Abstract
Single molecule force spectroscopy methods, such as optical and magnetic tweezers and atomic force microscopy, have opened up the possibility to study biological processes regulated by force, dynamics of structural conformations of proteins and nucleic acids, and load-dependent kinetics of molecular interactions. Among the various tools available today, optical tweezers have recently seen great progress in terms of spatial resolution, which now allows the measurement of atomic-scale conformational changes, and temporal resolution, which has reached the limit of the microsecond-scale relaxation times of biological molecules bound to a force probe. Here, we review different strategies and experimental configurations recently developed to apply and measure force using optical tweezers. We present the latest progress that has pushed optical tweezers’ spatial and temporal resolution down to today’s values, discussing the experimental variables and constraints that are influencing measurement resolution and how these can be optimized depending on the biological molecule under study.
Introduction
Force and mechanical stress act on biological systems at very different length scales, from cells, down to single molecules, and up to entire organisms. In the last few decades, force has emerged as a fundamental regulatory factor for cell life. It has been known for a long time that external forces acting on muscles regulate their contraction velocity and power output. Actually, it is well established that physical activity regulates expression of muscle proteins to adapt to changes in functional demands. More generally, emerging evidence indicates that factors such as the force applied or the rigidity of the extracellular matrix determine the shape and function of cells and organisms (1). Classically, the regulation of biological systems is described through a series of biochemical signals and enzymatic reactions that direct the processes and cell fate. However, mechanotransduction, i.e., the conversion of mechanical forces into biochemical signals, is at the base of many biological processes fundamental for the development and differentiation of cells (2), for their correct function and for the development of pathologies (3). At the molecular scale, force modulates enzymatic activity, induces structural changes in proteins and nucleic acids, alters kinetics of molecular bonds (4,5), regulates motions of molecular motors (6,7), and has a role in mechanical transduction and sensory functions (8). All these processes are ultimately related to the capacity of force to modulate lifetimes of molecular interactions and transition rates in biochemical reaction cycles that involve motion (9).
The study of the molecular mechanisms underlying force-dependent processes has been opened up by single molecule force spectroscopy techniques. Over the past two decades, a myriad of methods have been developed to apply and measure forces on single molecules. Among these, atomic force microscopes, optical tweezers, and magnetic tweezers have enabled the study of a wide range of molecular processes in which force plays a crucial role (10). The last few years have seen major improvements in spatial and temporal resolution of optical tweezers, which have extended the range of measurable quantities and the biological systems that can be interrogated with force.
After their invention by Ashkin et al. in 1986 (11), optical tweezers have become one of the most widely used single molecule tools in biology (12). The capacity to apply and measure forces from a few tens of femtoNewtons to ∼100 picoNewtons, which well overlaps with the range of forces experienced by biological molecules in their native environment, makes optical tweezers an ideal single-molecule tool for biologists. Optical tweezers are easily implemented in an optical microscope by focusing a laser beam through the microscope objective. Dielectric microparticles are stably trapped near the beam focus owing to the interaction between the laser light and the particle itself (Fig. 1 a). A polystyrene or silica microsphere is usually trapped and used as a handle to manipulate single molecules bound to it. The force experienced by the bead (F) points toward the trap center and grows with the distance of the bead center from the trap center (xbead). The trap behaves as a Hookean spring for small distances (F = −ktrapxbead, where ktrap is the trap stiffness). Beyond the linear region, a small zone of near-constant force follows at the border of the potential well, after which the force rapidly drops to zero (Fig. 1 b) (13). The position of the trapped bead can be measured using a variety of detection schemes and the applied force is obtained from the bead position and calibration of the trap stiffness. Several articles and reviews describe in detail the physics of optical tweezers, instrument design considerations, and calibration techniques (14–16).
Figure 1.
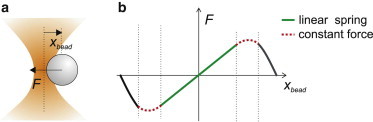
Force exerted by optical tweezers. (a) A dielectric microsphere is stably trapped near the laser beam focus. A lateral displacement of the bead (xbead) is opposed by a restoring force F. (b) For small displacements of the bead from the trap center (xbead), the force exerted by optical tweezers grows linearly with xbead. Beyond the linear region, a near-constant force region follows, after which the force rapidly drops to zero.
