Abstract
We have collected demographic and/or mutation information on a worldwide sample of 394 patients with type B Niemann-Pick disease (NPD). The disorder is panethnic, with the highest incidence occurring in individuals of Turkish, Arabic, and North African descent. Only five of the 394 patients were Ashkenazi Jewish, revealing that, unlike the type A form of NPD, type B NPD does not occur frequently within this population. Mutation analysis of the acid sphingomyelinase (ASM) gene (designated “SMPD1”) was performed on 228 patients (324 unique alleles), and several novel, “common” mutations were found. Among these were the L137P, fsP189, and L549P mutations, which accounted for ∼75% of the alleles in Turkish patients, the H421Y and K576N mutations, which accounted for ∼85% of the alleles in Saudi Arabian patients, the S379P, R441X, R474W, and F480L mutations, which accounted for ∼55% of the alleles in Portuguese/Brazilian patients, and the A196P mutation, which accounted for ∼42% of the alleles in Scottish/English patients. The previously reported ΔR608 mutation occurred on ∼12% of the alleles studied. Overall, a total of 45 novel mutations were found, and several new genotype/phenotype correlations were identified. In particular, the L137P, A196P, and R474W mutations were consistent with a less severe form of type B NPD, whereas the H421Y and K576N mutations led to an early-onset, more severe form that was specific to Saudi Arabia. These data provide the first extensive demographic assessment of this disorder and describe several new mutations that can be used to predict phenotypic outcome and to gain new insights into the structure and function of ASM.
Introduction
Types A and B Niemann-Pick disease (NPD [MIM 257200]) are lysosomal storage disorders resulting from the deficient activity of acid sphingomyelinase (ASM) and the subsequent accumulation of sphingomyelin, cholesterol, and other lipids within cells and tissues of affected individuals (Schuchman and Desnick 2001). Patients with type A NPD are usually diagnosed early in infancy with organomegaly and follow a rapid neurodegenerative course that leads to death by ∼3 years of age. In contrast, patients with type B NPD have little or no CNS involvement and often survive into adulthood. However, the type B form of NPD is clinically heterogeneous and can present with a variety of findings that may include hepatosplenomegaly, growth retardation, frequent respiratory infections, fatigue, and hematologic abnormalities such as high levels of LDL cholesterol and triglycerides, low levels of HDL cholesterol, and low numbers of platelets. In addition, several patients with type B NPD have been reported who had an intermediate phenotype that included neurodegeneration (see, e.g., Elleder and Cihula 1983; Elleder et al. 1986). Both forms of NPD are panethnic, although most reported cases of type A NPD occurred among Ashkenazi Jewish individuals.
The full-length cDNA and genomic sequences encoding human ASM have been isolated (Schuchman et al. 1991, 1992), and a total of 23 mutations causing type A or B NPD have been published (gene designation “SMPD1”). Of these, 15 are missense mutations (Ferlinz et al. 1991; Levran et al. 1991a, 1992; Takahashi et al. 1992a, 1992b; Schuchman 1995; Schuchman and Miranda 1997), 2 are nonsense mutations (Schuchman 1995; Schuchman and Miranda 1997), 4 are frameshift mutations (Levran et al. 1991a, 1993b), and 1 results from an in-frame 3-bp deletion that results in the removal of a single amino acid from the ASM polypeptide (Levran et al. 1991b). One splice-site alteration also has been described (Levran et al. 1993a). Notably, three common mutations account for >90% of the mutant alleles in patients of Ashkenazi Jewish ancestry who had type A NPD (Levran et al. 1991a, 1992, 1993b). Two are missense mutations, R496L and L302P, and the third is a 1-bp deletion that causes a frameshift and the introduction of a premature stop codon within the ASM open reading frame (fsP330). The combined carrier frequency for these mutations among Ashkenazi Jewish individuals is between 1/80 and 1/100 (Li et al. 1997). Only one other mutation has so far been identified in the Ashkenazi Jewish population with type A NPD (Schuchman 1995); this mutation, designated “N389T,” was identified as one of the mutant alleles in a single patient with type A NPD; the other allele was fsP330. In contrast to the Ashkenazi Jewish population, each of the non-Jewish patients with type A NPD studied has had unique, or “private,” ASM mutations.
