Abstract
Limb-girdle muscular dystrophy 1A (LGMD1A [MIM 159000]) is an autosomal dominant form of muscular dystrophy characterized by adult onset of proximal weakness progressing to distal muscle weakness. We have reported elsewhere a mutation in the myotilin gene in a large, North American family of German descent. Here, we report the mutation screening of an additional 86 families with a variety of neuromuscular pathologies. We have identified a new myotilin mutation in an Argentinian pedigree with LGMD1 that is predicted to result in the conversion of serine 55 to phenylalanine (S55F). This mutation has not been found in 392 control chromosomes and is located in the unique N-terminal domain of myotilin, only two residues from the T57I mutation reported elsewhere. Both T57I and S55F are located outside the α-actinin and γ-filamin binding sites within myotilin. The identification of two independent pedigrees with the same disease, each bearing a different mutation in the same gene, has long been the gold standard for establishing a causal relationship between defects in a gene and the resultant disease. As a description of the second known pedigree with LGMD1A, this finding constitutes that gold standard of proof that mutations in the myotilin gene cause LGMD1A.
The limb-girdle muscular dystrophies (LGMDs) include a clinically diverse group of dominant and recessive disorders characterized by proximal muscle weakness, elevated serum creatine kinase values, and absent or reduced deep-tendon reflexes. Among the autosomal recessive limb girdles, mutations in calpain 3 cause LGMD2A (Richard et al. 1995), mutations in dysferlin cause LGMD2B (Bashir et al. 1998), and mutations in telethonin cause LGMD2G (Moreira et al. 2000). LGMD2C through LGMD2F are all caused by defects in members of the dystrophin glycoprotein complex: γ-, α-, β-, and δ-sarcoglycan (Roberds et al. 1994; Bonnemann et al. 1995; Lim et al. 1995; Noguchi et al. 1995; Nigro et al. 1996). Mutations in TRIM32, a putative E3-ubiquitin-ligase gene, have been identified as the cause of LGMD2H (Frosk et al. 2002). Among the dominant forms, four have been mapped: LGMD1B to 1q11-21 (van der Kooi et al. 1997), LGMD1D to 6q23 (Messina et al. 1997), LGMD1E to 7q (Speer et al. 1999), and vocal cord and pharyngeal weakness with autosomal dominant distal myopathy to 5q (Feit et al. 1998). Caveolin 3 mutations have been described in LGMD1C (McNally et al. 1998; Minetti et al. 1998).
LGMD1A (MIM 159000) is characterized by onset of proximal muscle weakness at a mean age of 27 years, later progressing to include distal weakness. Approximately half of the affected individuals exhibit a distinctive nasal, dysarthric pattern of speech. Tightened heel cords and reduced knee and elbow deep-tendon reflexes are frequently seen. CK levels are elevated, ranging from 1.6-fold to 9-fold higher than the normal limit of 120 IU/l for males and 80 IU/l for females. Biopsy of affected individuals shows variations in fiber size, fiber splitting, and other hallmarks of degeneration; a large number of rimmed vacuoles; and patches of striking Z-line streaming, similar to that seen in nemaline myopathy (Hauser et al. 2000). Elsewhere, we have reported a mutation in the myotilin gene (TTID, MYOT [NCBI accession number NM_006790; MIM 604103]) in the only known LGMD1A pedigree, Duke family 39 (Hauser et al. 2000). Myotilin protein is localized to the Z-line and binds to α-actinin (Salmikangas et al. 1999) and γ-filamin (van der Ven et al. 2000b). The threonine 57 to isoleucine (T57I) mutation in family 39 occurs in a region of unknown function, so the mechanism by which it causes disease is not apparent.
