Abstract
Memory CD4+ T lymphocytes in peripheral blood that express integrins α4ß7 preferentially recirculate through gut-associated lymphoid tissue (GALT), a proposed site of significant HIV-1 replication. Tregs and activated CD4+ T cells in GALT could also be particularly susceptible to infection. We therefore hypothesized that infection of these subsets of memory CD4+ T cells may contribute disproportionately to the HIV-1 reservoir. A cross-sectional study of CD4+ T cell subsets of memory CD45RO+ cells in peripheral blood mononuclear cells (PBMCs) was conducted using leukapheresis from eight subjects with untreated chronic HIV-1 infection. Real-time polymerase chain reaction (PCR) was used to quantify total and integrated HIV-1 DNA levels from memory CD4+ T cells sorted into integrin β7+ vs. β7−, CD25+CD127low Treg vs. CD127high, and activated CD38+ vs. CD38−. More than 80% of total HIV-1 DNA was found to reside in the integrin β7-negative non-gut-homing subset of CD45RO+ memory CD4+ T cells. Less than 10% was found in highly purified Tregs or CD38+ activated memory cells. Similarly, integrated HIV-1 DNA copies were found to be more abundant in resting non-gut-homing memory CD4+ T cells (76%) than in their activated counterparts (23%). Our investigations showed that the majority of both total and integrated HIV-1 DNA was found within non-gut-homing resting CD4+ T cells.
Introduction
The human immunodeficiency virus type 1 (HIV-1) latent reservoir is a major obstacle to the eradication of HIV-1.1,2 Upon cessation of antiretroviral therapy (ART), virus rebound is rapid,3 most likely arising from latently infected long-lived cells, although their nature and location in the body are only incompletely understood.2,4 Memory CD4+ T cells have long been identified as significant contributors to the latent HIV-1 reservoir1 and their generation has been postulated to occur either through direct infection of resting CD4+ T cells5,6 or after the reversion of activated CD4+ T cells (containing replication-competent, integrated HIV-1 DNA) to a resting state.1,7 This reservoir is established early during primary HIV-1 infection (PHI)8 with therapy initiated during PHI only restricting the size of the reservoir to a limited degree.9–11 Tissues and cell types other than memory CD4+ T cells may also contain replication-competent HIV-1 provirus, including monocytes/macrophages, dendritic cells, and cells of the genitourinary tract, but their exact contribution to the viral reservoir remains to be determined.2,3
Much evidence indicates that gut-associated lymphoid tissue (GALT) plays a major role in the pathogenesis of progressive HIV-1 infection. GALT is believed to contain a large majority of the CD4+ T cells in the body,12 which are mostly CCR5+13 and in an activated state,14 making them highly susceptible to infection and depletion.15 Following this early depletion, chronic HIV-1 infection is believed to result in increased microbial translocation and increased activation, heightening susceptibility of more CD4+ T cells to infection and continuing depletion15; however, there are conflicting data regarding this theory.16 Furthermore, a recent report has suggested continuing replication of HIV-1 in GALT despite suppressive ART,17 and supporting this observation, treatment intensification has been reported to reduce viral replication in GALT tissue biopsies.18 It is therefore plausible to expect that a large number of memory CD4+ T cells containing HIV-1 DNA are present in cells trafficking through the GALT.
Resting memory CD4+ T cells have specialized migratory capacities determined by their expressed integrins.19 Those generated in GALT in the presence of metabolites of vitamin A express the integrin ß7,20 which is expressed in conjunction with α4.19,21–23 These cells recirculate through mucosal surfaces, such as the genitourinary tract and respiratory tree, and traffic through GALT from peripheral blood,20,24,25 via specific binding of integrin α4ß7 to MAdCAM-1, which is expressed on specialized endothelial cells in GALT20,26 and other mucosal surfaces involved in inflammation.26,27 Memory CD4+ T cells in peripheral blood can be subdivided into two main subsets based on integrin expression, gut-homing α4ß7+ and non-gut-homing α4ß1+ cells. The latter cells cannot access GALT since they cannot bind MAdCAM-1.20
T regulatory CD4+ cells (Tregs) reduce the effects of proinflammatory stimulus created by gut microbials.28,29 Hence microbial translocation occurring during chronic HIV-1 infection would likely increase Treg cells and possibly allow for their infection by HIV-1. Increases in Foxp3+ Tregs in mucosal tissue in chronic HIV-1 infection have been demonstrated.30,31 Tregs, originally defined as CD25high CD4+ T cells, have also been reported to be susceptible to HIV-1 infection in vitro.32 Currently there is little agreement in terms of their expansion or depletion during HIV-1 infection in vivo.33–41 With increased activation and infection of the remaining CD4+ T cells following the depletion of CD4+ T cells in the GALT, it is possible that Tregs are affected.15 However, their survival during HIV-1 disease progression and their presence in mucosal tissues42 suggest that they traffic through the GALT and hence may play a role as a viral reservoir.
Integrin ß7+ CD4+ T cells were sorted from peripheral blood mononuclear cells (PBMCs) of untreated, chronically HIV-infected subjects to determine whether they preferentially contained HIV-1 DNA. We have also used the improved cell surface definition of Tregs, CD25+CD127low, for purification of human Tregs from PBMCs by cell sorting,43 to allow us to better clarify the extent to which Tregs are infected in vivo with HIV-1 DNA. Finally, by sorting activated CD38+ memory CD4+ T cells, we have assessed whether there was preferential infection of these cells in chronic untreated HIV-1 infection.
Materials and Methods
Patients
Eight treatment-naive subjects with documented chronic HIV-1 infection (CHI) and relatively high CD4+ T cell counts in peripheral blood were included in this study (Table 1). Leukapheresis packs were collected between 1992 and 1994, and cells were cryopreserved as part of a study of adoptive immunotherapy.44 However, these samples preceded routine plasma viral load testing, and no results are available for HIV RNA copy numbers for the times of collection.
Table 1.
Subject Demographics and CD4+ T Cell Subsets as Percentage of CD3+CD4+ T Cells
|
Chronic HIV+ Median (IQR) |
|
|---|---|
| Number of subjects | 8 |
| Age | 34 (27–40) |
| Length of infection (years) | 5 (2.5–7.5) |
| CD4 cell count (cells/μl) | 622 (608–673) |
| Integrin β7+CD45RO+ % of CD3+CD4+ cells | 7.4 (6.0–8.9) |
| Integrin β7−CD45RO+ % of CD3+CD4+ cells | 40 (31–50) |
| Tregs % CD45RO+ | 8.2 (6.3–10.1) |
| CD127high % CD45RO+ | 74 (65–78) |
| CD38+CD45RO+ % of CD3+CD4+ | 3.9 (3.2–5.6) |
| CD38−CD45RO+ % of CD3+CD4+ | 42 (32–52) |
IQR, interquartile range.
In addition, gut-homing and non-gut-homing memory CD4+ T cell subsets were isolated from two PHI patients, both with HIV-1 viral load levels >750,000 copies/ml, as previously described,45 and two patients on cART for greater than 6 months, with plasma viral loads <50 copies/ml.46 Investigation of sorted CD4+ T cells was restricted to gut-homing and non-gut-homing memory CD4+ T cell subsets for these two groups of patients.