The first mutation (designated “ΔR608”) in a patient with type B NPD was found in a mildly affected Ashkenazi Jewish individual who carried a 3-bp deletion in exon 6 that predicted the removal of an arginine residue from position 608 of the ASM polypeptide (Levran et al. 1991b). This patient carried the R496L mutation on her other allele, suggesting that the presence of the ΔR608 mutation was “neuroprotective.” A second Ashkenazi Jewish type B patient was subsequently found to be heteroallelic for ΔR608 and a new mutation, designated “H575L.” In addition to these two patients, the ΔR608 mutation was frequently found in North African patients with type B NPD who originated from the Maghreb region (i.e., Tunisia, Algeria, and Morocco) (Vanier et al. 1993), where it may account for almost 90% of the mutant alleles. To date, only four other mutations have been reported in patients with type B NPD (Takahashi et al. 1992a, 1992b; Vanier et al. 1993).
In this article, we report demographic and/or molecular data on a worldwide sample of 394 patients with type B NPD. Acid sphingomyelinase mutation analysis was performed on 228 of these patients, and 45 novel mutations were identified, including several common mutations that could be used for genetic screening in specific populations. In addition, we have identified several new genotype/phenotype correlations for this disorder that should be useful to genetic counselors, physicians, and families affected with type B NPD.
Subjects and Methods
Subjects
A total of 394 patients with type B NPD were included in this study. Of these, 64 patients were evaluated at the Mount Sinai Clinical Research Center as part of an ongoing natural history study. Information on the remaining 330 patients was obtained from either the treating physicians or major diagnostic laboratories throughout the world. Blood was obtained from 228 of the 394 patients and was used to prepare DNA for mutation analysis (see below). For inclusion in the study, the diagnosis of NPD had to be confirmed by SMPD1 mutation analysis and/or by determining markedly reduced ASM activity (i.e., <5% of normal) in white blood cells or cultured skin fibroblasts. Cholesterol loading studies were performed on a small subset of patients to rule out type C NPD. Type C NPD was ruled out in the remaining patients by demonstrating either SMPD1 mutations or very low, residual ASM activity. Patients also needed to be ⩾3 years of age and without evidence of neurodegeneration (to rule out type A NPD).
SMPD1 Mutation Analysis
DNA was prepared from whole blood and was used for mutation analysis as described elsewhere (Schuchman and Miranda 1997). Putative mutations were confirmed by sequencing duplicate PCR products, restriction enzyme analysis, and by family studies whenever possible.