We report here the screening of 86 additional pedigrees to find additional myotilin mutations that might provide further insight into possible structure-function relationships in the myotilin protein. LGMDs are characterized by a large degree of variation in clinical presentation, between, and often within families. Individuals with novel myotilin mutations may well present with a significantly different set of symptoms from those previously seen, especially if the mutations are located in different protein domains. Therefore, mutation analysis was performed on at least one affected individual from each of 44 families with LGMD type 1 (autosomal dominant), 14 families with LGMD type 2 (autosomal recessive), 24 families with facioscapulohumeral muscular dystrophy, 2 families with scapuloperoneal muscular dystrophy, and 2 families with unclassified dominant myopathies. Informed consent was obtained from all human subjects.
Mutation analysis was performed on genomic DNA isolated from whole blood using the Puregene kit (Gentra Systems). The primers in table 1 were used to PCR amplify exons 2–9 and part of exon 10 (the full coding region) of the myotilin gene from genomic DNA in pools of five unrelated individuals. These PCR products were analyzed using the Transgenomic WAVE denaturing high-performance liquid chromatography system. Several different temperatures were tested for each PCR amplicon to optimize mutation detection sensitivity. All observed changes were confirmed by sequencing both DNA strands using the Beckman CEQ2000 capillary electrophoresis sequencer or the Beckman 3700 DNA sequencer using Big Dye chemistry.
Table 1.
Primers Used to Amplify Myotilin Coding Sequence
|
Primer |
|||
| Exon | Forward | Reverse | Product Size(bp) |
| 2a | CAGATCTGAAAGATGTCAAATAAACAA | GTTGTAACCCTTTGGCCTGG | 546 |
| 2b | CTCAACAAGGAAGAGCAGAC | TACTGCTATTGTAATCAGGC | 481 |
| 2c | GCTCCAGATTGCAGCCTCCT | CCAGTACCCTGGTTCAGCAT | 476 |
| 3 | ATTTGCAAAATGAGGCCAAG | GGGCCCAAATATTCCTTCTT | 373 |
| 4 | TGTCTCAATAAATTCTCTAAAGCG | GTGGATGGAACTGACCGACT | 273 |
| 5 | CTGGGCTTCTTGCTAGAGTGGTAG | GATCCTGGCTTATTTGACC | 462 |
| 6 | CTCCTGCCTTAGCCTCCTGAG | GGAGGATGGCAGAGCCAGAATT | 471 |
| 7 | TCTGCCATCTCCTTGTGTTTT | TGAAGTCTGCTGGGCTTTTC | 330 |
| 8 | GGTATAACAAAATAGTACTGCATGTC | AACTGGATTCACCCAAATAAAC | 380 |
| 9 | TGGTCAGAGACATCCACTTCA | TTTTATACTCTGCTGGGATTTTCA | 286 |
| 10a | CCAATTTGGTTAGAACAGGTTT | GTAGGCTTCACAAATCGGAG | 420 |
| 10b | TACCAACATTGGAAAACAG | TCATAGGTTTTGCTGAGTGGAG | 522 |
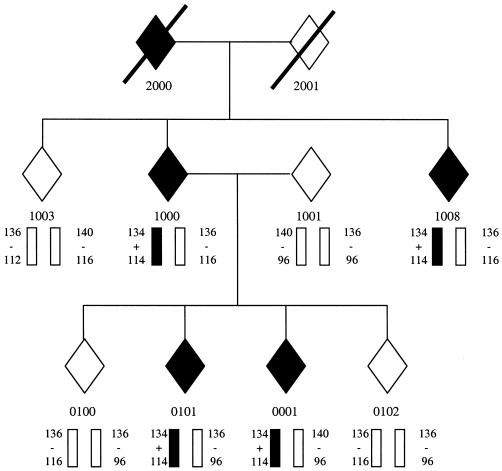
A single Argentinian sample exhibited a C444→T missense mutation in exon 2 of the myotilin gene, predicted to result in a change of residue 55 from serine to phenylalanine (S55F). This change is found in a single allele, consistent with the observed pattern of autosomal dominant inheritance, and was not detected in 392 control chromosomes. DNA samples from eight additional members of this Argentinian pedigree (family 2654) were then screened, and the missense mutation was found to segregate with disease status. The FASST method (Vance and Ben Othmane 1998) was used to genotype family 2654 with two polymorphic microsatellite repeat markers (D5S479 and D5S178) flanking the myotilin gene (fig. 1). These markers establish a disease-associated haplotype entirely different from that seen in our original LGMD1A pedigree (Duke family 39).