Several measurement strategies have been implemented to study very different biological systems. Here, we first report, to our knowledge, some of the prototypical optical tweezers configurations developed as of this writing, with particular emphasis on recent biological applications. We then discuss factors affecting spatial and temporal resolution in the different configurations of measurement and recent developments that allowed optical tweezers to reach unprecedented resolution.
Strategies to Apply and Measure Forces on Single Biological Molecules
Static geometries
In the simplest geometry (the single bead or single trap geometry), optical tweezers are kept at a fixed position and a trapped bead monitors conformational changes and movements of a protein that is linked to the bead (Fig. 2 a). This configuration has been largely used to investigate processive molecular motors such as conventional kinesin (17), which can move continuously along a microtubule for up to several microns. The force applied to the processive motor increases as the protein displaces the bead from the trap center while translocating along its track (F = −ktrapxbead). When the maximum force that the motor protein can develop is reached, the motor stalls. Stall forces of kinesin (18), RNA polymerase (19), and many other motors have been measured using the single bead geometry.
Figure 2.
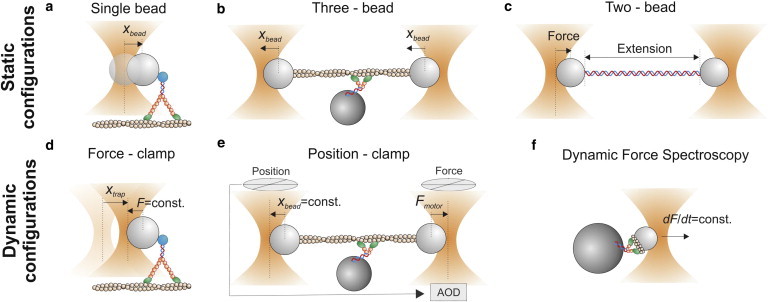
Configurations for the measurement of force and displacement with optical tweezers. (a) Single bead or single trap geometry. The trap is static and the bead displacement xbead measures protein displacement. (b) Three-bead assay. Both traps are static and the trapped beads displacement (xbead) measures protein displacement. (c) Two-bead or double trap assay. The left trap is stationary and measures the force applied to the polymer. The right bead moves in steps or ramps and, for each displacement, the forces applied to the polymer and its extension are measured. (d) Force-clamp or isotonic clamp. A feedback system moves the trap to keep force on the bead constant. Trap displacements (xtrap) measure protein displacements. (e) Position-clamp or isometric clamp. The left bead detects movements of the dumbbell (xbead), whereas the right bead moves using an AOD to oppose the detected movements. The right bead measures the force applied by the motor protein (Fmotor). (f) Dynamic force spectroscopy. The molecular bond is subjected to constant loading rates and rupture forces and bond lifetimes are measured.
A reversed configuration (the three-bead assay) was mainly used to investigate nonprocessive molecular motors such as skeletal muscle myosin, which briefly interacts with actin, displaces it by a ∼5-nm conformational change (working stroke), and unbinds (20–22). In the three-bead assay, an actin filament is suspended between two optically trapped beads, and a single myosin molecule is bound to a third bead stuck onto the coverslip (Fig. 2 b). The same assay was recently applied to investigate actin-binding proteins involved in mechanotransduction processes (23). By replacing the actin filament with a microtubule, the three-bead assay was adapted to study microtubule-based molecular motors such as cytoplasmic dynein (24,25) or ncd kinesin (26).
Double optical tweezers (the two-bead, double trap, or dumbbell assay) or a single trap together with a micropipette have been widely employed to investigate the mechanical properties (force-extension curves) of single dsDNA and ssDNA molecules (27), as well as single RNA molecules (28). In this configuration, one of the two traps is held at a fixed position and probes force applied to the polymer, while the second trap or the micropipette is displaced to stretch the polymer (Fig. 2 c). More recently, the same approach was applied to investigate protein folding by using dsDNA handles to tether the protein between the two beads (4,29,30). The same geometry was used to demonstrate that filamin A acts as a force-activatable mechanosensor (31).