Results
Distribution and Ethnicity of Type B NPD
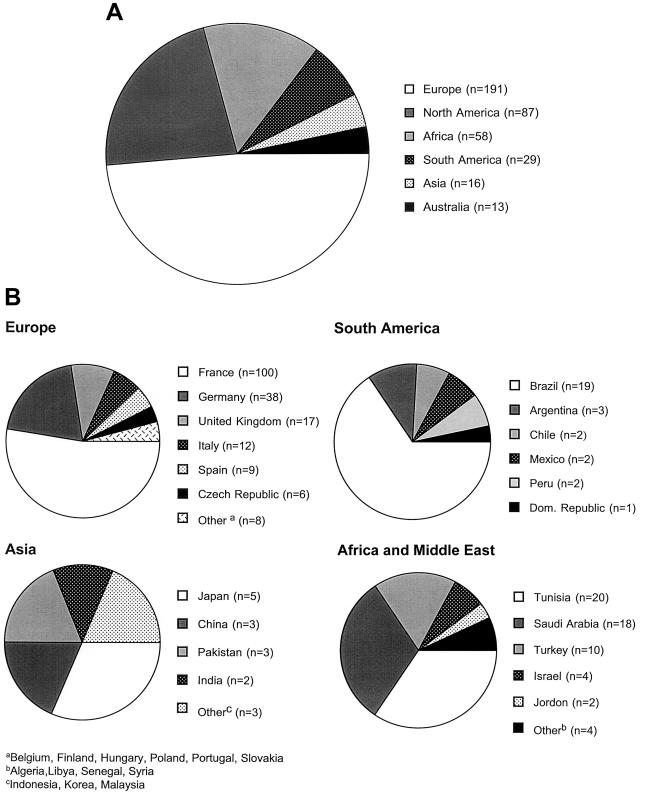
Information was collected on a worldwide sample of 394 patients with type B NPD. Mutation analysis was performed on 228 of these patients (see below). No patients were included in this study unless the diagnosis of NPD was confirmed by SMPD1 mutation analysis and/or ASM enzyme testing. The distribution of these patients by country is shown in figure 1. Of these patients, 87 were residing in the United States or Canada. Other countries with significant numbers of patients in this data set were France (100), Germany (38), Tunisia (20), Brazil (19), Saudi Arabia (18), the United Kingdom (17), Italy (12), and Turkey (10). In total, patients were identified in 29 countries. The most common ethnic group among these type B NPD patients was Turkish, followed by Arabic and Scottish. Only five of the patients were Ashkenazi Jewish.
Figure 1.
Worldwide distribution of patients with type B NPD. A, Distribution by continent of 394 patients with type B NPD. B, Distribution by country of patients with type B NPD. The distribution within North America is not shown, but 78 patients were residing in the United States, and 9 were residing in Canada.
Mutation Analysis and Genotype/Phenotype Correlations
Mutation analysis was carried out on 228 of the type B NPD patients (324 unique alleles), leading to the identification of 45 novel mutations (table 1). Among these were several “common” mutations that occurred within specific ethnic groups (table 2), including the L137P, fsP189, and L549P mutations, which accounted for ∼75% of the alleles among Turkish patients; the H421Y and K576N mutations, which accounted for ∼85% of the alleles among Saudi Arabian patients; the S379P, R441X, R474W, and F480L mutations, which accounted for ∼55% of the alleles among Portuguese/Brazilian patients; and the A196P mutation, which accounted for ∼42% of the alleles among Scottish/English patients. Overall, the previously reported ΔR608 mutation occurred on ∼12% of the NPD alleles studied (∼20% of the alleles among patients living in North America and Brazil).
Table 1.
Novel Mutations in the ASM Gene Causing Type B NPD
| Exon andMutation | cDNAAlterationa | GenomicAlterationb |
| 1: | ||
| G21X | C→T, nt 61 | nt 61 |
| D49V | A→T, nt 156 | nt 146 |
| C92W | C→G, nt 276 | nt 276 |
| 2: | ||
| L137P | T→C, nt 409 | nt 875 |
| C157R | T→C, nt 469 | nt 934 |
| fsD169 | delG, nt 507 | nt 970 |
| W174X | G→A, nt 522 | nt 986 |
| A196P | G→C, nt 586 | nt 1051 |
| R200C | C→T, nt 598 | nt 1063 |
| L225M | C→A, nt 673 | nt 1138 |
| R228C | C→T, nt 682 | nt 1147 |
| G232D | G→A, nt 695 | nt 1160 |
| G245S | G→A, nt 733 | nt 1198 |
| S248R | C→A, nt 744 | nt 1209 |
| R289H | G→A, nt 866 | nt 1331 |
| fsS321 | delG, nt 962 | nt 1427 |