Figure 1.
Segregation of 5q31 microsatellite markers and the C444T missense mutation in Family 2654. Affected individuals and the disease-bearing haplotype are indicated in solid black. From top to bottom, the alleles shown are for D5S479, the S55F mutation, and D5S178.
The identification of two independent pedigrees with the same disease, each bearing a different mutation in the same gene, has long been the gold standard for establishing a causal relationship between defects in a gene and the resultant disease. As a description of the second known pedigree with LGMD1A, this finding constitutes such gold-standard proof that mutations in the myotilin gene cause LGMD1A. The clinical presentation of affected individuals in family 2654 is quite similar to that of the LGMD1A pedigree we reported elsewhere. They exhibit proximal leg and arm weakness by 42–58 years of age, which later progresses to include distal weakness. Serum CK levels are elevated 5-fold to 15-fold. Two of the four living affected individuals exhibit a distinctive dysarthric pattern of speech. All members of family 2654 have been examined by two of the authors (A.L.R. and G.Z.). The similarity in clinical presentation in the two families may be related to the proximity of the observed mutations, which are located 2 aa apart in the unique N-terminal domain of the protein, immediately adjacent to a 20-aa hydrophobic stretch.
There are striking similarities between LGMD1A and the nemaline myopathies, suggesting possible mechanisms by which myotilin mutations may give rise to disease. Missense mutations in α-tropomyosin (NEM1) give rise to nemaline myopathy with Z-line streaming similar to that seen in LGMD1A (Laing et al. 1995), and both myotilin protein and α-tropomyosin bind to α-actinin. One of the primary roles of α-actinin is to tether actin filaments to the Z-line, and missense mutations in α-actin also give rise to nemaline myopathy (Nowak et al. 1999). These observations suggest that the observed myotilin mutations might act by disrupting the tethering of actin filaments through their interactions with α-actinin. However, we were unable to detect any effect of the T57I mutation on α-actinin binding by means of a yeast two-hybrid assay. Further, this assay identified a minimum α-actinin binding domain extending from residues 79–150, well separated from the observed mutations (Hauser et al. 2000).
It has also been reported that myotilin protein binds to γ-filamin, a form of filamin specifically expressed in striated muscle, and does so through a region spanning its 78-aa muscle-specific domain (van der Ven et al. 2000b). Several filamin isoforms bind to α-actin and are thought to regulate its polymerization (Wang and Singer 1977). This network of actin filaments is then anchored to the plasma membrane by filamin’s interactions with a number of other cytoskeletal proteins, including β1-integrin (Loo et al. 1998), β2-integrin (Sharma et al. 1995), and, in the case of γ-filamin, γ- and δ-sarcoglycan (Thompson et al. 2000). Further, γ-filamin colocalizes with α-actinin in the Z-disk in striated muscle and has been implicated in Z-disk assembly and myofibrillogenesis (van der Ven et al. 2000a). Through its binding interactions with both α-actinin and γ-filamin, myotilin protein may play a role in colocalizing α-actinin and γ-filamin to the Z-disk. This model is supported by the observation that transfection of differentiating C2C12 myoblasts with a truncated myotilin gene encoding just the C-terminal Ig domains disrupts Z-disk formation, whereas introduction of the entire myotilin gene does not (van der Ven et al. 2000b). This would seem to indicate that severing the connection between the N- and C-terminal domains creates a dominant negative protein in much the same way that the isolated Z1 and Z2 domains of titin have been reported to disrupt sarcomere assembly through their interactions with telethonin (Peckham et al. 1997; Mues et al. 1998). It is unlikely that the reported T57I and S55F myotilin mutations directly disrupt γ-filamin binding: yeast two-hybrid experiments have demonstrated that the γ-filamin binding site is located within the C-terminal Ig domains of myotilin protein (residues 215–493), well removed from the T57I and S55F mutations (van der Ven et al. 2000b). Although it is conceivable that the yeast two-hybrid system is not sufficiently sensitive to detect the subtle effects on tertiary structure caused by the myotilin mutations and that these mutations actually do disrupt binding with either α-actinin or γ-filamin, a more likely explanation is that there is another, as-yet-undetermined binding partner for myotilin protein and that the observed mutations disrupt this interaction or disrupt myotilin’s ability to colocalize this protein with γ-filamin.
