min | NO | NO2− | |
| DETA NONOate | 20 h | NO | NO2− | |
| GSNO | Spontaneous | NO/NO+ | GSH byproduct also drives glutathiolation; transnitrosylase | |
| MitoSNO | 23 h | NO | Transnitrosylase | |
| SNAP | 1–6 h | NO | ||
| SNO-cysteine | Spontaneous | NO/NO+ | Transnitrosylase | |
| Sodium nitroprusside | 2 min | NO+ | Cyanide byproduct | |
| Spermine NONOate | 39 min | NO | NO2− |
Assays for measuring SNO
To assess the physiological and pathological ramifications of protein SNO, it is important to have methods to detect and quantify this modification. However, the labile nature of SNO requires special experimental conditions. Addition of an alkylating agent to lysis buffer prevents oxidation or disulfide exchange between free thiols during homogenization (43) and also stabilizes SNO by inhibiting the activity of antioxidants, such as thioredoxin, which could potentially act as transnitrosylases or denitrosylases. Despite this precaution, loss of SNO from proteins during sample preparation remains a challenge, and this may be of greatest concern when examining the SNO signal within specific compartments that require extra purification steps.
The biotin switch assay has been used for the identification of SNO proteins (53). With this approach, reduced thiols are blocked with a methylthiolating agent, such as methyl methanethiosulfonate, or with N-ethylmaleimide, and SNO thiols are then reduced with ascorbate and labeled with biotin. SNO proteins can then be identified and quantified via western blot or with the use of mass spectrometry. The biotin switch assay is one of the most common techniques used for examining SNO, and many derivatives have been developed based upon this approach. These derivatives include two-dimensional fluorescence difference electrophoresis, which utilizes fluorescent maleimide dyes to label SNO cysteine residues (i.e., CyDyemaleimide, DyLightmaleimide) (67, 96, 107, 109). SNO-resin-assisted capture is another derivative of the biotin switch that utilizes a thiol-binding resin (i.e., thiopropylsepharose) to quantify SNO levels and identify specific SNO cysteine residues with mass spectrometry (30, 58, 59). Recently, Murray et al. utilized a modified biotin switch approach with cysteine-reactive tandem mass tags (cysTMT) to measure SNO levels in human pulmonary arterial endothelial cells with mass spectrometry (83). cysTMT labels confer the advantage of multiple isobaric tags, with reporter ions between 126 and 131 kDa, and this allows for sequential labeling and identification of more than one cysteine modification (59). We recently used cysTMT labeling to determine SNO occupancy (57). In the same sample, free thiols were labeled with one isobaric tag, while SNO thiols were subsequently labeled with a second isobaric tag. Free and SNO thiols were quantified using mass spectrometry, and SNO occupancy was calculated as the ratio of SNO thiols to total thiols (SNO+free). Using a similar approach, these labels could serve as an important tool for analyzing SNO within the context of other redox-based modifications. Additional variations of the biotin switch assay include SNO Site Identification (44) and the HIS-TAG switch method (14).
Although the biotin switch assay and variations thereof offer many advantages and are some of the most widely used methods for examining SNO, this approach is not without limitations. The major issue with this methodology results from the nonspecificity of ascorbate, which has been reported to reduce other cysteine-based modifications (34, 50, 64), although these claims have been recently refuted (28, 29); however, it is critically important to implement the proper controls when utilizing a biotin switch-based approach. There are some alternative methods for examining SNO that are not derivatives of the biotin switch and do not require the use of ascorbate. Mass spectrometry has been utilized to directly identify SNO cysteine residues, but these studies have been limited to purified proteins (78, 115). This approach remains a challenge, as the experimental conditions required for mass spectrometry may lead to the degradation of protein SNO.
Another alternative method developed by Doulias et al., utilized a phenylmercury- based capture approach to examine protein SNO (23). With this approach, the SNO modification is reacted with phenylmercury to form a stable thiol-mercury bond. Labeled peptides are then captured with the use of an organomercury resin and examined via mass spectrometry. This method avoids the use of ascorbate, but the specificity of the phenylmercury compound still remains a potential issue.
There are also several commercially available antibodies for the detection of SNO-modified proteins. However, preserving SNO during cell lysis and gel electrophoresis or during cell fixation presents a challenge and care must be taken to keep samples in the dark, free of reducing agents, and free of exogenous oxidants throughout the procedure. As with any antibody, specificity can be a potential issue with this approach. In particular, many SNO-specific antibodies are raised to nitrosylated serum albumin, and these epitopes may not correspond to those found in a complex homogenate. To address this, a reducing agent, such as ascorbate, should be used in negative controls to reverse SNO and prevent antibody recognition. Antibody peptide competition using either SNO bovine serum albumin or GSNO may also serve as a negative control and help to identify any nonspecific binding (38). Positive controls can also be generated by treating samples with NO donors. It is interesting to note that SNO-specific antibodies have been detected in patients with human African trypanosomiasis (101).
There are many different methods that have been developed for the detection and quantification of the SNO modification, and all have distinct advantages and disadvantages. The approaches utilized for a particular study will vary depending on experimental objectives (i.e., targeted vs. high-throughput), but in the end, a multifaceted approach will yield more robust results, and avoid the common pitfalls of each of the individual methods, while reducing the probability of false-positive identifications.
Interaction of SNO with Nonoxidative Post-Translational Modifications
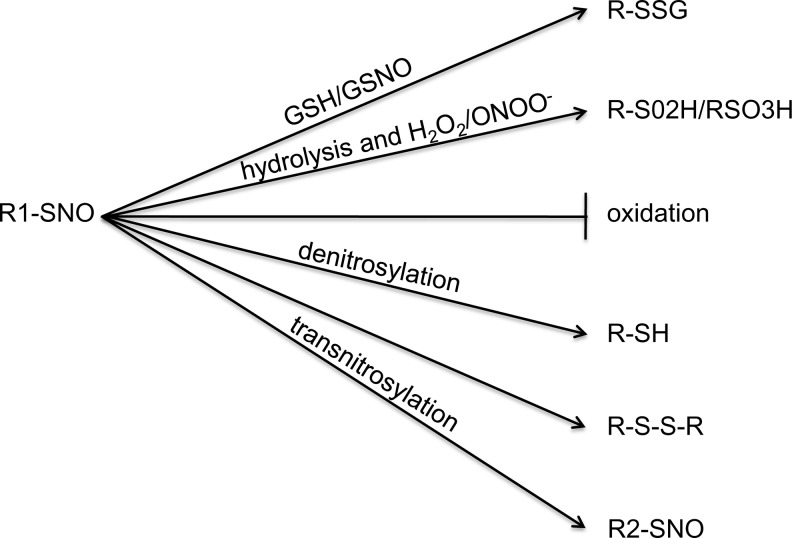
SNO serves a dual role as both a protective modification and as an intermediate to further oxidation, such as glutathiolation and sulfinic and sulfonic acid modifications (9). SNO of proteins responsible for maintaining acylation, phosphorylation, ubiquitination, and palmitoylation has also emerged as a method through which SNO can diversely signal [See Hess and Stamler (47) for discussion of these pathways). Additionally, it is now recognized that SNO modifications can act as an intermediate for the formation of secondary post-translational modifications, such as palmitoylation (3) or ubiquitination (55), either on the SNO-modified cysteine or on surrounding amino acids. By serving as a platform for further oxidation, the potential role of SNO intermediates in the propagation of signaling is enhanced (Fig. 2).