| P323A | C→G, nt 967 | nt 1432 |
| P330R | C→G, nt 989 | nt 1454 |
| A357D | C→A, nt 1070 | nt 1535 |
| 3: | ||
| fsF366 | insT, nt 1096 | nt 2621 |
| R376H | G→A, nt 1128 | nt 2652 |
| R376L | G→T, nt 1128 | nt 2652 |
| S379P | T→C, nt 1135 | nt 2661 |
| fsL380 | delC, nt 1140 | nt 2663 |
| 4: | ||
| A413V | C→T, nt 1239 | nt 2762 |
| H421Y | C→T, nt 1261 | nt 3014 |
| C431R | T→C, nt 1291 | nt 3045 |
| L432P | T→C, nt 1295 | nt 3049 |
| W435C | G→C, nt 1305 | nt 3059 |
| 5: | ||
| A452V | C→T, nt 1355 | nt 3311 |
| G456D | G→A, nt 1367 | nt 3323 |
| R474W | C→T, nt 1420 | nt 3376 |
| P475L | C→T, nt 1424 | nt 3380 |
| F480L | C→A, nt 1440 | nt 3396 |
| A485V | C→T, nt 1454 | nt 3410 |
| Y488N | T→A, nt 1462 | nt 3432 |
| 6: | ||
| G494S | G→A, nt 1480 | nt 3603 |
| R496C | C→T, nt 1486 | nt 3609 |
| H514Q | T→G, nt 1542 | nt 3665 |
| E515V | A→T, nt 1544 | nt 3667 |
| fsI530 | delA, nt 1589 | nt 3702 |
| W533R | T→C, nt 1609 | nt 3709 |
| L549P | T→C, nt 1666 | nt 3758 |
| K576N | G→C, nt 1728 | nt 3840 |
| R600H | G→A, nt 1799 | nt 3911 |
| R600P | G→C, nt 1799 | nt 3911 |
Table 2.
Common Type B NPD Mutations in Specific Ethnic Groups
| Ethnic Group (No. ofNPD Alleles Studied)and Mutation | AlleleFrequency(%) |
| Turkish (20): | |
| L137P | 37 |
| L549P | 20 |
| fsP189 | 16.6 |
| Saudi Arabian (28): | |
| H421Y | 71.4 |
| K576N | 13.2 |
| Scottish/British (30): | |
| A196P | 42.3 |
| Brazilian/Portuguese (24): | |
| ΔR608 | 20.8 |
| S379P | 16.6 |
| R441X | 12.5 |
| R474W | 12.5 |
| F480L | 12.5 |
| Other (222): | |
| ΔR608 | 20 |
On the basis of these molecular studies, several new genotype/phenotype correlations were identified. In particular, the L137P, A196P, and R474W mutations were associated with a mild disease course (table 3), whereas the H421Y and K576N mutations were consistent with an early onset form of type B NPD that led to childhood death. Several other frame-shift or termination mutations were also found that predicted a severe disease course when homoallelic or present in combination with another such “null” mutation (see table 1).
Table 3.
Genotype/Phenotype Correlations among Patients with Type B NPD Carrying Common ASM Mutations
|
Finding for Phenotypea |
|||||
| Genotype | No. ofPatients | Average Age(years) | NeurodegenerativeCourse | Organomegaly | PulmonaryInvolvement |
| L137P/L137P | 4 | 14 | − | + | +/− |
| L137P/fsP189 | 2 | 20 | − | + | +/− |
| L549/L549 | 3 | 5 | − | ++ | ++ |
| fsP189/fsP189 | 1 | 3 | − | ++ | ++ |
| H421Y/H421Y | 11 | 5 | +/− | ++ | ++ |
| K576N/K576N | 2 | 6 | +/− | ++ | ++ |
| A196P/A196P | 2 | 37 | − | + | +/− |
| A196P/*b | 8 | 30 | − | + | +/− |
| ΔR474W/*c | 6 | 18 | − | + | +/− |
| ΔR608/ΔR608 | 9 | 17 | − | + | +/− |
| ΔR608/*d | 21 | 19 | − | + | +/− |
+ = present; − = absent; ++ = severe; +/− = mild.