The N-terminal region of myotilin protein that contains the T57I and S55F mutations is not homologous to any other protein. It includes a 23-residue hydrophobic domain and a region rich in serine (27/96 residues). Although the function of this domain is unknown, it is possible that the hydrophobic stretch mediates the localization of small amounts of myotilin protein to the sarcolemmal membrane. Both of the observed myotilin mutations would have the effect of elongating this hydrophobic stretch, possibly disturbing its interactions with the sarcolemmal membrane or with a novel protein-binding partner. A better understanding of the biological function of the N-terminus will be necessary to fully appreciate the effects of the two observed myotilin mutations. Further, mutations in a different portion of the protein might well result in a significantly different phenotype. For example, myotilin protein is expressed at relatively high levels in the heart, and, although there is no evidence of a cardiac defect in either known pedigree, novel mutations might give rise to a cardiac phenotype. We are currently testing this hypothesis by expressing synthetic myotilin mutations in transgenic mouse models. These experiments may suggest alternate groups of human patients who could be screened for defects in myotilin.
Acknowledgments
We acknowledge Richard Tim, P. Craig Gaskell, Jeffrey Stajich, Kathryn North, Angel Alonso, Melissa Lees, Rup Tandan, and Allessandra Renieri for contribution of patient samples. This work is supported by grants from the Muscular Dystrophy Association (to M.A.H. and M.C.S.) and from National Institutes of Health grant NS26630. Work in the lab of A.L.R. is supported by CONICET, SETCIP, and Fundacion Antorchas (Argentina), the International Program of the Howard Hughes Medical Institute, and the Association Française contre les Myopathies (AFM).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/ (for myotilin, TTID, or MYOT [accession number NM_006790])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for LGMD1A [MIM 159000])
References
- Bashir R, Britton S, Strachan T, Keers S, Vafiadaki E, Lako M, Richard I, Marchand S, Bourg N, Argov Z, Sadeh M, Mahjneh I, Marconi G, Passos-Bueno MR, de Sa Moreira E, Zatz M, Beckmann JS, and Bushby K (1998) A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat Genet 20:37–42 [DOI] [PubMed] [Google Scholar]
- Bonnemann CG, Modi R, Noguchi S, Mizuno Y, Yoshida M, Gussoni E, McNally EM, Duggan DJ, Angelini C, Hoffman EP, Ozawa E, Kunkel LM (1995) β-sarcoglycan (A3b) mutations cause autosomal recessive muscular dystrophy with loss of the sarcoglycan complex. Nat Genet 11:266–272 [DOI] [PubMed] [Google Scholar]
- Feit H, Silbergleit A, Schneider LB, Gutierrez JA, Fitoussi R-P, Reyes C, Rouleau GA, Brais B, Jackson CE, Beckmann JS, Seboun E (1998) Vocal cord and pharyngeal weakness with autosomal dominant distal myopathy: clinical description and gene localization to 5q31. Am J Hum Genet 63:1732–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosk P, Weiler T, Nylen E, Sudha T, Greenberg CR, Morgan K, Fujiwara TM, Wrogemann K (2002) Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am J Hum Genet 70:663–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MA, Horrigan SK, Salmikangas P, Viles KD, Tim RW, Torian UM, Taivainen U, Bartoloni L, Dancel R, Gilchrist JM, Stajich JM, Gaskell PC, Gilbert JR, Vance JM, Pericak-Vance MA, Carpen O, Westbrook CA, Speer MC (2000) Myotilin is mutated in limb girdle muscular dystrophy 1A. Hum Mol Genet 9:2141–2147 [DOI] [PubMed] [Google Scholar]