Purification of memory CD4+ T cell subsets
Following thawing of leukapheresis packs, cell viability was routinely >80%. After enrichment with Dynabeads Untouched Human CD4 T Cell kit (Invitrogen, Oslo, Norway), 1–2×108 enriched CD4+ T cells were stained using the following three panels of monoclonal antibody cocktails into 14 subsets: CD3-PerCP-Cy5.5, CD4+-PE-Cy7 (BD Biosciences, San Jose, CA), and CD45RO-ECD (Beckman Coulter, Hialeah, FL) were used in all three panels to define CD4+ T cells and to mark memory phenotype.
In panel 1, CD27-FITC was added and combined with CD45RO-ECD to gate naive cells. Integrin β7-PE (BD Biosciences) was added to collect gut-homing (β7+) and non-gut-homing (β7−) memory cells. CD25-APC (BD Biosciences) and CD127-eFluor450 (eBioscience) were combined for sorting naive and memory regulatory T cells (CD25+CD127low) and CD127+ cells.
In panel 2, CCR7-PE (purified CCR7 IgM, BD Biosciences plus anti-IgM-PE (antimouse μ chain-specific, Jackson ImmunoResearch, West Grove, PA) and CD27-FITC (BD Biosciences) were used to define central memory (CD45RO+CCR7+ or CD45RO+CD27+), effector memory (CD45RO+CCR7−CD27− or CD45RO+CD27−), transitional memory (CD45RO+CCR7−CD27+), and naive and terminally differentiated (CD45RO−CD27−) CD4 cells.
In panel 3, CD38-APC (BD Biosciences) was used to collect two memory populations (CD45RO+CD38+ and CD45RO+CD38−). Cell–antibody mixtures were incubated at room temperature for 15 min then washed with PBA (DPBS with 10% bovine serum albumin and 2% azide) and fixed with 0.5% PFA. Cell suspensions were filtered through a 70-μm cell strainer and adjusted to a density of 20×106/ml ready for sorting. CD3+CD4+CD45RO+ memory cells were sorted into subsets using a FACSAria (Becton Dickinson) with >96% purity. A minimum of 200,000 purified cells was obtained for each subset.
Total and integrated HIV-1 DNA real-time quantification
Total DNA was extracted from sorted CD4+ subsets using the Qiagen DNeasy Blood and Tissue kit (Hilden, Germany) and total HIV-1 DNA was quantified by a real-time polymerase chain reaction (PCR) assay specific for HIV-gag47 with a lower limit of detection (LLOD) of 10 copies and a CV of 11%.10 A standard dilution curve of pNL4-3 plasmid from 107 through to 101 copies was used as an internal control. Briefly, 800 nM of the sense primer SK145 5′-AGTGGGGGGACATCAAGCAGCCATGCAAAT-3′ and the antisense primer SKCC1B 5′-TACTAGTAGTTCCTGCTATGTCACTTCC-3′ was used in conjunction with 200 nM of the dual-labeled fluorogenic TaqMan locked nucleic acid probe SKLNA2-3 5′-6-FAM AT[C]A[A]T[G]AGGAA[G]CT[G]C-BHQ-1-3′. For a 25 μl reaction, 12.5 μl of iQ Supermix (Bio-Rad Laboratories, Hercules, CA) was used with 5 μl of DNA template. PCR conditions consisted of one cycle of 95°C for 3 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.
Integrated HIV-1 DNA was quantified using real-time PCR with an LLOD of eight copies and a CV of 26% as previously described.10 In brief, an ACH2 cell line was used for the standard curve. For the first round 300 nM of Alu 1 and 2 primers was used in conjunction with 100 nM of the lambda heel primer L-M667 on an Eppendorf thermocycler (Hamburg, Germany). PCR conditions were one cycle of 94°C for 7 min, 12 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 3 min and one cycle of 72°C for 7 min. In the second round 300 nM of primers Lambda T and AA55M were used with 200 nM of a dual-labeled fluorogenic probe, LTR FL, on a Bio-Rad iQ-5 real-time PCR machine. PCR conditions consisted of one cycle of 5 min at 95°C, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. Samples were run in quadruplicate for the first round PCR and performed in duplicate for the second round PCR.
Total and integrated HIV-1 DNA was compared and normalized with genomic DNA, determined by β-actin detection using the Applied Biosystems (Carlsbad, CA) TaqMan β-actin detection reagents with a FAM-labeled probe. For the β-actin assay, 12.5 μl of iQ Supermix was used in a 25 μl reaction with 180 nM of sense and antisense primers and 120 nM of probe. PCR conditions were as per each assay. The standard curve was constructed from purified human buffy coat DNA and results were expressed as copies/500 ng DNA. Where DNA yield in a subset was inadequate for quantification, these patients were omitted. Therefore the number of patients compared for each subset (n) is given on the corresponding graphs.
Statistical analysis
Percentages of subsets of memory cells within CD4+ T cells were expressed as medians and compared between the HIV-infected subject group by the Mann–Whitney nonparametric test (GraphPad Software, San Diego, CA). Absolute total and integrated HIV-1 DNA copy numbers/500 ng DNA for each subset were expressed as medians and compared. To calculate the relative contribution of each subset to the copy number in memory CD4 T cells, for each individual subject, the absolute HIV-1 DNA copy number median/500 ng DNA for the subset of interest was adjusted to take into account the relative proportion of that subset, as determined by flow cytometric phenotyping. For example, in one subject, ß7+ cells contained 1,166 copies HIV DNA/500 ng DNA. Using the conversion that 500 ng DNA is equivalent to 80,000 cells, then each ß7+ cell contained 0.0146 copies, whereas in the same patient, ß7− cells contained 535/80,000=0.0066875 copies per cell. By flow cytometry, ß7+ cells were 10.5% of CD45RO+ cells, while ß7− cells were 85% of CD45RO+ cells. Therefore for every 100 CD45RO+ CD4+ T cells, ß7+ cells contributed 10.5×0.0146=0.1533 copies, while ß7− cells contributed 85×0.0066875=0.568 copies. This corresponds to ß7+ contributing 0.1533/0.7213=21% of copies and ß7− cells contributing 0.568/0.7213=79% of copies. These calculations were performed for each individual subject, for each pair of subsets of CD45RO+ cells, namely ß7+ vs. ß7−, Treg vs. CD127high, and CD38+ vs. CD38− for each individual. Two-tailed Wilcoxon matched-pairs signed rank tests were used to compare between paired subsets for all individuals (GraphPad Software, San Diego, CA).
Results
Immunophenotyping of subsets of memory CD4+ T cells
The eight CHI subjects included in this study were treatment naïve and infected for a median of 5 years of infection since diagnosis with relatively high CD4+ T cell numbers (Table 1). No HIV-1 plasma viral load data were available for these patients at the time of collection, although their median CD4+ cell count of 622 cells/μl was significantly lower than the median for HIV-1-uninfected controls (1053 cells/μl, p<0.001), clearly indicating early progressive disease.