Heterozygous for one of the following mutations: D49V, C157R, G245S, R289H, R376H, and C431R.
Heterozygous for one of the following mutations: fsL260, S379P, A485V, or R496L.
Heterozygous for one of the following mutations: Q21X, D49V, L225M, G245S, S379P, R441X, Y446C, P475L R494S, R496L, H575L, or R600H.
ASM Structure/Function Insights
Several of the novel SMPD1 point mutations revealed new structure/function insights (table 4). Among these, the G21X mutation was particularly interesting, since it occurred before the second in-frame translation start site in the ASM coding region. There are two in-frame start codons in the ASM sequence (Schuchman et al. 1991), and this mutation reveals that the first ATG must be used in vivo. In addition, the H421Y mutation was interesting, because it occurs within the putative active-site region of ASM and at a histidine residue that might be involved in zinc binding (He et al. 1999). There are also three mutations occurring at cystine residues that might be involved in intra- or intermolecular disulfide bond formation (C92W, C157R, and C431R). Two of these cystine residues are within the saposin domain of ASM and are conserved among all four of the saposin polypeptides. Finally, two independent mutations occurred at R376 (R376H and R376L), a residue that is within a putative sphingomyelin binding domain and is conserved between ASM and the sphingomyelin binding protein, lysenin (Yamaji et al. 1998).
Table 4.
Structure/Function Analysis of Several ASM Mutations
| Residue | Structure/Function Implications |
| G21X | Reveals that first in-frame ATG is functional |
| C92W | May affect disulfide bond formation; within saposin domain (conserved aa with saposins A–D) |
| C157R | May affect disulfide bond formation; within saposin domain (conserved aa with saposins A–D) |
| R376H and R376L | Within putative sphingomyelin binding domaina |
| H421Y | Within zinc binding domain; within putative active site regionb |
| C431R | May affect disulfide bond formation |
Putative sphingomyelin binding domain based on homology to the sphingomyelin binding protein, lysenin.
Putative active site region based on homologies to phospholipases C and A2.
Discussion
Several conclusions can be drawn from the data presented in this manuscript. First, type B NPD is clearly a panethnic disorder with a worldwide distribution. Most of the patients were living in the United States or Western Europe, where the enzymatic diagnosis can be readily obtained, or in countries with high rates of consanguinity, such as Saudi Arabia, Turkey, and Tunisia. However, patients with type B NPD were identified in 29 different countries, confirming that it is a disorder affecting many distinct populations. Of the 394 patients analyzed in this study, family histories were available for 228. Only 5 of these patients were Ashkenazi Jewish, revealing that unlike the type A form of this disorder, type B NPD does not have an Ashkenazi Jewish predilection. Interestingly, however, each of these five patients carried one copy of a common Ashkenazi Jewish type A mutation (L302P, R496L, or fsP330) in combination with ΔR608. This is the same genotype combination found previously in Ashkenazi Jewish type B patients (Levran et al. 1991b), and it is consistent with past “mixing” of Jewish and North African populations (since ΔR608 represents almost 90% of the type B NPD alleles in the Maghreb region; see Vanier et al. 1993). In contrast to the Ashkenazi Jewish population, in which the disease is quite rare, more than half (∼54%) of the 228 patients studied who had type B NPD were of Turkish, Arabic, or North African ancestry.
Another observation made apparent from this study is that the presentation of type B NPD is remarkably heterogeneous, making its clinical diagnosis particularly difficult. Most of the referring physicians were working within specialized metabolic disease centers, although a significant number were private practice endocrinologists or cardiologists who were managing the patients for growth retardation or abnormal plasma lipids, respectively. The diagnosis was often not made for several years after presentation, and in at least seven documented cases type B NPD patients were misdiagnosed with type 1 Gaucher disease. Two others were misdiagnosed with type C NPD. Thus, it is not possible to accurately estimate the incidence of type B NPD because many patients remain undiagnosed either because of the lack of enzyme testing, variability in presenting symptoms, and/or lack of knowledge about type B NPD among the treating physicians.