Dynamic geometries
A more sophisticated geometry is the so-called force-clamp or isotonic clamp (6,32) (Fig. 2 d). In active force-clamps, the distance between the bead and the trap (xbead) is continuously monitored through a position detector. As the distance changes owing, for example, to the movements of a motor protein, a feedback system rapidly moves the trap to keep the distance, and thus the force, constant (xbead = const. → F = −ktrapxbead = const.). In this configuration, trap movements (xtrap) accurately probe protein movements. Active force clamps were extensively applied in a single bead geometry to study processive molecular motors such as kinesin (32), myosin Va (6), myosin VI (33), and RNA polymerase (34), and in a double trap configuration, in which one trap was stationary and the other one was force-clamped, to study nucleic acids or protein folding (28,29,31,35). Despite the higher complexity of implementing force-clamps, single molecule data collected in this configuration allow a direct measurement of force-dependence of conformational transition rates, thus enabling the reconstruction of the energy landscapes of molecular structural states. Force-clamp geometries also open up the possibility of measuring the mechanical output of molecular motors, such as the working stroke and the average distance traveled before dissociating (run length), as a function of load. This configuration also eliminates the complication of correcting the measured distances taking into account motor and linkage compliances, which remain at a fixed length under force-clamp (see next section and Eq. 3). Piezo- or motorized mirrors can be used to steer the laser beam and move the trap, although much faster response (μs) can be reached using acousto-optic deflectors (AODs) (36) or electro-optic deflectors (37).
The limited bandwidth of the feedback loop can sometimes be restrictive (38), depending on the time constant of the process under study relative to the response time of the feedback loop. Alternative configurations were developed to overcome such limitation. For example, Nambiar et al. (39) created a one-dimensional region of constant force extending over several micrometers by rapidly line-scanning the trapping light and simultaneously modulating its intensity. Greenleaf et al. (13) developed a passive all-optical force-clamp exploiting the constant-force region of optical tweezers near the border of the potential well (Fig. 1 b). Such a clamp was successfully employed to measure the force-velocity relationship for transcription by RNA polymerase (40), as well as folding trajectories of nucleic acids under constant force (41,42).
Opposite to force-clamp, where force on the protein is kept constant and protein movements are measured, the position-clamp (also called the isometric clamp) prevents protein movements and measures the force that is developed. Molecular isometric clamp was first developed by Finer et al. (20) to measure forces exerted by single actomyosin complexes in the three-bead geometry and subsequently refined by Takagi et al. (43). In this latter implementation, one of the two beads, termed the transducer bead, detected movements of the dumbbell whereas the other bead, the motor bead, was moved using an AOD to oppose the detected movements and maintain the bead-actin-bead assembly at its initial position (Fig. 2 e). Myosin isometric force measured from these experiments was ∼9 pN, close to that estimated from high-resolution fiber mechanics studies (44). The same assay recently allowed the measurement of force-dependent interaction between actin and different isoforms of myosin I (8,45,46).
Another strategy to probe the effect of force on molecular bonds is dynamic force spectroscopy (DFS), in which the distribution of rupture forces of molecular bonds is measured at different loading rates (47). Constant loading rates were applied by moving the trapped bead at constant velocity (dF/dt = −ktrap ⋅ (dxbead/dt) = −ktrap ⋅ vbead) (48–53) (Fig. 2 f), or by clamping the position of the bead relative to the optical trap (xbead = const.) and increasing the optical power at a constant rate (dF/dt = −(dktrap/dt) ⋅ xbead) (54). Data collected with DFS can be converted, using appropriate models, to get the kinetics of bond detachment under constant forces (47,55). Such an approach is commonly used for weak bonds, in which the kinetics are too fast to effectively work in a force-clamp geometry; however, a proper conversion relies on the accuracy of model assumptions. Among the many reports, DFS was applied to study the interaction between actin and skeletal muscle myosin (48,49) or titin (52), and between lipid bilayers and myosin I (53). The binding of the glycoprotein Ibα (GPIbα) subunit to the A1 domain of von Willebrand factor, which mediates the formation of platelet plugs for arterioles and is associated with the bleeding disorder von Willebrand disease, was recently studied by DFS (50,56).