In initial cell sorting experiments, the rate of infection of the CD45RO− subset was substantially lower (≈10 fold) compared to the CD45RO+ subset, consistent with previous reports.45,48,49 Therefore, staining panels were rationalized to allow for a better understanding of subsets thought to contribute most substantially to HIV-1 viral reservoirs. As a result, the final panel included CD45RO+β7+ (gut-homing) vs. CD45RO+β7− (non-gut-homing) cells, CD45RO+CD25+CD127low (Tregs) vs. CD45RO+CD127high cells, and CD45RO+CD38+ (activated memory) vs. CD45RO+CD38− (resting memory) cells.
The phenotyping results showed that gut-homing ß7+ cells, Tregs, and activated cells are a minority of CD45RO+ memory CD4+ T cells in peripheral blood. Gut-homing ß7+ cells are outnumbered greater than 5-fold by non-gut-homing ß7− memory CD4+ T cells, Tregs outnumbered over 9-fold by CD127high memory CD4+ T cells, and CD38+ activated cells outnumbered over 11-fold by resting CD38− memory CD4+ T cells, respectively (Table 1).
Gut-homing versus non-gut-homing CD4+ T cells
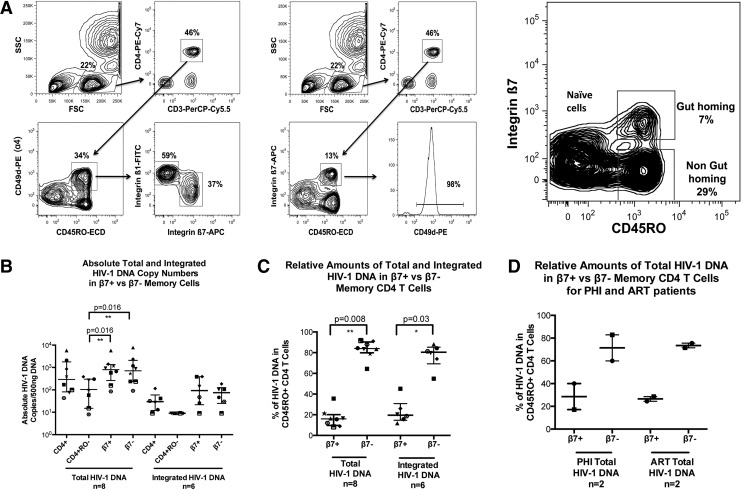
CD4+ memory T cells that express integrin α4 reciprocally express either β1 or β7 integrin (Fig. 1A). Therefore we identified gut-homing memory CD4+ T cells as CD45RO+ integrin ß7+, and non-gut-homing memory CD4+ T cells as CD45RO+ integrin ß7−(Fig. 1A).
FIG. 1.
Gut-homing memory CD4+ T cell population in antiretroviral treatment (ART)-suppressed primary HIV-1 infection (PHI) and chronic HIV-1 infection (CHI) patients. Memory gut-homing CD4+ T cells were identified by expression of CD45RO and integrin β7. (A) The first panel shows the clearly defined β1+ and β7+ populations from the CD3+CD4+CD45RO+α4+(CD49d) cells within peripheral blood mononuclear cells (PBMCs). The second panel depicts sorting based on CD3+CD4+CD45RO+β7+ cells. These cells are clearly associated with α4, with 98% of the β7+ cells shown to be positive for α4. The presence of β7+ cells within CD45RO+CD4+ T cells is shown for one representative subject studied in the third panel. The proportions of memory gut-homing and memory non-gut-homing CD3+CD4+ T cell subsets are shown with the majority of cells demonstrated to be non-gut-homing β7−CD4+ T cells. (B) Scatter plots of medians and interquartile range (IQR) of absolute total and integrated HIV-1 DNA copies/500 ng DNA are shown for CD3+CD4+, CD3+CD4+CD45RO−, CD3+CD4+CD45RO+β7+, and CD3+CD4+CD45RO+β7− T cell subsets from CHI patients. Each individual patient is represented by a unique symbol. Both the gut-homing and non-gut-homing memory CD4+ T cell subsets contained a significantly greater number of total HIV-1 DNA copies/500 ng DNA than the naive CD4+ T cell subsets (p=0.016 for both comparisons). (C) Scatter plots of the median percentage and IQR of total and integrated HIV-1 DNA in either integrin β7+CD45RO+CD4+ T cells (16% and 19.5%, respectively) or β7−CD45RO+CD4+ T cells (84% and 80.5%, respectively) are shown for CHI patients with each individual patient represented by a unique symbol. (D) Corresponding scatter plots of medians and IQR of absolute total HIV-1 DNA copies/500 ng DNA are shown for ART-suppressed and PHI patients.
Total and integrated HIV-1 DNA were quantified for purified CD4+ T cells, naive CD4+ T cells (CD3+CD4+CD45RO−), gut-homing memory CD4+ T cells (CD3+CD4+CD45RO+β7+), and non-gut-homing memory CD4+ T cells (CD3+CD4+CD45RO+β7−) for CHI patients (Fig. 1B). Absolute copy number medians showed that there was no preferential infection of the gut-homing CD4+ T cells compared to their non-gut-homing counterparts. In contrast, the naive CD4+ T cell population median was lower in both DNA species than either the CD4+ T cell or memory population medians, and was significantly lower upon comparison of total HIV-1 DNA contained within either β7 subset (p=0.016).
However, when cell proportions were taken into account, the gut-homing memory CD4+ T cell subset contributed only 16% of all total HIV-1 DNA copies and 19.5% of all integrated HIV-1 DNA copies in memory CD4+ T cells (Fig. 1C), significantly different from non-gut-homing memory CD4+ T cells (p=0.008 and p=0.03 for total and integrated HIV-1 DNA, respectively). A similar distribution of total HIV-1 DNA was found in both acute untreated PHI patients and the CHI patients on suppressive ART (Fig. 1D).
Tregs versus long-term memory CD4+ T cells
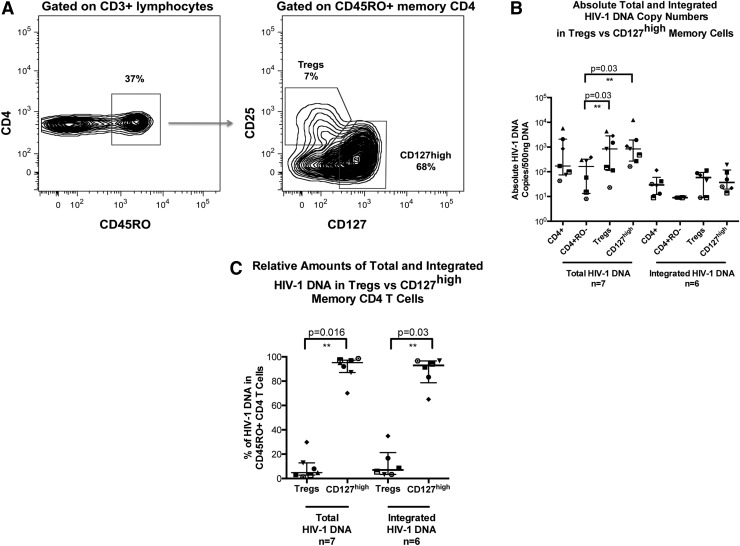
Memory CD4+ T cells were stained and sorted for memory Treg phenotype (CD45RO+CD25+CD127dim) and compared to memory CD127high CD4+ T cells (CD45RO+CD127high), as shown in Fig. 2A. The memory CD127high CD4+ T cells were shown to be 10 times more abundant than the memory Treg CD4+ T cells.