Mutation analysis was carried out on 228 of the type B NPD patients. Not surprisingly, within the Turkish and Saudi Arabian populations there were a small number of common ASM mutations that accounted for most of the patients. Three mutations were responsible for >70% of the Turkish NPD alleles, and one mutation accounted for ∼70% of the alleles in Saudi Arabia. We also identified a number of type B NPD patients with Scottish heritage. These patients resided in the United States, the United Kingdom, or Canada, and all carried the same point mutation (A196P). Patients carrying at least one copy of this mutation, even when in combination with a termination or other presumably “null” mutation, had an adult form of type B NPD that followed a fairly mild course. Thus, like ΔR608, the A196P mutation appears to predict a less severe form of type B NPD. Similar genotype/phenotype analysis suggested that three other mutations (L137P, R474W, and R600H) were also consistent with less severe forms of type B NPD, but more patients carrying these mutations will have to be identified and studied for these observations to be confirmed.
In contrast to these mutations, the H421Y mutation, which occurred on >70% of the type B NPD alleles in Saudi Arabia, was predictive of an early onset, severe form of type B NPD. This is a particularly interesting mutation, since H421 is conserved in several zinc metalloenzymes, including a cGMP specific phosphodiesterase (Francis et al. 1994), and is also conserved among the human, mouse, and Caenorhabditis elegans ASM sequences (Lin et al. 1998). Intriguingly, the C. elegans sequence in which H421 is conserved is zinc activated, whereas the other C. elegans ASM sequence is not (Lin et al. 1998). These observations suggest that H421 is a critical residue in the ASM active site and that the severe H421Y mutation may affect ASM function by disrupting zinc binding. Studies to confirm this observation are currently underway.
It is also notable that three ASM mutations were identified at cystine residues, since previous studies using sulfhydryl reducing reagents had shown that intra- or intermolecular disulfide bonds were important for ASM structure and function (He et al. 1999). Indeed, each of these cystine residues were conserved in both of the C. elegans ASM sequences, and two (C92 and C157) occurred within the previously identified saposin homology domain (Linke et al. 2001). It is also of interest that the C157R mutation had been previously found in an ASM cDNA clone obtained from a commercial, placental cDNA library (Ida et al. 1993). Expression studies using that cDNA sequence had revealed that this mutation led to an inactive ASM polypeptide.
Thus, with these data, we report the first extensive demographic analysis of type B NPD. Forty-five new ASM mutations were identified, including several common mutations occurring within specific ethnic groups. This information should be useful to genetics counselors, physicians, and/or families who are considering prenatal testing for NPD and/or genetic screening to identify carrier individuals. In addition, several new genotype/phenotype correlations were identified. Most important among these is the fact that patients carrying at least one copy of the A196P mutation will develop a less severe form of type B NPD than patients homoallelic for the H421Y mutation, who will develop an early-onset, severe form of the disease. The identification of these new mutations has also provided new structure/function insights about ASM.
These results also have important implications for the design of upcoming enzyme-replacement therapy trials for type B NPD, since several of the mutations identified (e.g., fsP189 and R441X) are frame-shift or termination mutations that will likely lead to the production of rapidly degraded proteins. Thus, individuals carrying these mutations may experience more immunologic reactions to the infused enzyme than those carrying point mutations expressing residual activity (e.g., ΔR608, A196P) (Brooks et al. 1997).
Finally, the data also highlight the fact that type B NPD remains a disorder that is particularly difficult for clinicians to diagnose and suggests that screening programs might be useful in identifying affected patients who might benefit from future therapies. In this regard, it might be useful to screen in heart disease clinics for patients with very low HDL cholesterol levels, since this is a common finding in almost all patients with type B NPD, or in endocrinology clinics, where patients may be seen for growth retardation. The availability of several common mutations should facilitate these screening efforts.