Optical Tweezers with Ångstrom Spatial Resolution
One of the key features of optical tweezers is the capability to measure small conformational changes and displacements produced by single biological molecules. Such movements range from several nanometers for cytoskeletal molecular motors, down to one basepair (3.6 Å) for DNA and RNA processing enzymes (57,58). High spatial resolution is, thus, a fundamental requirement for the study of a wide range of biological molecules. Molecular displacements can be measured through different position detection systems that monitor the position of the trapped bead. To date, the most sensitive detectors, which are based on interferometry (17,59,60), allow the detection of Ångstrom movements on a timescale of 1 ms or better. However, different sources of noise deteriorate localization accuracy. A source of noise that cannot be eliminated arises from Brownian fluctuations of the trapped bead owing to thermal forces, which are inherent in single molecule experiments performed in liquid solution. Thermal noise, thus, sets fundamental limits on single molecule force and position measurements.
Thermal noise
Understanding which parameters affect thermal noise is crucial to optimizing spatial resolution in position measurements with optical tweezers. Thermal noise amplitude can be calculated from its frequency distribution, which depends on the system stiffness and viscosity. In the single bead geometry depicted in Fig. 2 a, a trapped bead is linked to a motor protein, which is moving on its track bound to the coverslip. The system has a combined stiffness k, which comprises the trap stiffness (ktrap) and the stiffness of the molecule and linkages connecting the molecule to the bead and to the coverslip surface (kmotor); and a drag coefficient mainly due to the solution viscosity and the bead (γbead, Fig. 3 a). The power spectrum (i.e., the squared magnitude of the Fourier transform) of thermal motion for a sphere of radius r constrained by an elastic element of stiffness k, immersed in a fluid of viscosity η at temperature T, is given by (10,61)
| (1) |
where γbead = 6πηr and fC = k/2πγbead is the cutoff frequency of the motion. The power spectrum is constant for low frequencies (f << fC) and decreases as 1/f2 for high frequencies (f >> fC) (Fig. 3 c). The position variance σx2 (the square of the position fluctuation amplitude) is the area under the power spectrum, which is given by σx2 = kBT/k. However, the bead position is always measured over a finite frequency interval or bandwidth Δf, and the position variance is the area under the power spectrum within the interval Δf (for example, the turquoise area in Fig. 3 c). This area can be easily derived from Eq. 1 in the low-frequency approximation:
| (2) |
Stiffer systems and smaller beads, therefore, reduce the position noise at a given bandwidth. When the bead is linked to a stiffer element (Fig. 3, b and c, red curve), the total amplitude of thermal noise is reduced. On the other hand, a smaller bead does not change the total amplitude of thermal motion, but changes its spectral distribution, leaving less noise at the low frequencies in which measurements are usually made (Fig. 3 c, green curve). Because molecular stiffness is usually nonlinear and increases with the applied tension, from Eq. 2 we also deduce that applying higher force to a molecule is advantageous because it reduces the fluctuations by increasing the system stiffness.
Figure 3.
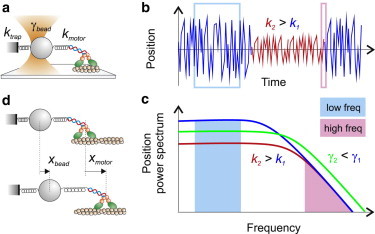
Parameters affecting thermal noise and spatial resolution. (a) ktrap is the trap stiffness and kmotor comprises the motor protein stiffness in series with the stiffness of the linkages connecting the protein to the bead and the coverslip surface. γbead is the bead viscous drag coefficient. (b) Position fluctuations. Drawing of the position signal of a bead linked to a compliant (k1) or stiff system (k2). (c) Power density spectrum of the bead position. When the stiffness of the system increases (k2 > k1), the noise amplitude decreases at low frequencies (f << fC) (turquoise area), but it is unchanged at high frequencies (pink area). When the viscous drag decreases (γ2 < γ1), the low-frequency noise decreases, whereas the high-frequency noise increases. The area under the power spectrum stays constant (d) The measured bead displacement (xbead) depends on the motor protein displacement (xmotor) and on the values of the trap and protein stiffness.