FIG. 2.
Memory T regulatory and memory CD127high CD4+ T cell subsets in CHI. Memory Treg CD3+CD4+ cells were identified within the CD45RO+ population by high expression of CD25 and dim expression of CD127. (A) Representative histograms for one CHI subject are shown. The percentages for each population are shown. (B) Scatter plots of medians and IQR of total and integrated HIV-1 DNA copies/500 ng DNA are shown for CD3+CD4+, CD3+CD4+CD45RO−, CD3+CD4+CD45RO+CD25+CD127dim, and CD3+CD4+CD45RO+CD127high T cell subsets from CHI patients. Each individual patient is represented by a unique symbol. Total HIV-1 DNA copies/500 ng of DNA significantly resided within the Treg and CD127high memory CD4+ T cell subsets compared to the naive CD4+ T cell subset (p=0.03 for both comparisons). (C) Scatter plots of median percentage and IQR of total and integrated HIV-1 DNA in either Treg CD25+CD127dim (5% and 7%, respectively) or CD127high (95% and 93%, respectively) subsets of memory CD4+ T cells are shown for CHI patients. Each individual patient is represented by a unique symbol.
Absolute total and integrated HIV-1 DNA quantification showed a similar distribution of HIV-1 DNA copies/500 ng between the two populations, with no evidence of preferential infection of Tregs (Fig. 2B). However, both populations significantly contained greater levels of total HIV-1 DNA compared to naive CD4+ T cells (p=0.03). Memory Treg CD4+ T cells were shown to contribute a median of 5% of total HIV-1 copies, compared to 95% contributed by CD127high memory CD4+ T cells (p=0.016; Fig. 2C). Similarly, only 7% of integrated HIV-1 DNA copies were contained within memory Tregs, compared to 93% of copies found in the CD127high memory CD4+ T cell population (p=0.03, Fig. 2C).
Activated CD38+ memory CD4+ T cells versus resting CD38− memory CD4+ T cells
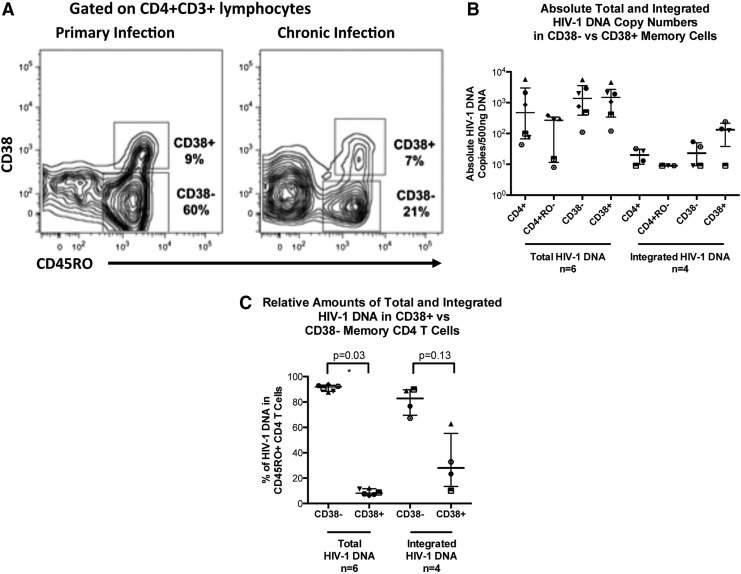
Memory CD4+ T cells were stained and sorted based on the activation marker CD38+, as shown for PHI and CHI patients, respectively, in Fig. 3A. During PHI the level of activation is increased (as we previously reported50) but eventually plateaus at around 10% in untreated CHI, similar to donor 2 for this study (Fig. 3A).
FIG. 3.
Activated memory cells identified by their expression of CD38. (A) Representative histograms for a PHI subject and a CHI subject gated on activated memory CD4+ T cells are shown. Percentages for each population are also shown. (B) Scatter plots of medians and IQR of total and integrated HIV-1 DNA copies/500 ng DNA are shown for CD3+CD4+CD45RO−, CD3+CD4+CD45RO+CD38−, and CD3+CD4+CD45RO+CD38+ T cell subsets from CHI patients. Each individual patient is represented by a unique symbol. (C) Scatter plots of median percentage and IQR of total and integrated HIV-1 DNA in CD38+ (92% and 72%, respectively) or CD38− (8% and 28%, respectively) subsets of memory CD45RO+CD4+ T cells are shown for CHI patients. Each individual patient is represented by a unique symbol.
Quantification of total HIV-1 DNA in activated CD38+ memory CD4+ T cells showed that there was no preferential infection of this subset (Fig. 3B). Taking into account the proportional differences, we found that the majority of total HIV-1 DNA was significantly contained within the resting CD38− population (p=0.03; Fig. 3C). A median of 92% of all CD4+ T cell total HIV-1 DNA was within this population, with a median of only 8% within the activated CD38+ population.
Integrated HIV-1 DNA copies in the activated CD38+ memory CD4+ T cell population were higher than in the resting CD38− memory CD4+ T cells in the four samples quantified. The median absolute copy numbers were 3.5-fold higher in the activated CD38+ memory CD4+ T cells than in the resting CD38− memory CD4+ T cells (Fig. 3B). Even so, upon consideration of cell numbers, the CD38− population was still shown to comprise the majority of integrated HIV-1 DNA, containing a median of 76% (Fig. 3C); however, due to the low sample number these differences in integrated HIV-1 DNA were not shown to be significant (p=0.13).
Discussion
To rationally design therapies that target the HIV-1 reservoir, it is essential to improve our understanding of which cell subsets contain proviral HIV-1 DNA and to further delineate the mechanisms of how this reservoir is generated and maintained. Recently, progress has been made in studies characterizing the infection of central versus effector memory CD4+ T cells in peripheral blood.49 However, to date, the contribution of circulating gut-homing memory CD4+ T cells in peripheral blood to the HIV-1 reservoir has been studied only in vitro.25 Similarly, study of infection of resting cells has generally used the lack of expression of HLA-DR, CD25, and CD69 as markers of activation.51 Infection of activated CD38+ CD4+ T cells in PBMCs in vitro and lymphoid tissue in vivo has been investigated52; however, circulating CD38+ activated memory CD4+ T cells, which are pathognomonic of progressive chronic HIV-1 infection,53 have not been studied in detail.