FIG. 2.
SNO dynamically regulates other oxidative post-translational modifications. SNO-cysteines can serve as intermediates for glutathiolation, sulfinic/sulfonic acid formation, or disulfide bond formation. SNO can also protect cysteines from further oxidation and later be reduced by denitrosylases to restore protein function. SNO modifications can be passed between proteins by transnitrosylation, where SNO can serve as an intermediate or protective modification on a different protein, propagating the SNO signal.
SNO as a platform for secondary oxidation
SNO can serve as an intermediate for the formation of sulfinic or sulfonic acid. The mechanism of this oxidation appears to be due to the high on/off rate of the NO group, allowing for hydrolysis of SNO to free thiol and subsequent formation of a relatively unstable sulfenic acid adduct that can then lead to further oxidation to sulfinic or sulfonic acid (46, 65, 91). Increased peroxynitrite production due to either overproduction of ROS or decreased antioxidant activity has been implicated in the formation of these irreversible modifications (114). For example, Gu et al. demonstrate that matrix metalloproteinase-9 (MMP-9) can be SNO in the zinc-coordinating domain, allowing MMP-9 to dissociate from inhibitory zinc to become active in its uncleaved form (40). However, under conditions of increased oxidative stress, such as following ischemia/reperfusion, MMP-9 is activated through sulfonic acid formation at the same site in a SNO-dependent manner. In this instance, SNO seems to serve as a temporary activator of MMP-9, while sulfonation leads to prolonged activation, which is associated with coronary artery disease (94), diabetic retinopathy (61), and idiopathic dilated cardiomyopathy (94).
In contrast, the role of SNO as a protective modification has been explored in several contexts, including that of cardiac ischemic preconditioning (58, 67, 107) and immune response (45). In this context, SNO either prevents the formation of higher order, irreversible modifications, such as sulfinic and sulfonic acid, or acts as an intermediate to other reversible modifications, such as glutathiolation, that can also prevent oxidation. In the case of glutathiolation, SNO primes the cysteine for a thiol exchange reaction wherein the sulfur of a GSH displaces the covalent NO bond of the SNO, resulting in the formation of an RSSG (7), which can occupy the cysteine site and prevent further oxidation. Like SNO, glutathiolation modifications also act to regulate protein function (2, 12, 63). Similarly SNO itself can prevent further oxidation of occupied cysteine sites. The chemistry underlying this protection is context driven and may be unique to individual proteins; however, it is likely that SNO can inhibit oxidation through a number of different pathways. In some proteins, SNO is known to alter protein conformation, effectively shielding a thiol from oxidants (88). It is also likely that SNO inhibits further oxidation of specific cysteines by occupying the cysteine thiol and decreasing its reactivity with surrounding oxidants. This may be a particularly important mechanism of protection for reactive cysteines that exist in a thiolate anion state and are thus, more readily oxidized than in the SNO-bound state.
As an example, let us consider protein tyrosine phosphatase-1B (PTP1B). PTP1B is a nonreceptor phosphotyrosine phosphatase that negatively regulates insulin signaling through dephosphorylation of the insulin receptor. Specific hydrogen peroxide-induced irreversible oxidation of PTP1B at cysteine-215 plays an important role in the inhibition of PTP1B and the promigratory behavior of cancer cells (72); however, pretreatment of PTP1B with a NO donor prevents hydrogen peroxide-induced sulfonic acid formation at this site (16). Mass spectrometric measurements indicate that formation of SNO at cysteine-215, but not at other cysteine residues, prevents the reversible oxidation seen under conditions of oxidative stress and preserves PTP1B catalytic function (16). Cysteine-215 is a highly reactive cysteine that can be sulfenated, glutathiolated (8), nitrosylated (16), or irreversibly oxidized (16, 72). SNO at this site appears to shield the cysteine from further oxidation, preserving the thiol for reversible, regulatory oxidation.
SNO as a regulator of nonoxidative post-translational modifications
The crosstalk between SNO and other post-translational modifications has recently become an area of interest, and SNO is known to competitively inhibit some post-translational modifications. For example, palmitoylation of the neuronal scaffolding protein, PSD-95, is prevented by SNO adduct formation at the same sites in response to NO treatment (48). Similarly, SNO is inversely correlated with phosphorylation of eNOS at serine-1179, a site that is associated with eNOS activation (24). The nitrosylation and denitrosylation of eNOS is dependent on intracellular targeting and lends insight into the regulatory mechanisms underlying dynamic SNO. The mechanism of SNO inhibition of post-translational modifications has not been fully explored, but likely shares many characteristics of SNO-mediated protection from oxidation (i.e., changing protein conformation or altering the redox environment).
SNO is also a mediator of protein degradation pathways. Broadly, SNO of E3 ligases is linked to decreased global ubiquitination (20, 84); however, direct interaction of SNO and ubiquitination has been observed on an individual protein level and is known to both enhance and inhibit protein ubiquitination (6, 55, 62, 95). A direct correlation between SNO and degradation is demonstrated through the SNO-mediated enhancement of ubiquitination that has been documented for phosphatase and tensin homology (PTEN) in the brain. SNO of the PI3K-inhibiting protein, PTEN, at cysteine-124 leads to its ubiquitination after treatment of cultured cortical neurons with a NO donor. Mutation of PTEN cysteine-124 prevents ubiquitination and enhances protein stability, inhibiting removal of PTEN from the cells and preventing PI3K signaling (62). It is reasonable to assume that SNO-mediated regulation of ubiquitination may be present in other proteins, and indeed, it has been suggested in the regulation of iron homeostasis (55), apoptosis (6), and Alzheimer's Disease (95). Dynamic regulation between SNO and other post-translational modifications likely will be uncovered as the topic is explored in greater depth.