Care must be taken when measuring molecular movements, because bead movements (xbead) correctly report molecular movements (xmotor) only when ktrap << kmotor. In fact, referring to Fig. 3 d,
| (3) |
and the signal-to-noise ratio (SNR) is given by
| (4) |
From Eqs. 2–4 we see that increasing trap stiffness reduces thermal noise, but it also reduces movements of the probe relative to the real movements of the molecule, so that the SNR is not changed. Eq. 4 is also valid in the force-clamp geometry (Fig. 2 d), where the trap stiffness is zero for a trap that applies a constant force (61).
In the two-bead assay, a single molecule is tethered between two microbeads trapped in optical tweezers (Fig. 2 c). The added Brownian noise due to the second microsphere deteriorates the SNR, which is always smaller compared to the single-bead assay (62). However, Moffitt et al. (62) demonstrated that, by detecting the positions of both trapped microspheres (x1bead and x2bead), correlations in their motions could be exploited to maximize the SNR. In fact, the difference coordinate (x− = x1bead – x2bead), with appropriate choice of experimental parameters, displays a SNR that is always superior to that of the single-trap geometry (62).
Instrumental noise
Thermal fluctuations set a lower limit for high-resolution position measurements, but many sources of instrumental noise deteriorate spatial resolution in optical tweezers. Instrumental noise originates from electronic noise in the position detector, from mechanical oscillations, from thermal expansion and contraction in the microscope (thermal drifts), and from laser pointing and intensity instabilities. Several strategies have been developed in the last decade to limit instrumental noise and reach spatial resolutions near the thermal noise limit, either using active or passive stabilization strategies.
High-resolution measurements in surface-coupled geometries (Fig. 2, a, b, and d–f) require stabilization of both the trapping laser and the microscope stage. Active stabilization of the microscope stage was obtained through a feedback system that monitors movements of the stage using a fiducial mark attached to the coverslip surface and compensates such movements using piezo translators. Simple video microscopy was used to monitor movements of a bead stuck to a microscope coverslip and compensate thermal drifts to <1 nm in three dimensions (63). Nanometer stability of the trapping laser was achieved with a simple design to minimize air turbulence and laser intensity fluctuations (63,64). Steffen et al. (65) developed such a nanometer-stabilized system to reveal the distribution of binding sites of a single myosin molecule along an actin filament. A much more complex approach is required to reach Ångstrom stabilization, which is necessary to measure single basepair movements of nucleic acid processing enzymes. Carter et al. (66) used a second laser source and back-focal-plane interferometry to monitor the movements of a fiducial mark microfabricated onto the chamber surface and thereby correct thermal drifts using a piezo stage down to ∼0.1 nm. Stabilization of the trapping laser was achieved using an optical fiber to convert laser pointing noise into intensity noise and actively stabilizing it with a feedback loop involving an acousto-optic modulator. Combining stabilization of the sample and the trapping laser, they demonstrated sensitivity to single basepair steps in a surface-coupled DNA assay (Fig. 4, a–c) (66,67).
Figure 4.