Research has focused on HIV-1 replication and CD4+ T cell depletion in GALT as one of the major drivers of HIV-1 pathogenesis and subsequent immunodeficiency.15,54 Experimental evidence strongly suggests that CD4+ T cells from GALT and other mucosal surfaces mostly recirculate through efferent lymph to the thoracic duct and thence back into the circulation, before reentering GALT via cell surface expression of integrins α4ß7.20,55 We therefore tested the hypothesis that the HIV-1 reservoir is, to a significant degree, established and maintained in memory CD4+ T cells associated with GALT, particularly in those cells that express CD38 during untreated chronic HIV-1 infection. We expected these memory CD4+ T cells in peripheral blood to contain higher levels of HIV-1 DNA compared to non-gut-homing memory CD4+ T cells. In particular, we expected that the phenotyping of recently activated CD38+ cells as well as Treg cells may further narrow the definition of the cells that contain a large part of the reservoir within the circulating CD4+ T cell pool.
Surprisingly, our results suggest that the HIV-1 reservoir within circulating memory CD4+ T cells is not preferentially generated by recent activation of these cells within GALT, since only a small minority of HIV-1 DNA was contributed by memory CD4+ T cells that were either gut-homing or that were expressing CD38 and therefore recently activated. Our finding that CD38+ and CD38− memory CD4+ T cells had similar per cell levels of HIV-1 DNA is consistent with our previous findings in PHI,45 and reports in SIV infection,56,57 but differs from two other studies of PBMCs and one study of lymph node cells where activated cells were reported to contain more HIV-1 DNA.51,52,58 In those latter studies, activated CD4+ T cells were compared to nonactivated CD4+ T cells, which presumably contained substantial proportions of naive CD4+ T cells, which contain approximately 10-fold less HIV-1 DNA than memory CD4+ T cells.45,59 One study defined HLA-DR−CD25−CD69− as resting cells,51 but we have found that this subset contains CD38+ cells, which were also Ki-67+, consistent with very recent activation and proliferation (data not shown), and consistent with the fact that CD38 elevation on its own represents an independent marker for progressive disease.53
One possible alternative explanation for our findings is that infection of α4ß7+ cells results in loss of cell surface expression of these integrins, leading to an apparent α4ß7− phenotype. However, we were able to confirm that the reciprocal α4ß1+ subset was clearly infected (data not shown), and since these cells were unlikely to have previously been α4ß7+ a possible loss of α4ß7 expression appears less likely. It would also be conceivable that a significant fraction of HIV-1 DNA containing CD4+ T cells is selectively retained in GALT. However, compelling data from other studies indicate that the initial rise in CD4+ T cell counts after initiation of ART is, to a significant degree, due to the release of memory CD4+ T cells retained in sites such as peripheral lymph nodes,60 and it seems reasonable to expect that the same mechanism applies to all secondary lymphoid tissues.
In a recent analysis of patients who have initiated therapy with raltegravir-containing ART during either primary or chronic HIV-1 infection we did not observe significant changes in the proportions of circulating α4ß7+ memory CD4+ T cells after commencing therapy, indicating no preferential retention of these cells subset.10 It has also been reported that HIV-1 uses a tripeptide motif in the V2 region of Env to engage α4ß7 on the surface of CCR5+ CD4+ T cells, increasing entry into target cells in vitro,61 particularly during PHI. However, these data have been recently contested with tropism shown to be restricted to a limited number of quasispecies.62 Our findings suggest that this mechanism does not significantly contribute to the relative amounts of HIV-1 DNA found in peripheral blood subsets.
Our direct comparison of gut-homing to non-gut-homing memory CD4+ T cells showed no significant difference between these two subsets on a per-cell basis similar to one other study in vitro.25 Other studies, however, have indicated relatively high levels of HIV-1 DNA in CD4+ T cells from GALT biopsies compared to PBMCs,14,17,63 presumably due to the presence of activated CCR5+ CD4+ T cells as target cells in GALT biopsies.14 However, earlier studies of CD4+ T cells isolated from peripheral lymph nodes also showed high levels of activated CD4+ T cells64 as well as high levels of HIV-1 DNA,65 relative to PBMCs, similar in magnitude to the GALT versus PBMC comparisons.
Our study confirms and extends our previous finding that most of the HIV-1 DNA in PBMCs is found in CD4+ T cells that are CD127high,45 which is consistent with the recent suggestion that interleukin-7 (IL-7) signaling is a major mechanism of maintenance of the reservoir.49 In contrast, in Tregs, which are CD127low, we confirmed infection with HIV-1, but not preferential infection, since this subset did not contain increased amounts of HIV-1 DNA compared to CD127high cells; however, both subsets contained approximately a log greater number of HIV-1 DNA copies/500 ng DNA when compared to CD45RO− cells. We were able to detect HIV-1 DNA in highly purified CD45RO+ memory Tregs, but not in the CD45RA+ naive Tregs subset36 (data not shown). This is consistent with low, but detectable expression of CCR5 on a small subset of memory Tregs (data not shown). It is still debatable whether Tregs are relatively expanded or depleted in CHI,66 but our current data would suggest that there is not preferential depletion due to infection.
Taken together the results from our study suggest that the exact contribution of the GALT to the HIV-1 reservoir will require further exploration. It is likely that the ability of these subsets to produce infectious HIV-1 upon reactivation will likely differ, which is beyond the scope of this study. However, it was recently shown that despite the role of GALT as an HIV-1 viral reservoir, it was not the major source of rebounding virus67; nevertheless, further investigations are required, especially if reactivation of the viral reservoir is to be incorporated into therapeutic interventions. We did not find preferential infection of, or higher levels of, HIV-1 DNA in peripheral gut-homing memory CD4+ T cells. Recently, Ganusov and De Boer have suggested that GALT may contain only a minority of all the memory CD4+ T cells in the body at any one time.68 Therefore, if the infection rate in memory CD4+ T cells in GALT is comparable to the infection rate in other lymphoid tissues, as supported by our results, it would be plausible that only a small fraction of the HIV-1 reservoir exists in GALT at any one time. To further clarify this point, direct quantification of HIV-1 DNA in these CD4+ T cell subsets from gut mucosa would be highly desirable; however, in our experience, the accurate quantification of total HIV-1 DNA in gut mucosa is problematic, as the minimum cell numbers required for quantification from sorted cells are difficult to attain from standard pinch biopsies (unpublished data). Other studies nevertheless have shown a decreased number of CD4+ T cells compared to peripheral blood but a higher proviral burden.17,69,70 Others again have also found correlations between HIV DNA burden in mucosal tissues and PBMCs, in those on HAART,71 although this is also a point of divergence.63
There are several limitations associated with this study, which should be noted. First the lack of HIV-1 plasma viral load (pVL) did not allow for correlations between the level of viral DNA within a subset and HIV-1 pVL to be made. While memory stem cells are now thought to be an important contributor to the HIV-1 viral reservoir,72 these cells were described only during the conduct of the study outlined above and hence for reasons of consistency, the panel remained unchanged and does not include these cells. A further restraint was the fact these patients were untreated and were not subsequently followed over the course of infection; we were therefore unable to confirm if the differences seen between the HIV-1 DNA levels within subsets were maintained during therapy. The HIV-1 DNA real-time PCRs used within this study are capable only of quantifying HIV-1 DNA and cannot distinguish between HIV-1 DNA that is replication competent or incompetent; however, it is also clear that outgrowth assays may not detect all replication-competent virus.73 This study also did not explore the quantification of HIV-1 DNA within memory CD4+ T cells in patient tissue, due to the lack of access of such material determined by the period of sampling.