A barcode of post-translational modifications
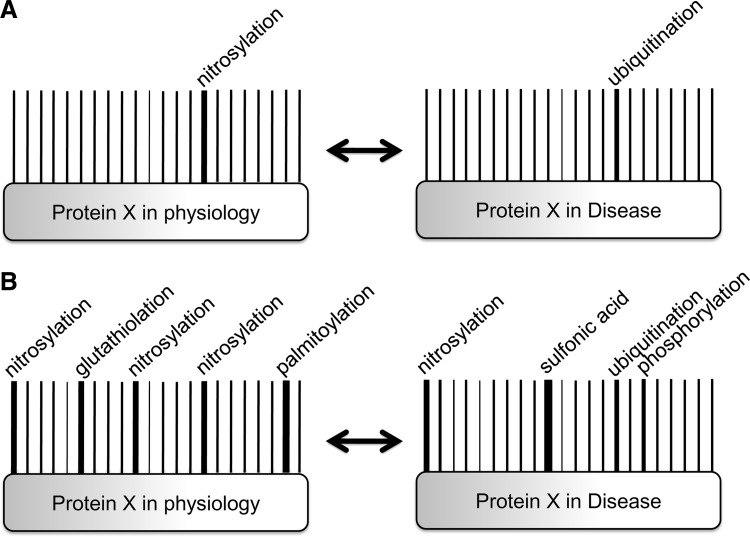
As the role of SNO in the modulation of other post-translational modifications becomes clearer, it may be important to ask how these post-translational modifications interact on proteins. The barcode concept suggests that proteins can be regulated by a post-translational modification at a single site or by the interplay among several different modifications, each of which plays an important role in altering protein chemistry and function. Much like the individual lines on a barcode, each combination of modifications results in a different outcome; thus, the individual contribution of any given post-translational modification is contextual. A phosphorylation barcode has been demonstrated in the beta(2)-adrenergic receptor, where the pattern of phosphorylation differs depending on which G-protein-coupled receptor kinases are active. Each distinct pattern of phosphorylation alters the functional capability of beta-arrestin (86). In considering more than one modification, there are a vast number of potential post-translational modification arrays, and these arrays could produce changes in protein–protein interaction, induce protein translocation, or alter protein activity (Fig. 3).
FIG. 3.
A barcode of post-translational modifications. (A) The traditional approach to post-translational modifications focuses on changes in a single amino acid. In this example, protein X is SNO at a specific cysteine under physiological conditions, but loss of SNO at that site leads to increased ubiquitination in disease. (B) The barcode approach shows all modifications of protein X under physiological conditions. As opposed to the traditional approach shown in (A), the barcode approach considers the whole protein and shows that the change in protein X from physiology to disease requires increased ubiquitination, loss of specific SNO sites, loss of glutathiolation, increased oxidation and phosphorylation, and decreased palmitoylation, all of which may coordinate to affect protein function.
As an example, let us consider post-translational regulation of the N-methyl-D-aspartate (NMDA) receptor. The NMDA receptor is a heterotetrameric protein complex expressed in the brain, which acts as a voltage-gated, nonselective ion channel important for postsynaptic second messenger signaling (22). The NMDA receptor has been reported to be modified by SNO (111), phosphorylation, ubiquitination, and palmitoylation (35), each of which is said to play a unique role in altering protein function or localization, and some aspects of the interplay between these modifications have been explored. Takahashi et al. demonstrated that the NMDA receptor can be SNO at cysteine-744 and 798 of the NR1 subunit under conditions of hypoxia (111). Although SNO of these cysteines has no direct functional consequence, these sites act as a thiol sensor, selectively enhancing SNO susceptibility at other cysteine sites that can then inhibit NMDA receptor function. In contrast, SNO of cysteine-87, 320, and 399 on the NR2A subunit appear to directly inhibit NMDA receptor activity (18), confirming that SNO on multiple sites is required for modulation of function. Cysteine-744 and 798 of the NR1 subunit and cysteine-87 and 320 of the NR2A subunit can also be oxidized to form disulphide bonds in the presence of ROS or following SNO, which has been shown to decrease NMDA receptor activity (17, 18). These data are consistent with a SNO barcode.
In considering this small subset of post-translational modifications on the NMDA receptor, it becomes clear that multiple post-translation modification sites have overlapping and/or interacting functionalities. However, the status of post-translational modifications on the whole protein at any given time has not been explored and will most likely be experimentally challenging. Understanding the patterns of regulation for individual post-translational modifications, as was done for SNO on the NMDA receptor (111) or for phosphorylation on the beta(2)-adrenergic receptor (86), may be a more feasible step. Regardless, it is likely that the interplay of post-translational modifications contributes to dynamic protein regulation and that further analysis of these interactions will lend insight into cellular regulatory mechanisms.
SNO as a molecular switch
While many post-translational modifications are thought to act as on/off switches that either increase or decrease protein function, this may not be true in all cases. For example, dynamic regulation of disulfide states has been linked to changes in protein folding, trafficking, and localization in addition to the prototypical protein inhibition and activation (119). Following from this concept, dynamic oxidation of proteins may alter cellular function. OxyR, a transcription factor that can be activated by both SNO and oxidation, has distinct graded responses to different modifications at the same cysteine (56). Circular dichroism analysis indicates that different modifications at this site alter protein structure (i.e., alpha helical content) and seem to change DNA affinity and cooperativity between subunits. Further studies indicate that SNO-OxyR and oxidized OxyR have both overlapping and distinct gene transcriptional targets (102). Interestingly, many of the genes regulated by SNO-OxyR are regulators of nitrosative stress, implying a cysteine-specific redox-derived molecular switch.
Conclusions
SNO is a dynamic and diverse oxidative post-translational modification that has important implications for the regulation of protein function. While many SNO sites have been discovered on a variety of proteins, the importance of SNO in mediating other post-translational modifications and in altering protein function has not been fully explored. Future work should focus not only on identifying SNO sites, but also on understanding the mechanism of SNO action and developing methods that will allow for studies of the dynamic interplay between post-translational modifications.