Acknowledgments
The authors acknowledge the many patients and families throughout the world who have contributed valuable time and materials for these studies. In addition, we acknowledge the physicians and scientists who have referred these patients and provided demographic information. In particular, we acknowledge Drs. Aida Al-Aqeel (Saudi Arabia), Guy Besley (United Kingdom), Bruno Bembi (Italy), Eugen Mengel (Germany), Pinar Ozand (Saudi Arabia), Roberto Giugliani (Brazil), Figan Gurakan (Turkey), David Wenger (U.S.A.), W. Ed Wraith (United Kingdom), and Marie Vanier (France). We also thank Ms. Charlene Polin for coordinating the data collection. These studies were supported by a research grant (1 RO1 HD28607) from the National Institutes of Health, a grant (1 P30 HD28822) for the Mount Sinai Child Health Research Center from the National Institutes of Health, and a grant (5 MO1 RR00071) for the Mount Sinai General Clinical Research Center from the National Center for Research Resources at the National Institutes of Health. M.W. is the recipient of a Mentored Patient-Oriented Research Career Development Award (K23 RR16052-01) from the National Institutes of Health.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih/gov/Omim/ (for NPD [MIM 257200]) [PubMed]
References
- Brooks DA, King BM, Crawley AC, Byers S, Hopwood JJ (1997) Enzyme replacement therapy in mucopolysaccharidosis VI: evidence for immune responses and altered efficacy of treatment in animal models. Biochim Biophys Acta 1361:203–216 [DOI] [PubMed] [Google Scholar]
- Elleder M, Cihula J (1983) Niemann-Pick disease (variation in the sphingomyelinase deficient group): neurovisceral phenotype (A) with an abnormally protracted clinical course and variable expression of neurological symptomatology in three siblings. Eur J Pediatr 140:323–328 [DOI] [PubMed] [Google Scholar]
- Elleder M, Nevoral J, Spicakova V, Hyniova H, Kraus J, Krasny J, Vanier MT (1986) A new variant of sphingomyelinase deficiency (Niemann-Pick): visceromegaly, minimal neurological lesions and low in vivo degradation rate of sphingomyelin. J Inherit Metab Dis 9:357–366 [DOI] [PubMed] [Google Scholar]
- Ferlinz K, Hurwitz R, Sandhoff K (1991) Molecular basis of acid sphingomyelinase deficiency in a patient with Niemann-Pick disease type A. Biochem Biophys Res Comm 179:1187–1191 [DOI] [PubMed] [Google Scholar]
- Francis SH, Colbran JL, McAllister-Lucas LM, Corbin JD (1994) Zinc interactions and conserved motifs of the cGMP-binding CGMP-specific phosphodiesterase suggest that it is a zinc hydrolase. J Biol Chem 269:22477–22480 [PubMed] [Google Scholar]
- He X, Miranda SRP, Xiong X, Dagan A, Gatt S, Schuchman EH (1999) Characterization of human acid sphingomyelinase purified from the media of overexpressing Chinese hamster ovary cells. Biochim Biophys Acta 1432:251–264 [DOI] [PubMed] [Google Scholar]
- Ida H, Rennert OM, Eto Y, Chan WY (1993) Cloning of a human acid sphingomyelinase cDNA with a new mutation that renders the enzyme inactive. J Biochem 114:15–20 [DOI] [PubMed] [Google Scholar]
- Levran O, Desnick RJ, Schuchman EH (1991a) Niemann-Pick disease: a frequent missense mutation in the acid sphingomyelinase gene of Ashkenazi Jewish type A and B patients. Proc Natl Acad Sci USA 88:3748–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1991b) Niemann-Pick type B disease: identification of a single codon deletion in the acid sphingomyelinase gene and genotype/phenotype correlations in type A and B patients. J Clin Invest 88:806–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1992) A common missense mutation (L302P) in Ashkenazi Jewish type A Niemann-Pick disease patients. Transient expression studies demonstrate the causative nature of the two common Ashkenazi Jewish Niemann-Pick disease mutations. Blood 80:2081–2087 [PubMed] [Google Scholar]