Other sites of infection may indeed be secondary lymphoid tissues, which contain large amounts of virus attached to follicular dendritic cells, as well as many productively infected cells, as shown by PCR and in situ hybridization.74 In particular, the spleen can contain more than 107 infected cells at any one time,75 with estimates of HIV-1 DNA detected in up to 10% of CD4+ T cells in lymphoid tissue.74 Together with increased expression of proinflammatory cytokines in HIV-infected lymph nodes76,77 these tissues may contribute to the maintenance of the pool of latently infected resting CD4+ memory T cells, which recirculate into the periphery. To this end, exposure to IL-778 may promote infection of resting CCR5+CD127+ CD4+ memory T cells, or extravasating lymphocytes may be infected following stimulation with chemokines as they enter lymph nodes.6 In considering possible interventions to target the HIV-1 reservoir, it will be important to analyze all secondary lymphoid tissues.
Acknowledgments
The authors would like to thank the Australian Immunovirology Research Network for provision of leukopheresis packs. The Kirby Institute is supported by funding from the Australian Government Department of Health and Ageing. This work has been supported by NHMRC Project Grant 510325 (J.Z., J.M., and K.S.) and NHMRC Program Grant 510488 (D.A.C., A.D.K., J.Z., N.S., and J.M.).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Blankson JN. Persaud D. Siliciano RF. The challenge of viral reservoirs in HIV-1 infection. Annu Rev Med. 2002;53:557–593. doi: 10.1146/annurev.med.53.082901.104024. [DOI] [PubMed] [Google Scholar]
- 2.Geeraert L. Kraus G. Pomerantz RJ. Hide-and-seek: The challenge of viral persistence in HIV-1 infection. Annu Rev Med. 2008;59:487–501. doi: 10.1146/annurev.med.59.062806.123001. [DOI] [PubMed] [Google Scholar]
- 3.Davey RT., Jr Bhat N. Yoder C, et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci USA. 1999;96(26):15109–15114. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joos B. Fischer M. Kuster H, et al. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci USA. 2008;105(43):16725–16730. doi: 10.1073/pnas.0804192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agosto LM. Yu JJ. Dai J, et al. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology. 2007;368:60–72. doi: 10.1016/j.virol.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron PU. Saleh S. Sallmann G, et al. Establishment of HIV-1 latency in resting CD4+ T cells depends on chemokine-induced changes in the actin cytoskeleton. Proc Natl Acad Sci USA. 2010;107(39):16934–16939. doi: 10.1073/pnas.1002894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun TW. Finzi D. Margolick J, et al. In vivo fate of HIV-1-infected T cells: Quantitative analysis of the transition to stable latency. Nat Med. 1995;1(12):1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 8.Chun TW. Engel D. Berrey MM, et al. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci USA. 1998;95(15):8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaunders JJ. Cunningham PH. Kelleher AD, et al. Potent antiretroviral therapy of primary human immunodeficiency virus type 1 (HIV-1) infection: Partial normalization of T lymphocyte subsets and limited reduction of HIV-1 DNA despite clearance of plasma viremia. J Infect Dis. 1999;180(2):320–329. doi: 10.1086/314880. [DOI] [PubMed] [Google Scholar]
- 10.Koelsch KK. Boesecke C. McBride K, et al. Impact of treatment with raltegravir during primary or chronic HIV infection on RNA decay characteristics and the HIV viral reservoir. AIDS. 2011;25(17):2069–2078. doi: 10.1097/QAD.0b013e32834b9658. [DOI] [PubMed] [Google Scholar]
- 11.Strain MC. Little SJ. Daar ES, et al. Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J Infect Dis. 2005;191:1414–1418. doi: 10.1086/428777. [DOI] [PubMed] [Google Scholar]
- 12.Haase AT. Perils at mucosal front lines for HIV and SIV and their hosts. Nat Rev Immunol. 2005;5(10):783–792. doi: 10.1038/nri1706. [DOI] [PubMed] [Google Scholar]
- 13.Brenchley JM. Schacker TW. Ruff LE, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200(6):749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehandru S. Poles MA. Tenner-Racz K, et al. Mechanisms of gastrointestinal CD4+ T-cell depletion during acute and early human immunodeficiency virus type 1 infection. J Virol. 2007;81(2):599–612. doi: 10.1128/JVI.01739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenchley JM. Price DA, et al. HIV disease: Fallout from a mucosal catastrophe? Nat Immunol. 2006;7:235–239. doi: 10.1038/ni1316. [DOI] [PubMed] [Google Scholar]
- 16.Redd AD. Gray RH. Quinn TC. Is microbial translocation a cause or consequence of HIV disease progression? J Infect Dis. 2011;203(5):744–745. doi: 10.1093/infdis/jiq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun T-W. Nickle DC. Justement JS, et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis. 2008;197:714–720. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- 18.Yukl SA. Shergill AK. McQuaid K, et al. Effect of raltegravir-containing intensification on HIV burden and T-cell activation in multiple gut sites of HIV-positive adults on suppressive antiretroviral therapy. AIDS. 2010;24(16):2451–2460. doi: 10.1097/QAD.0b013e32833ef7bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schweighoffer T. Yoshiya T. Tidswell M, et al. Selective expression of integrin α4β7 on a subset of human CD4+ memory T cells with hallmarks of gut-tropism. J Immunol. 1993;151(2):717–729. [PubMed] [Google Scholar]
- 20.von Andrian UH. Mackay CR. T-cell function and migration. Two sides of the same coin. N Engl J Med. 2000;343(14):1020–1034. doi: 10.1056/NEJM200010053431407. [DOI] [PubMed] [Google Scholar]
- 21.Chan BM. Elices MJ. Murphy E, et al. Adhesion to vascular cell adhesion molecule 1 and fibronectin. Comparison of alpha 4 beta 1 (VLA-4) and alpha 4 beta 7 on the human B cell line JY. J Biol Chem. 1992;267(12):8366–8370. [PubMed] [Google Scholar]
- 22.Holzmann B. Weissman IL. Peyer's patch-specific lymphocyte homing receptors consist of a VLA-4-like α chain associated with either of two integrin β chains, one of which is novel. EMBO J. 1989;8(6):1735–1741. doi: 10.1002/j.1460-2075.1989.tb03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruegg C. Postigo A. Sikorski E, et al. Role of integrin alpha 4 beta 7/alpha 4 beta P in lymphocyte adherence to fibronectin and VCAM-1 and in homotypic cell clustering. J Cell Biol. 1992;117(1):179–189. doi: 10.1083/jcb.117.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farstad IN. Halstensen TS. Kvale D, et al. Topographic distribution of homing receptors on B and T cells in human gut-associated lymphoid tissue: Relation of L-selectin and integrin α4β7 to naive and memory phenotypes. Am J Pathol. 1997;150(1):187–199. [PMC free article] [PubMed] [Google Scholar]