Abbreviations Used
- cysTMT
cysteine-reactive tandem mass tags
- DEA
diethylamine
- DETA
diethylenetriamine
- eNOS
endothelial NOS
- GSH
reduced glutathione
- GSNO
S-nitrosoglutathione
- MMP-9
matrix metalloproteinase-9
- NMDA
N-methyl-D-aspartate
- NO
nitric oxide
- NO−
nitroxyl anion
- NO+
nitrosonium
- NOS
nitric oxide synthase
- ONOO−
peroxynitrite
- PTEN
phosphatase and tensin homology
- PTP1B
protein tyrosine phosphatase-1B
- ROS
reactive oxygen species
- RS−
thiyl radical
- S−
thiolate anion
- SNAP
S-nitroso-N-acetyl-d,l-penicillamine
- SNO
S-nitrosylation
- SO2H
sulfinic acid
- SO3H
sulfonic acid
- SOH
sulfenic acid
Acknowledgments
The authors acknowledge the support of the National Institutes of Health (Z01HL006059 and Z01HL002066, A.M.E., E.M.) and the American Heart Association (12BGIA11780030, M.J.K.).
References
- 1.Abrams AJ. Farooq A. Wang G. S-nitrosylation of ApoE in Alzheimer's disease. Biochemistry. 2011;50:3405–3407. doi: 10.1021/bi200266v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi T. Weisbrod RM. Pimentel DR. Ying J. Sharov VS. Schoneich C. Cohen RA. S-glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat Med. 2004;10:1200–1207. doi: 10.1038/nm1119. [DOI] [PubMed] [Google Scholar]
- 3.Adam L. Bouvier M. Jones TL. Nitric oxide modulates beta(2)-adrenergic receptor palmitoylation and signaling. J Biol Chem. 1999;274:26337–26343. doi: 10.1074/jbc.274.37.26337. [DOI] [PubMed] [Google Scholar]
- 4.Arnelle DR. Stamler JS. NO+, NO, and NO- donation by S-nitrosothiols: implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch Biochem Biophys. 1995;318:279–285. doi: 10.1006/abbi.1995.1231. [DOI] [PubMed] [Google Scholar]
- 5.Atar S. Ye Y. Lin Y. Freeberg SY. Nishi SP. Rosanio S. Huang MH. Uretsky BF. Perez-Polo JR. Birnbaum Y. Atorvastatin-induced cardioprotection is mediated by increasing inducible nitric oxide synthase and consequent S-nitrosylation of cyclooxygenase-2. Am J Physiol Heart Circ Physiol. 2006;290:H1960–H1968. doi: 10.1152/ajpheart.01137.2005. [DOI] [PubMed] [Google Scholar]
- 6.Azad N. Vallyathan V. Wang L. Tantishaiyakul V. Stehlik C. Leonard SS. Rojanasakul Y. S-nitrosylation of Bcl-2 inhibits its ubiquitin-proteasomal degradation. J Biol Chem. 2006;281:34124–34134. doi: 10.1074/jbc.M602551200. [DOI] [PubMed] [Google Scholar]
- 7.Baba SP. Wetzelberger K. Hoetker JD. Bhatnagar A. Posttranslational glutathiolation of aldose reductase (AKR1B1): a possible mechanism of protein recovery from S-nitrosylation. Chem Biol Interact. 2009;178:250–258. doi: 10.1016/j.cbi.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett WC. DeGnore JP. König S. Fales HM. Keng Y-F. Zhang Z-Y. Yim MB. Chock PB. Regulation of PTP1B via glutathionylation of the active site cysteine 215. Biochemistry. 1999;38:6699–6705. doi: 10.1021/bi990240v. [DOI] [PubMed] [Google Scholar]
- 9.Becker K. Savvides SN. Keese M. Schirmer RH. Karplus PA. Enzyme inactivation through sulfhydryl oxidation by physiologic NO-carriers. Nat Struct Biol. 1998;5:267–271. doi: 10.1038/nsb0498-267. [DOI] [PubMed] [Google Scholar]
- 10.Beigi F. Gonzalez DR. Minhas KM. Sun QA. Foster MW. Khan SA. Treuer AV. Dulce RA. Harrison RW. Saraiva RM. Premer C. Schulman IH. Stamler JS. Hare JM. Dynamic denitrosylation via S-nitrosoglutathione reductase regulates cardiovascular function. Proc Natl Acad Sci U S A. 2012;109:4314–4319. doi: 10.1073/pnas.1113319109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benhar M. Forrester MT. Hess DT. Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berndt C. Lillig CH. Holmgren A. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. Am J Physiol Heart Circ Physiol. 2007;292:H1227–H1236. doi: 10.1152/ajpheart.01162.2006. [DOI] [PubMed] [Google Scholar]
- 13.Brown-Steinke K. deRonde K. Yemen S. Palmer LA. Gender differences in S-nitrosoglutathione reductase activity in the lung. PLoS ONE. 2010;5:e14007. doi: 10.1371/journal.pone.0014007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camerini S. Polci ML. Restuccia U. Usuelli V. Malgaroli A. Bachi A. A novel approach to identify proteins modified by nitric oxide: the HIS-TAG switch method. J Proteome Res. 2007;6:3224–3231. doi: 10.1021/pr0701456. [DOI] [PubMed] [Google Scholar]
- 15.Canton M. Menazza S. Sheeran FL. Polverino de Laureto P. Di Lisa F. Pepe S. Oxidation of myofibrillar proteins in human heart failure. J Am Col Cardiol. 2011;57:300–309. doi: 10.1016/j.jacc.2010.06.058. [DOI] [PubMed] [Google Scholar]
- 16.Chen YY. Chu HM. Pan KT. Teng CH. Wang DL. Wang AH. Khoo KH. Meng TC. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J Biol Chem. 2008;283:35265–35272. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi Y. Chen HV. Lipton SA. Three pairs of cysteine residues mediate both redox and Zn2+ modulation of the nmda receptor. J Neurosci. 2001;21:392–400. doi: 10.1523/JNEUROSCI.21-02-00392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi YB. Tenneti L. Le DA. Ortiz J. Bai G. Chen HS. Lipton SA. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nat Neurosci. 2000;3:15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- 19.Chouchani ET. Hurd TR. Nadtochiy SM. Brookes PS. Fearnley IM. Lilley KS. Smith RA. Murphy MP. Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem J. 2010;430:49–59. doi: 10.1042/BJ20100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung KK. Thomas B. Li X. Pletnikova O. Troncoso JC. Marsh L. Dawson VL. Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 21.Crow JP. Spruell C. Chen J. Gunn C. Ischiropoulos H. Tsai M. Smith CD. Radi R. Koppenol WH. Beckman JS. On the pH-dependent yield of hydroxyl radical products from peroxynitrite. Free Radic Biol Med. 1994;16:331–338. doi: 10.1016/0891-5849(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 22.Dingledine R. Borges K. Bowie D. Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 23.Doulias PT. Greene JL. Greco TM. Tenopoulou M. Seeholzer SH. Dunbrack RL. Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci U S A. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erwin PA. Lin AJ. Golan DE. Michel T. Receptor-regulated dynamic S-nitrosylation of endothelial nitric-oxide synthase in vascular endothelial cells. J Biol Chem. 2005;280:19888–19894. doi: 10.1074/jbc.M413058200. [DOI] [PubMed] [Google Scholar]