- Laing NG, Wilton SD, Akkari PA, Dorosz S, Boundy K, Kneebone C, Blumbergs P, White S, Watkins H, Love DR, Haan E (1995) A mutation in the alpha tropomyosin gene TPM3 associated with autosomal dominant nemaline myopathy. Nat Genet 9:75–79 [DOI] [PubMed] [Google Scholar]
- Lim LE, Duclos F, Brouz O, Bourg N, Sunada Y, Allamand V, Meyer J, Richard I, Moomaw C, Slaughter C, Tome FMS, Fardeau M, Jackson CE, Beckmann JS, Campbell KP (1995) β-sarcoglycan: characterization and role in limb-girdle muscular dystrophy linked to 4q12. Nat Genet 11:257–264 [DOI] [PubMed] [Google Scholar]
- Loo DT, Kanner SB, Aruffo A (1998) Filamin binds to the cytoplasmic domain of the beta 1-integrin: identification of amino acids responsible for this interaction. J Biol Chem 273:23304–23312 [DOI] [PubMed] [Google Scholar]
- McNally EM, de Sa Moreira E, Duggan DJ, Bonnemann CG, Lisanti MP, Lidov HG, Vainzof M, Passos-Bueno MR, Hoffman EP, Zatz M, Kunkel LM (1998) Caveolin-3 in muscular dystrophy. Hum Mol Genet 7:871–877 [DOI] [PubMed] [Google Scholar]
- Messina DN, Speer MC, Pericak-Vance MA, McNally EM (1997) Linkage of familial dilated cardiomyopathy with conduction defect and muscular dystrophy to chromosome 6q23. Am J Hum Genet 61:909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minetti C, Sotgia F, Bruno C, Scartezzini P, Broda P, Bado M, Masetti E, Mazzocco M, Egeo A, Donati MA, Volonte D, Galbiati F, Cordone G, Bricarelli FD, Lisanti MP, Zara F (1998) Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat Genet 18:365–368 [DOI] [PubMed] [Google Scholar]
- Moreira ES, Wiltshire TJ, Faulkner G, Nilforoushan A, Vainzof M, Suzuki OT, Valle G, Reeves R, Zatz M, Passos-Bueno MR, Jenne DE (2000) Limb-girdle muscular dystrophy type 2G is caused by mutations in the gene encoding the sarcomeric protein telethonin. Nat Genet 24:163–166 [DOI] [PubMed] [Google Scholar]
- Mues A, van der Ven PFM, Young P, Fürst DO, Gautel M (1998) Two immunoglobulin-like domains of the Z-disc portion of titin interact in a conformation-dependent way with telethonin. FEBS Lett 428:111–114 [DOI] [PubMed] [Google Scholar]
- Nigro V, Moreira E, Piluso G, Vanzof M, Belsito A, Politano L, Puca AA, Passos-Bueno MR, Zatz M (1996) Autosomal recessive limb-girdle muscular dystrophy, LGMD2F, is caused by a mutation in the δ-sarcoglycan gene. Nat Genet 14:195–198 [DOI] [PubMed] [Google Scholar]
- Noguchi S, McNally EM, Ben Othmane K, Hagiwara Y, Mizuno Y, Yoshida M, Yamamoto H, Bonnemann CG, Gussoni E, Denton PH, Kyriakides T, Middleton L, Hentati F, Ben Hamida M, Nonaka I, Vance JM, Kunkel LM, Ozawa E (1995) Mutations in the dystrophin-associated protein-gamma-sarcoglycan in chromosome 13 muscular dystrophy. Science 270:819–822 [DOI] [PubMed] [Google Scholar]