- 25.Monteiro P. Gosselin A. Wacleche VS, et al. Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin β7. J Immunol. 2011;186(8):4618–4630. doi: 10.4049/jimmunol.1004151. [DOI] [PubMed] [Google Scholar]
- 26.Erle DJ. Briskin MJ. Butcher EC, et al. Expression and function of the MAdCAM-1 receptor, integrin alpha 4 beta 7, on human leukocytes. J Immunol. 1994;153(2):517–528. [PubMed] [Google Scholar]
- 27.Berlin C. Berg EL. Briskin MJ, et al. α4β7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell. 1993;74(1):185–195. doi: 10.1016/0092-8674(93)90305-a. [DOI] [PubMed] [Google Scholar]
- 28.Benson MJ. Pino-Lagos K. Rosemblatt M, et al. All-trans retinoic acid mediates enhanced Treg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med. 2007;204(8):1765–1774. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh B. Read S. Asseman C, et al. Control of intestinal inflammation by regulatory T cells. Immunol Rev. 2001;182:190–200. doi: 10.1034/j.1600-065x.2001.1820115.x. [DOI] [PubMed] [Google Scholar]
- 30.Andersson J. Boasso A. Nilsson J, et al. The prevalence of regulatory T cells in lymphoid tissue is correlated with viral load in HIV-infected patients. J Immunol. 2005;174(6):3143–3147. doi: 10.4049/jimmunol.174.6.3143. [DOI] [PubMed] [Google Scholar]
- 31.Epple HJ. Loddenkemper C. Kunkel D, et al. Mucosal but not peripheral FOXP3+ regulatory T cells are highly increased in untreated HIV infection and normalize after suppressive HAART. Blood. 2006;108(9):3072–3078. doi: 10.1182/blood-2006-04-016923. [DOI] [PubMed] [Google Scholar]
- 32.Moreno-Fernandez ME. Zapata W. Blackard JT, et al. Human regulatory T cells are targets for human immunodeficiency virus (HIV) infection, and their susceptibility differs depending on the HIV type 1 strain. J Virol. 2009;83(24):12925–12933. doi: 10.1128/JVI.01352-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seddiki N. Sasson SC. Santner-Nanan B, et al. Proliferation of weakly suppressive regulatory CD4+ T cells is associated with over-active CD4+ T-cell responses in HIV-positive patients with mycobacterial immune restoration disease. Eur J Immunol. 2009;39(2):391–403. doi: 10.1002/eji.200838630. [DOI] [PubMed] [Google Scholar]
- 34.Chen X. Oppenheim JJ. Winkler-Pickett RT, et al. Glucocorticoid amplifies IL-2-dependent expansion of functional FoxP3+CD4+CD25+ T regulatory cells in vivo and enhances their capacity to suppress EAE. Eur J Immunol. 2006;36(8):2139–2149. doi: 10.1002/eji.200635873. [DOI] [PubMed] [Google Scholar]
- 35.Douek DC. Betts MR. Hill BJ, et al. Evidence for increased T cell turnover and decreased thymic output in HIV infection. J Immunol. 2001;167(11):6663–6668. doi: 10.4049/jimmunol.167.11.6663. [DOI] [PubMed] [Google Scholar]
- 36.Seddiki N. Santner-Nanan B. Tangye SG, et al. Persistence of naive CD45RA+ regulatory T cells in adult life. Blood. 2006;107(7):2830–2838. doi: 10.1182/blood-2005-06-2403. [DOI] [PubMed] [Google Scholar]
- 37.Aandahl EM. Michaëlsson J. Moretto WJ, et al. Human CD4+CD25+ regulatory T cells control T-cell responses to human immunodeficiency virus and cytomegalovirus antigens. J Virol. 2004;78(5):2454–2459. doi: 10.1128/JVI.78.5.2454-2459.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersson J. Boasso A. Nilsson J, et al. Cutting edge: The prevalence of regulatory T cells in lymphoid tissues is correlated with viral load in HIV-infected patients. J Immunol. 2005;174(6):3143–3147. doi: 10.4049/jimmunol.174.6.3143. [DOI] [PubMed] [Google Scholar]
- 39.Kinter AL. Hennessey M. Bell A, et al. CD25+CD4+ regulatory T cells from the peripheral blood of asymptomatic HIV-infected individuals regulate CD4+ and CD8+ HIV-specific T cell immune responses in vitro and are associated with favorable clinical markers of disease status. J Exp Med. 2004;200(3):331–343. doi: 10.1084/jem.20032069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakaguchi S. Regulatory T cells: Mediating compromises between host and parasite. Nat Immunol. 2003;4:10–11. doi: 10.1038/ni0103-10. [DOI] [PubMed] [Google Scholar]
- 41.Weiss L. Donkova-Petrini V. Caccavelli L, et al. Human immunodeficiency virus-driven expansion of CD4+CD25+ regulatory T cells, which suppress HIV-specific CD4 T-cell responses in HIV-infected patients. Blood. 2004;104(10):3249–3256. doi: 10.1182/blood-2004-01-0365. [DOI] [PubMed] [Google Scholar]
- 42.Nilsson J. Boasso A. Velilla PA, et al. HIV-1-driven regulatory T-cell accumulation in lymphoid tissues is associated with disease progression in HIV/AIDS. Blood. 2006;108(12):3808–3817. doi: 10.1182/blood-2006-05-021576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seddiki N. Santner-Nanan B. Martinson J, et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med. 2006;203(7):1693–1700. doi: 10.1084/jem.20060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trickett A. Dwyer J. Tedla N, et al. Safety and feasibility of harvesting cells for adoptive immunotherapy from patients with asymptomatic HIV infection. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;12(5):523–524. doi: 10.1097/00042560-199608150-00014. [DOI] [PubMed] [Google Scholar]
- 45.Zaunders JJ. Ip S. Munier ML, et al. Infection of CD127+ (interleukin-7 receptor+) CD4+ cells and overexpression of CTLA-4 are linked to loss of antigen-specific CD4 T cells during primary human immunodeficiency virus type 1 infection. J Virol. 2006;80(20):10162–10172. doi: 10.1128/JVI.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin A. Smith DE. Carr A, et al. Reversibility of lipoatrophy in HIV-infected patients 2 years after switching from a thymidine analogue to abacavir: The MITOX Extension Study. AIDS. 2004;18(7):1029–1036. doi: 10.1097/00002030-200404300-00011. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki K. Shijuuku T. Fukamachi T, et al. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J RNAi Gene Silencing. 2005;1(2):66–78. [PMC free article] [PubMed] [Google Scholar]
- 48.Brenchley JM. Hill BJ. Ambrozak DR, et al. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: Implications for HIV pathogenesis. J Virol. 2004;78:1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chomont N. El-Far M. Ancuta P, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaunders J. Carr A. McNally L, et al. Effects of primary HIV-1 infection on subsets of CD4+ and CD8+ T lymphocytes. AIDS. 1995;9:561–566. doi: 10.1097/00002030-199506000-00005. [DOI] [PubMed] [Google Scholar]