- 25.Evangelista AM. Rao VS. Filo AR. Marozkina NV. Doctor A. Jones DR. Gaston B. Guilford WH. Direct regulation of striated muscle myosins by nitric oxide and endogenous nitrosothiols. PLoS ONE. 2010;5:e11209. doi: 10.1371/journal.pone.0011209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrer-Sueta G. Manta B. Botti H. Radi R. Trujillo M. Denicola A. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem Res Toxicol. 2011;24:434–450. doi: 10.1021/tx100413v. [DOI] [PubMed] [Google Scholar]
- 27.Ferrer-Sueta G. Radi R. Chemical biology of peroxynitrite: kinetics, diffusion, and radicals. ACS Chem Biol. 2009;4:161–177. doi: 10.1021/cb800279q. [DOI] [PubMed] [Google Scholar]
- 28.Forrester MT. Foster MW. Benhar M. Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic Biol Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forrester MT. Foster MW. Stamler JS. Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 30.Forrester MT. Thompson JW. Foster MW. Nogueira L. Moseley MA. Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foster MW. Stamler JS. New insights into protein S-nitrosylation. J Biol Chem. 2004;279:25891–25897. doi: 10.1074/jbc.M313853200. [DOI] [PubMed] [Google Scholar]
- 32.Friederich JA. Butterworth JF., 4th Sodium nitroprusside: twenty years and counting. Anesth Analg. 1995;81:152–162. doi: 10.1097/00000539-199507000-00031. [DOI] [PubMed] [Google Scholar]
- 33.Furchgott RF. Jothianandan D. Endothelium-dependent and -independent vasodilation involving cyclic GMP: relaxation induced by nitric oxide, carbon monoxide and light. Blood Vessels. 1991;28:52–61. doi: 10.1159/000158843. [DOI] [PubMed] [Google Scholar]
- 34.Giustarini D. Dalle-Donne I. Colombo R. Milzani A. Rossi R. Is ascorbate able to reduce disulfide bridges? A cautionary note. Nitric Oxide. 2008;19:252–258. doi: 10.1016/j.niox.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Gladding CM. Raymond LA. Mechanisms underlying NMDA receptor synaptic/extrasynaptic distribution and function. Mol Cell Neurosci. 2011;48:308–320. doi: 10.1016/j.mcn.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez DR. Treuer AV. Castellanos J. Dulce RA. Hare JM. Impaired S-nitrosylation of the ryanodine receptor caused by xanthine oxidase activity contributes to calcium leak in heart failure. J Biol Chem. 2010;285:28938–28945. doi: 10.1074/jbc.M110.154948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gow AJ. Buerk DG. Ischiropoulos H. A novel reaction mechanism for the formation of S-nitrosothiol in vivo. J Biol Chem. 1997;272:2841–2845. doi: 10.1074/jbc.272.5.2841. [DOI] [PubMed] [Google Scholar]
- 38.Gow AJ. Davis CW. Munson D. Ischiropoulos H. Immunohistochemical detection of S-nitrosylated proteins. Methods Mol Biol. 2004;279:167–172. doi: 10.1385/1-59259-807-2:167. [DOI] [PubMed] [Google Scholar]
- 39.Greco TM. Hodara R. Parastatidis I. Heijnen HF. Dennehy MK. Liebler DC. Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci U S A. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu Z. Kaul M. Yan B. Kridel SJ. Cui J. Strongin A. Smith JW. Liddington RC. Lipton SA. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- 41.Guo G. Bhat NR. Hypoxia/reoxygenation differentially modulates NF-kappaB activation and iNOS expression in astrocytes and microglia. Antioxid Redox Signal. 2006;8:911–918. doi: 10.1089/ars.2006.8.911. [DOI] [PubMed] [Google Scholar]
- 42.Han W. Fu S. Wei N. Xie B. Li W. Yang S. Li Y. Liang Z. Huo H. Nitric oxide overproduction derived from inducible nitric oxide synthase increases cardiomyocyte apoptosis in human atrial fibrillation. Int J Cardiol. 2008;130:165–173. doi: 10.1016/j.ijcard.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 43.Hansen RE. Winther JR. An introduction to methods for analyzing thiols and disulfides: reactions, reagents, and practical considerations. Anal Biochem. 2009;394:147–158. doi: 10.1016/j.ab.2009.07.051. [DOI] [PubMed] [Google Scholar]
- 44.Hao G. Derakhshan B. Shi L. Campagne F. Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernansanz-Agustin P. Izquierdo-Alvarez A. Garcia-Ortiz A. Ibiza S. Serrador JM. Martinez-Ruiz A. Nitrosothiols in the immune system: signalling and protection. Antioxid Redox Signal. 2013;18:288–308. doi: 10.1089/ars.2012.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hess DT. Matsumoto A. Kim SO. Marshall HE. Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 47.Hess DT. Stamler JS. Regulation by S-nitrosylation of protein post-translational modification. J Biol Chem. 2012;287:4411–4418. doi: 10.1074/jbc.R111.285742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ho Gary PH. Selvakumar B. Mukai J. Hester Lynda D. Wang Y. Gogos Joseph A. Snyder Solomon H. S-nitrosylation and S-palmitoylation reciprocally regulate synaptic targeting of PSD-95. Neuron. 2011;71:131–141. doi: 10.1016/j.neuron.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hogg N. The kinetics of S-transnitrosation—a reversible second-order reaction. Anal Biochem. 1999;272:257–262. doi: 10.1006/abio.1999.4199. [DOI] [PubMed] [Google Scholar]