- Nowak KJ, Wattanasirichaigoon D, Goebel HH, Wilce M, Pelin K, Donner K, Jacob RL, et al. (1999) Mutations in the skeletal muscle α-actin gene in patients with actin myopathy and nemaline myopathy. Nat Genet 23:208–212 [DOI] [PubMed] [Google Scholar]
- Peckham M, Young P, Gautel M (1997) Constitutive and variable regions of Z-disk titin/connectin in myofibril formation: a dominant-negative screen. Cell Struct Funct 22:95–101 [DOI] [PubMed] [Google Scholar]
- Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C, Pasturaud P, Roudaut C, Hillaire D, Passos-Bueno MR, Zatz M, Tischfield JA, Fardeau M, Jackson CE, Cohen D, Beckmann JS (1995) Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell 81:27–40 [DOI] [PubMed] [Google Scholar]
- Roberds SL, Leturcq F, Allamand V, Piccolo F, Jeanpierre M, Anderson RD, Lim LE, Lee JC, Tome FMS, Romero NB, Fardeau M, Beckmann JS, Kaplan JC, Campbell KP (1994) Missense mutations in the adhalin gene linked to autosomal recessive muscular dystrophy. Cell 78:625–633 [DOI] [PubMed] [Google Scholar]
- Salmikangas P, Mykkänen OM, Grönholm M, Heiska L, Kere J, Carpén O (1999) Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum Mol Genet 8:1329–1336 [DOI] [PubMed] [Google Scholar]
- Sharma CP, Ezzell RM, Arnaout MA (1995) Direct interaction of filamin (ABP-280) with the β2-integrin subunit CD18. J Immunol 154:3461–3470 [PubMed] [Google Scholar]
- Speer MC, Vance JM, Grubber JM, Graham FL, Stajich JM, Viles KD, Rogala A, McMichael R, Chutkow J, Goldsmith C, Tim RW, Pericak-Vance MA (1999) Identification of a new autosomal dominant limb-girdle muscular dystrophy locus on chromosome 7. Am J Hum Genet 64:556–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson TG, Chan Y-M, Hack AA, Brosius M, Rajala M, Lidov HGW, McNally EM, Watkins S, Kunkel LM (2000) Filamin 2 (FLN2): a muscle-specific sarcoglycan interacting protein. J Cell Biol 148:115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kooi AJ, van Meegen M, Ledderhof TM, McNally EM, de Visser M, Bolhuis PA (1997) Genetic localization of a newly recognized autosomal dominant limb-girdle muscular dystrophy with cardiac involvement (LGMD1B) to chromosome 1q11-21. Am J Hum Genet 60:891–895 [PMC free article] [PubMed] [Google Scholar]
- van der Ven PF, Obermann WM, Gautel M, Gautel M, Weber K, Fürst DO (2000a) Characterization of muscle filamin isoforms suggests a possible role of γ-filamin/ABP-L in sarcomeric z-disc formation. Cell Motil Cytoskeleton 45:149–162 [DOI] [PubMed] [Google Scholar]
- van der Ven PF, Wiesner S, Salmikangas P, Auerbach D, Himmel M, Kempa S, Hayess K, Pacholsky D, Taivainen A, Schröder R, Carpén O, Fürst DO (2000b) Indications for a novel muscular dystrophy pathway: γ-filamin, the muscle-specific filamin isoform, interacts with myotilin. J Cell Biol 151:235–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JM, Ben Othmane K (1998) Methods of genotyping. In: Haines JL, Pericak-Vance MA (eds) Approaches to gene mapping in complex human diseases. Wiley-Liss, New York, pp 213–228 [Google Scholar]
- Wang K, Singer SJ (1977) Interaction of filamin with F-actin in solution. Proc Natl Acad Sci USA 74:2021–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]