- ——— (1993a) Identification of a 3′ acceptor splice site mutation (g2610c) in the acid sphingomyelinase gene of patients with Niemann-Pick disease. Hum Mol Genet 2:205–206 [DOI] [PubMed] [Google Scholar]
- ——— (1993b) Type A Niemann-Pick disease: a frameshift mutation in the acid sphingomyelinase gene (fsP330) occurs in Ashkenazi Jewish patients. Hum Mut 2:317–319 [DOI] [PubMed] [Google Scholar]
- Li L, Caggana M, Robinowitz J, Shabeer J, Desnick RJ, Eng CM (1997) Prenatal screening in the Ashkenazi Jewish population: a pilot program of multiple option testing for five disorders. Am J Hum Genet Suppl 61:A24 [Google Scholar]
- Lin X, Hengartner R, Kolesnick R (1998) Caenorhabditis elegans contains two distinct acid sphingomyelinases. J Biol Chem 273:14374–14379 [DOI] [PubMed] [Google Scholar]
- Linke T, Wilkening G, Lansmann S, Moczall H, Bartelsen O, Weisgerber J, Sandhoff K (2001) Stimulation of acid sphingomyelinase activity by lysosomal lipids and sphingolipid activator proteins. J Biol Chem 382:283–290 [DOI] [PubMed] [Google Scholar]
- Schuchman EH, Desnick RJ (2001) Niemann-Pick disease types A and B: acid sphingomyelinase deficiencies. In: Schriver CR, Beaudet AL, Valle D, Sly WS (eds) The metabolic and molecular bases of inherited disease. McGraw Hill, New York [Google Scholar]
- Schuchman EH, Levran O, Pereira LV, Desnick RJ (1992) Structural organization and complete nucleotide sequence of the gene encoding human acid sphingomyelinase (SMPD1). Genomics 12:197–205 [DOI] [PubMed] [Google Scholar]
- Schuchman EH, Miranda SRP (1997) Niemann-Pick disease: Mutation update, genotype/phenotype correlations, and prospects for genetic testing. Genet Test 1:13–19 [DOI] [PubMed] [Google Scholar]
- Schuchman EH, Suchi M, Takahashi T, Sandhoff K, Desnick RJ (1991) Human acid sphingomyelinase: isolation, nucleotide sequence, and expression of the full-length and alternatively spliced cDNA. J Biol Chem 266:8531–8539 [PubMed] [Google Scholar]
- Schuchman EH (1995) Two new mutations in the acid sphingomyelinase gene causing type A Niemann-pick disease: N389T and R441X. Hum Mut 6:352–354 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Desnick RJ, Takada G, Schuchman EH (1992a) Identification of a missense mutation (S436R) in the acid sphingomyelinase gene from a Japanese patient with type B Niemann-Pick disease. Hum Mutat 1:70–71 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Suchi M, Desnick RJ, Takada G, Schuchman EH (1992b) Identification and expression of five mutations in the human acid sphingomyelinase gene causing types A and B Niemann-Pick disease: molecular evidence for genetic heterogeneity in the neuronopathic and non-neuronopathic forms. J Biol Chem 267:12552–12558 [PubMed] [Google Scholar]
- Vanier MT, Ferlinz K, Rousson R, Duthel S, Louisot P, Sandhoff K, Suzuki K (1993) Deletion of arginine (608) in acid sphingomyelinase is the prevalent mutation among Niemann-Pick disease type B patients from northern Africa. Hum Genet 92:325–330 [DOI] [PubMed] [Google Scholar]
- Yamaji A, Sekizawa Y, Emoto K, Sakuraba H, Inoue K, Kobayashi H, Umeda M (1998) Lysenin, a novel sphingomyelin-specific binding protein. J Biol Chem 273:5300–5306 [DOI] [PubMed] [Google Scholar]