- 50.Huang B. Chen C. An ascorbate-dependent artifact that interferes with the interpretation of the biotin switch assay. Free Radic Biol Med. 2006;41:562–567. doi: 10.1016/j.freeradbiomed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 51.Huges MN. Wimbledon PE. The chemisry of trioxodinitrates. Part I. Decompostion of sodium trioxodinitrate (Angeli's salt) in aqueous solution. J Chem Soc Dalton Trans. 1976:703–707. [Google Scholar]
- 52.Iwakiri Y. Satoh A. Chatterjee S. Toomre DK. Chalouni CM. Fulton D. Groszmann RJ. Shah VH. Sessa WC. Nitric oxide synthase generates nitric oxide locally to regulate compartmentalized protein S-nitrosylation and protein trafficking. Proc Natl Acad Sci U S A. 2006;103:19777–19782. doi: 10.1073/pnas.0605907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaffrey SR. Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:PL1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 54.Ji Y. Akerboom TP. Sies H. Thomas JA. S-nitrosylation and S-glutathiolation of protein sulfhydryls by S-nitroso glutathione. Arch Biochem Biophys. 1999;362:67–78. doi: 10.1006/abbi.1998.1013. [DOI] [PubMed] [Google Scholar]
- 55.Kim S. Wing SS. Ponka P. S-nitrosylation of IRP2 regulates its stability via the ubiquitin-proteasome pathway. Mol Cell Biol. 2004;24:330–337. doi: 10.1128/MCB.24.1.330-337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim SO. Merchant K. Nudelman R. Beyer WF., Jr. Keng T. DeAngelo J. Hausladen A. Stamler JS. OxyR: a molecular code for redox-related signaling. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 57.Kohr MJ. Aponte A. Sun J. Gucek M. Steenbergen C. Murphy E. Measurement of S-nitrosylation occupancy in the myocardium with cysteine-reactive tandem mass tags. Circ Res. 2012;111:1308–1312. doi: 10.1161/CIRCRESAHA.112.271320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohr MJ. Aponte AM. Sun J. Wang G. Murphy E. Gucek M. Steenbergen C. Characterization of potential S-nitrosylation sites in the myocardium. Am J Physiol Heart Circ Physiol. 2011;300:H1327–H1335. doi: 10.1152/ajpheart.00997.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kohr MJ. Sun J. Aponte A. Wang G. Gucek M. Murphy E. Steenbergen C. Simultaneous measurement of protein oxidation and S-nitrosylation during preconditioning and ischemia/reperfusion injury with resin-assisted capture. Circ Res. 2011;108:418–426. doi: 10.1161/CIRCRESAHA.110.232173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kornberg MD. Sen N. Hara MR. Juluri KR. Nguyen JV. Snowman AM. Law L. Hester LD. Snyder SH. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kowluru RA. Kanwar M. Oxidative stress and the development of diabetic retinopathy: contributory role of matrix metalloproteinase-2. Free Radic Biol Med. 2009;46:1677–1685. doi: 10.1016/j.freeradbiomed.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kwak YD. Ma T. Diao S. Zhang X. Chen Y. Hsu J. Lipton SA. Masliah E. Xu H. Liao FF. NO signaling and S-nitrosylation regulate PTEN inhibition in neurodegeneration. Mol Neurodegener. 2010;5:49. doi: 10.1186/1750-1326-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lancel S. Zhang J. Evangelista A. Trucillo MP. Tong X. Siwik DA. Cohen RA. Colucci WS. Nitroxyl activates SERCA in cardiac myocytes via glutathiolation of cysteine 674. Circ Res. 2009;104:720–723. doi: 10.1161/CIRCRESAHA.108.188441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Landino LM. Koumas MT. Mason CE. Alston JA. Ascorbic acid reduction of microtubule protein disulfides and its relevance to protein S-nitrosylation assays. Biochem Biophys Res Commun. 2006;340:347–352. doi: 10.1016/j.bbrc.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 65.Leonard SE. Carroll KS. Chemical ‘omics' approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 2011;15:88–102. doi: 10.1016/j.cbpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 66.Lima B. Lam GK. Xie L. Diesen DL. Villamizar N. Nienaber J. Messina E. Bowles D. Kontos CD. Hare JM. Stamler JS. Rockman HA. Endogenous S-nitrosothiols protect against myocardial injury. Proc Natl Acad Sci U S A. 2009;106:6297–6302. doi: 10.1073/pnas.0901043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin J. Steenbergen C. Murphy E. Sun J. Estrogen receptor-beta activation results in S-nitrosylation of proteins involved in cardioprotection. Circulation. 2009;120:245–254. doi: 10.1161/CIRCULATIONAHA.109.868729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lipton SA. Choi Y-B. Pan Z-H. Lei SZ. Chen H-SV. Sucher NJ. Loscalzo J. Singel DJ. Stamler JS. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 69.Lipton SA. Singel DJ. Stamler JS. Nitric oxide in the central nervous system. Prog Brain Res. 1994;103:359–364. doi: 10.1016/s0079-6123(08)61149-8. [DOI] [PubMed] [Google Scholar]
- 70.Liu L. Hausladen A. Zeng M. Que L. Heitman J. Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 71.Liu L. Yan Y. Zeng M. Zhang J. Hanes MA. Ahearn G. McMahon TJ. Dickfeld T. Marshall HE. Que LG. Stamler JS. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell. 2004;116:617–628. doi: 10.1016/s0092-8674(04)00131-x. [DOI] [PubMed] [Google Scholar]
- 72.Lou YW. Chen YY. Hsu SF. Chen RK. Lee CL. Khoo KH. Tonks NK. Meng TC. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. FEBS J. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- 73.Mallis RJ. Thomas JA. Effect of S-nitrosothiols on cellular glutathione and reactive protein sulfhydryls. Arch Biochem Biophys. 2000;383:60–69. doi: 10.1006/abbi.2000.2048. [DOI] [PubMed] [Google Scholar]
- 74.Marino SM. Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Melillo G. Taylor LS. Brooks A. Cox GW. Varesio L. Regulation of inducible nitric oxide synthase expression in IFN-gamma-treated murine macrophages cultured under hypoxic conditions. J Immunol. 1996;157:2638–2644. [PubMed] [Google Scholar]