- 51.Chun TW. Nickle DC. Justement JS, et al. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J Clin Invest. 2005;115(11):3250–3255. doi: 10.1172/JCI26197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meditz AL. Haas MK. Folkvord JM, et al. HLA-DR+CD38+CD4+ T lymphocytes have elevated CCR5 expression and produce the majority of R5-tropic HIV-1 RNA in vivo. J Virol. 2011;85(19):10189–10200. doi: 10.1128/JVI.02529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Z. Cumberland WG. Hultin LE, et al. Elevated CD38 antigen expression on CD8+ T cells is a stronger marker for the risk of chronic HIV disease progression to AIDS and death in the Multicenter AIDS Cohort Study than CD4+ cell count, soluble immune activation markers, or combinations of HLA-DR and CD38 expression. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;16(2):83–92. doi: 10.1097/00042560-199710010-00003. [DOI] [PubMed] [Google Scholar]
- 54.Mehandru S. Dandekar S. Role of the gastrointestinal tract in establishing infection in primates and humans. Curr Opin HIV AIDS. 2008;3(1):22–27. doi: 10.1097/COH.0b013e3282f331b0. [DOI] [PubMed] [Google Scholar]
- 55.Klonowski KD. Williams KJ. Marzo AL, et al. Dynamics of blood-borne CD8 memory T cell migration in vivo. Immunity. 2004;20(5):551–562. doi: 10.1016/s1074-7613(04)00103-7. [DOI] [PubMed] [Google Scholar]
- 56.Li Q. Duan L. Estes JD, et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434(7037):1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 57.Nishimura Y. Sadjadpour R. Mattapallil JJ, et al. High frequencies of resting CD4+ T cells containing integrated viral DNA are found in rhesus macaques during acute lentivirus infections. Proc Natl Acad Sci USA. 2009;106(19):8015–8020. doi: 10.1073/pnas.0903022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maenetje P. Riou C. Casazza JP, et al. A steady state of CD4+ T cell memory maturation and activation is established during primary subtype C HIV-1 infection. J Immunol. 2010;184(9):4926–4935. doi: 10.4049/jimmunol.0903771. [DOI] [PubMed] [Google Scholar]
- 59.Brenchley JM. Karandikar NJ. Betts MR, et al. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 2003;101(7):2711–2720. doi: 10.1182/blood-2002-07-2103. [DOI] [PubMed] [Google Scholar]
- 60.Bucy RP. Hockett RD. Derdeyn CA, et al. Initial increase in blood CD4(+) lymphocytes after HIV antiretroviral therapy reflects redistribution from lymphoid tissues. J Clin Invest. 1999;103(10):1391–1398. doi: 10.1172/JCI5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arthos J. Cicala C. Martinelli E, et al. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9(3):301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 62.Parrish NF. Wilen CB. Banks LB, et al. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin α4β7. PLoS Pathog. 2012;8(5):e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yukl SA. Gianella S. Sinclair E, et al. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis. 2010;202(10):1553–1561. doi: 10.1086/656722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tedla N. Dwyer J. Truskett P, et al. Phenotypic and functional characterization of lymphocytes derived from normal and HIV-1-infected human lymph nodes. Clin Exp Immunol. 1999;117(1):92–99. doi: 10.1046/j.1365-2249.1999.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pantaleo G. Graziosi C. Butini L, et al. Lymphoid organs function as major reservoirs for human immunodeficiency virus. Proc Natl Acad Sci USA. 1991;88(21):9838–9842. doi: 10.1073/pnas.88.21.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seddiki N. Kelleher AD. Regulatory T cells in HIV infection: Who's suppressing what? Curr Infect Dis Rep. 2008;10(3):252–258. doi: 10.1007/s11908-008-0041-8. [DOI] [PubMed] [Google Scholar]
- 67.Lerner P. Guadalupe M. Donovan R, et al. The gut mucosal viral reservoir in HIV-infected patients is not the major source of rebound plasma viremia following interruption of highly active antiretroviral therapy. J Virol. 2011;85(10):4772–4782. doi: 10.1128/JVI.02409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ganusov VV. De Boer RJ. Do most lymphocytes in humans really reside in the gut? Trends Immunol. 2007;28(12):514–518. doi: 10.1016/j.it.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 69.Macal M. Sankaran S. Chun T-W, et al. Effective CD4+ T-cell restoration in gut-associated lymphoid tissue of HIV-infected patients is associated with enhanced Th17 cells and polyfunctional HIV-specific T-cell responses. Mucosal Immunol. 2008;1(6):475–488. doi: 10.1038/mi.2008.35. [DOI] [PubMed] [Google Scholar]
- 70.Schneider T. Jahn H-U. Schmidt W, et al. Loss of CD4 T lymphocytes in patients infected with human immunodeficiency virus type 1 is more pronounced in the duodenl mucosa than in the peripheral blood. Gut. 1995;37:524–529. doi: 10.1136/gut.37.4.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anton PA. Mitsuyasu RT. Deeks SG, et al. Multiple measures of HIV burden in blood and tissue are correlated with each other but not with clinical parameters in aviremic subjects. AIDS. 2003;17(1):53–63. doi: 10.1097/00002030-200301030-00008. [DOI] [PubMed] [Google Scholar]
- 72.Buzon M. Seiss K. Li C, et al. T memory stem cells: A long-term reservoir for HIV-1. IDWeek 2012 Meeting. Vol Paper #594; Oct 17–21;; San Diego, California. 2012. [Google Scholar]
- 73.Ho Y-C. Shan L. Wang J, et al. Vol Paper #. Characterization of non-induced HIV-1 proviruses dampens the hope for HIV-1 eradication. CROI—Conference on Retroviruses and Opportunistic Infections; Atlanta, Georgia. 2013. [Google Scholar]
- 74.Haase AT. Population biology of HIV-1 infection: Viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu Rev Immunol. 1999;17:625–656. doi: 10.1146/annurev.immunol.17.1.625. [DOI] [PubMed] [Google Scholar]
- 75.McIlroy D. Autran B. Cheynier R, et al. Infection frequency of dendritic cells and CD4+ T lymphocytes in spleens of human immunodeficiency virus-positive patients. J Virol. 1995;69(8):4737–4745. doi: 10.1128/jvi.69.8.4737-4745.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gavin M. Rudensky A. Control of immune homeostasis by naturally arising regulatory CD4+ T cells. Curr Opin Immunol. 2003;15:690–696. doi: 10.1016/j.coi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 77.Fehérvari Z. Sakaguchi S. CD4+ Tregs and immune control. J Clin Invest. 2004;114(9):1209–1217. doi: 10.1172/JCI23395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sasson SC. Zaunders JJ. Kelleher AD. The IL-7/IL-7 receptor axis: Understanding its central role in T-cell homeostasis and the challenges facing its utilization as a novel therapy. Curr Drug Targets. 2006;7(12):1571–1582. doi: 10.2174/138945006779025365. [DOI] [PubMed] [Google Scholar]