- 76.Miller MR. Megson IL. Recent developments in nitric oxide donor drugs. Br J Pharmacol. 2007;151:305–321. doi: 10.1038/sj.bjp.0707224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miranda KM. Katori T. Torres de Holding CL. Thomas L. Ridnour LA. McLendon WJ. Cologna SM. Dutton AS. Champion HC. Mancardi D. Tocchetti CG. Saavedra JE. Keefer LK. Houk KN. Fukuto JM. Kass DA. Paolocci N. Wink DA. Comparison of the NO and HNO donating properties of diazeniumdiolates: primary amine adducts release HNO in vivo. J Med Chem. 2005;48:8220–8228. doi: 10.1021/jm050151i. [DOI] [PubMed] [Google Scholar]
- 78.Mirza UA. Chait BT. Lander HM. Monitoring reactions of nitric oxide with peptides and proteins by electrospray ionization-mass spectrometry. J Biol Chem. 1995;270:17185–17188. doi: 10.1074/jbc.270.29.17185. [DOI] [PubMed] [Google Scholar]
- 79.Mitchell DA. Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat Chem Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- 80.Moon KH. Abdelmegeed MA. Song BJ. Inactivation of cytosolic aldehyde dehydrogenase via S-nitrosylation in ethanol-exposed rat liver. FEBS Lett. 2007;581:3967–3972. doi: 10.1016/j.febslet.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murphy E. Steenbergen C. Gender-based differences in mechanisms of protection in myocardial ischemia–reperfusion injury. Cardiovasc Res. 2007;75:478–486. doi: 10.1016/j.cardiores.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 82.Murray CI. Kane LA. Uhrigshardt H. Wang S-B. Van Eyk JE. Site-mapping of in vitro S-nitrosation in cardiac mitochondria: implications for cardioprotection. Mol Cell Proteomics. 2011;10:M110.004721. doi: 10.1074/mcp.M110.004721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murray CI. Uhrigshardt H. O'Meally RN. Cole RN. Van Eyk JE. Identification and quantification of S-nitrosylation by cysteine reactive tandem mass Tag switch assay. Mol Cell Proteomics. 2011;11:M111.013441. doi: 10.1074/mcp.M111.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakamura T. Wang L. Wong CC. Scott FL. Eckelman BP. Han X. Tzitzilonis C. Meng F. Gu Z. Holland EA. Clemente AT. Okamoto S. Salvesen GS. Riek R. Yates JR., 3rd Lipton SA. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nguyen TT. Stevens MV. Kohr M. Steenbergen C. Sack MN. Murphy E. Cysteine 203 of cyclophilin D is critical for cyclophilin D activation of the mitochondrial permeability transition pore. J Biol Chem. 2011;286:40184–40192. doi: 10.1074/jbc.M111.243469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nobles KN. Xiao K. Ahn S. Shukla AK. Lam CM. Rajagopal S. Strachan RT. Huang T-Y. Bressler EA. Hara MR. Shenoy SK. Gygi SP. Lefkowitz RJ. Distinct phosphorylation sites on the {beta}2-adrenergic receptor establish a barcode that encodes differential functions of {beta}-arrestin. Sci Signal. 2011;4:ra51. doi: 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ovadia H. Haim Y. Nov O. Almog O. Kovsan J. Bashan N. Benhar M. Rudich A. Increased adipocyte S-nitrosylation targets anti-lipolytic action of insulin. J Biol Chem. 2011;286:30433–30443. doi: 10.1074/jbc.M111.235945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paige JS. Xu G. Stancevic B. Jaffrey SR. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pautz A. Art J. Hahn S. Nowag S. Voss C. Kleinert H. Regulation of the expression of inducible nitric oxide synthase. Nitric Oxide. 2010;23:75–93. doi: 10.1016/j.niox.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 90.Permyakov SE. Zernii EY. Knyazeva EL. Denesyuk AI. Nazipova AA. Kolpakova TV. Zinchenko DV. Philippov PP. Permyakov EA. Senin II. Oxidation mimicking substitution of conservative cysteine in recoverin suppresses its membrane association. Amino Acids. 2012;42:1435–1442. doi: 10.1007/s00726-011-0843-0. [DOI] [PubMed] [Google Scholar]
- 91.Poole LB. Formation and functions of protein sulfenic acids. Curr Protoc Toxicol. 2004;1 doi: 10.1002/0471140856.tx1701s18. Chapter 17: Unit17. [DOI] [PubMed] [Google Scholar]
- 92.Poole LB. Karplus PA. Claiborne A. Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 93.Prime TA. Blaikie FH. Evans C. Nadtochiy SM. James AM. Dahm CC. Vitturi DA. Patel RP. Hiley CR. Abakumova I. Requejo R. Chouchani ET. Hurd TR. Garvey JF. Taylor CT. Brookes PS. Smith RA. Murphy MP. A mitochondria-targeted S-nitrosothiol modulates respiration, nitrosates thiols, and protects against ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2009;106:10764–10769. doi: 10.1073/pnas.0903250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reinhardt D. Sigusch HH. Hensse J. Tyagi SC. Korfer R. Figulla HR. Cardiac remodelling in end stage heart failure: upregulation of matrix metalloproteinase (MMP) irrespective of the underlying disease, and evidence for a direct inhibitory effect of ACE inhibitors on MMP. Heart. 2002;88:525–530. doi: 10.1136/heart.88.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Riederer IM. Schiffrin M. Kovari E. Bouras C. Riederer BM. Ubiquitination and cysteine nitrosylation during aging and Alzheimer's disease. Brain Res Bull. 2009;80:233–241. doi: 10.1016/j.brainresbull.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 96.Santhanam L. Gucek M. Brown TR. Mansharamani M. Ryoo S. Lemmon CA. Romer L. Shoukas AA. Berkowitz DE. Cole RN. Selective fluorescent labeling of S-nitrosothiols (S-FLOS): a novel method for studying S-nitrosation. Nitric Oxide. 2008;19:295–302. doi: 10.1016/j.niox.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Santhanam L. Tuday EC. Webb AK. Dowzicky P. Kim JH. Oh YJ. Sikka G. Kuo M. Halushka MK. Macgregor AM. Dunn J. Gutbrod S. Yin D. Shoukas A. Nyhan D. Flavahan NA. Belkin AM. Berkowitz DE. Decreased S-nitrosylation of tissue transglutaminase contributes to age-related increases in vascular stiffness. Circ Res. 2010;107:117–125. doi: 10.1161/CIRCRESAHA.109.215228. [DOI] [PubMed] [Google Scholar]
- 98.Schafer FQ. Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 99.Scharfstein JS. Keaney JF. Slivka A. Welch GN. Vita JA. Stamler JS. Loscalzo J. In vivo transfer of nitric oxide between a plasma protein-bound reservoir and low molecular weight thiols. J Clin Investig. 1994;94:1432–1439. doi: 10.1172/JCI117480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schreiter ER. Rodriguez MM. Weichsel A. Montfort WR. Bonaventura J. S-nitrosylation-induced conformational change in blackfin tuna myoglobin. J Biol Chem. 2007;282:19773–19780. doi: 10.1074/jbc.M701363200. [DOI] [PubMed] [Google Scholar]
- 101.Semballa S. Geffard M. Daulouede S. Malvy D. Veyret B. Lemesre JL. Holzmuller P. Mnaimneh S. Vincendeau P. Antibodies directed against nitrosylated neoepitopes in sera of patients with human African trypanosomiasis. Trop Med Int Health. 2004;9:1104–1110. doi: 10.1111/j.1365-3156.2004.01305.x. [DOI] [PubMed] [Google Scholar]
- 102.Seth D. Hausladen A. Wang Y-J. Stamler JS. Endogenous protein S-nitrosylation in E. coli: regulation by OxyR. Science. 2012;336:470–473. doi: 10.1126/science.1215643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Singh RJ. Hogg N. Joseph J. Kalyanaraman B. Mechanism of nitric oxide release from S-nitrosothiols. J Biol Chem. 1996;271:18596–18603. doi: 10.1074/jbc.271.31.18596. [DOI] [PubMed] [Google Scholar]
- 104.Stamler JS. Jaraki O. Osborne J. Simon DI. Keaney J. Vita J. Singel D. Valeri CR. Loscalzo J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc Natl Acad Sci U S A. 1992;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stamler JS. Simon DI. Jaraki O. Osborne JA. Francis S. Mullins M. Singel D. Loscalzo J. S-nitrosylation of tissue-type plasminogen activator confers vasodilatory and antiplatelet properties on the enzyme. Proc Natl Acad Sci U S A. 1992;89:8087–8091. doi: 10.1073/pnas.89.17.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stamler JS. Simon DI. Osborne JA. Mullins ME. Jaraki O. Michel T. Singel DJ. Loscalzo J. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci U S A. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sun J. Kohr MJ. Nguyen T. Aponte AM. Connelly PS. Esfahani SG. Gucek M. Daniels MP. Steenbergen C. Murphy E. Disruption of caveolae blocks ischemic preconditioning-mediated S-nitrosylation of mitochondrial proteins. Antioxid Redox Signal. 2012;16:45–56. doi: 10.1089/ars.2010.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. This reference has been deleted.
- 109.Sun J. Morgan M. Shen RF. Steenbergen C. Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res. 2007;101:1155–1163. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 110.Sun J. Picht E. Ginsburg KS. Bers DM. Steenbergen C. Murphy E. Hypercontractile female hearts exhibit increased S-nitrosylation of the L-type Ca2+ channel α1 subunit and reduced ischemia/reperfusion injury. Circ Res. 2006;98:403–411. doi: 10.1161/01.RES.0000202707.79018.0a. [DOI] [PubMed] [Google Scholar]
- 111.Takahashi H. Shin Y. Cho SJ. Zago WM. Nakamura T. Gu Z. Ma Y. Furukawa H. Liddington R. Zhang D. Tong G. Chen HS. Lipton SA. Hypoxia enhances S-nitrosylation-mediated NMDA receptor inhibition via a thiol oxygen sensor motif. Neuron. 2007;53:53–64. doi: 10.1016/j.neuron.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Villanueva C. Giulivi C. Subcellular and cellular locations of nitric oxide synthase isoforms as determinants of health and disease. Free Radic Biol Med. 2010;49:307–316. doi: 10.1016/j.freeradbiomed.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Villanueva S. Céspedes C. González AA. Vio CP. Velarde V. Effect of ischemic acute renal damage on the expression of COX-2 and oxidative stress-related elements in rat kidney. Am J Physiol Renal Physiol. 2007;292:F1364–F1371. doi: 10.1152/ajprenal.00344.2006. [DOI] [PubMed] [Google Scholar]
- 114.Viner RI. Williams TD. Schoneich C. Peroxynitrite modification of protein thiols: oxidation, nitrosylation, and S-glutathiolation of functionally important cysteine residue(s) in the sarcoplasmic reticulum Ca-ATPase. Biochemistry. 1999;38:12408–12415. doi: 10.1021/bi9909445. [DOI] [PubMed] [Google Scholar]
- 115.Wang Y. Liu T. Wu C. Li H. A strategy for direct identification of protein S-nitrosylation sites by quadrupole time-of-flight mass spectrometry. J Am Soc Mass Spectrom. 2008;19:1353–1360. doi: 10.1016/j.jasms.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Winterbourn CC. Metodiewa D. The reaction of superoxide with reduced glutathione. Arch Biochem Biophys. 1994;314:284–290. doi: 10.1006/abbi.1994.1444. [DOI] [PubMed] [Google Scholar]
- 117.Xue Y. Liu Z. Gao X. Jin C. Wen L. Yao X. Ren J. GPS-SNO: Computational Prediction of protein S-nitrosylation sites with a modified GPS algorithm. PLoS ONE. 2010;5:e11290. doi: 10.1371/journal.pone.0011290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang Y. Loscalzo J. S-nitrosoprotein formation and localization in endothelial cells. Proc Natl Acad Sci U S A. 2005;102:117–122. doi: 10.1073/pnas.0405989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang Y. Song Y. Loscalzo J. Regulation of the protein disulfide proteome by mitochondria in mammalian cells. Proc Natl Acad Sci U S A. 2007;104:10813–10817. doi: 10.1073/pnas.0702027104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang H. Xu Y. Joseph J. Kalyanaraman B. Influence of intramolecular electron transfer mechanism in biological nitration, nitrosation, and oxidation of redox-sensitive amino acids. Methods Enzymol. 2008;440:65–94. doi: 10.1016/S0076-6879(07)00804-X. [DOI] [PubMed] [Google Scholar]