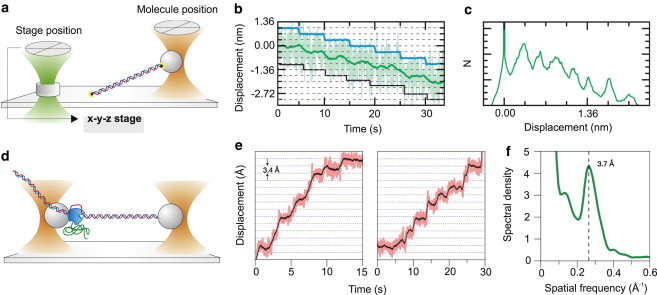
Strategies to reduce instrumental noise. (a) Active stabilization of a surface coupled assay (67). A laser beam (green) was focused on a fiducial mark (transparent cylinder) bound to the coverslip to measure stage drifts, which were compensated by a feedback system driving a piezo stage. (b) To demonstrate the resolution of the system illustrated in panel a, the stage was moved in 0.34-nm increments (blue) such that the apparent DNA contour length changed. A step-fitting algorithm found steps (black) at 0.33 ± 0.08 nm. Data filtered at 5 Hz (light green) and 0.2 Hz (dark green). (c) A pairwise distance distribution of the 0.2-Hz data from panel b show a peak at 0.31 ± 0.09 nm. (d) Passive stabilization via a suspended double-trap assay. The picture represents the experiment by Abbondanzieri et al. (40), in which a single molecule of RNA polymerase (blue) was attached to the left bead held in the trap and tethered via the upstream DNA to the right bead held in a second trap. (e) Representative records from Abbondanzieri et al. (40) for single molecules of RNAP transcribing <18 pN of assisting load, median-filtered at 50 ms (pink) and 750 ms (black). Horizontal lines (dotted) are spaced at 3.4 Å intervals. (f) In Abbondanzieri et al. (40), the position histograms for 37 segments derived from transcription records for 28 individual RNAP molecules were computed and the autocorrelation function was calculated for each of these. The power spectral density of the averaged autocorrelation function showed a peak at the dominant spatial frequency, corresponding to the inverse of the fundamental step size, 3.7 ± 0.6 Å.
An alternative approach consists of decoupling the biological system under study from the coverslip surface by suspending it between two optical tweezers, which are stabilized to the required accuracy level. Abbondanzieri et al. (40) reduced laser pointing noise induced by air turbulence by enclosing the laser path within a closed chamber filled with a gas at low refractive index and temperature-stabilizing the experimental apparatus down to 0.1 K variation. Reduction of instrumental noise to < 1Å usi this approach allowed them to probe single basepair stepping by RNA polymerase (Fig. 4, d–f). Using similar double optical tweezers, Cheng et al. (68) could measure single-basepair unwinding of double-stranded RNA by the hepatitis C virus NS3 helicase. In this experiment, the setup was temperature-stabilized, employed a low-noise laser (69) and the differential detection described in the previous section (62).
Optical Tweezers with Microsecond Temporal Resolution
Optical tweezers are affected by various limitations in their capacity to temporally resolve events; the term “temporal resolution” can, thus, refer to different physical quantities and vary depending on the molecule under investigation. Position data can be sampled at high frequencies (hundreds of kiloHertz), limited by the temporal response of the photodetector and electronics. Temporal resolution of the position detector, however, is rarely the limiting factor in optical tweezers measurements.
Relaxation time
A lower limit on temporal resolution of optical tweezers is given by the relaxation time of the system. Let us consider again the arrangement depicted in Fig. 3 a, where a micrometer-sized bead is trapped in optical tweezers and linked to a biological molecule, immersed in a viscous solution. When this system is perturbed from equilibrium, for example by protein conformational changes or by trap displacements, it moves exponentially to a new equilibrium position with a time constant (relaxation time) τ = γ/k where γ is the viscous drag coefficient and k the stiffness of the system (9). Therefore, systems with high stiffness attached to small probes exhibit fast responses to perturbations. If the perturbation develops faster than τ, the bead moves with the same relaxation time τ, filtering out all the movements that occur on shorter timescales.
For a surface-coupled geometry, the stiffness of the surface-coupled molecule and the stiffness of the optical tweezers act in parallel and sum with each other (k = ktrap + kmotor in Fig. 3 a). Therefore, rigid molecules also display fast temporal responses even in the presence of weak traps. In principle, relaxation times of ∼2 μs can be reached on rigid molecules (1 pN/nm) using small beads (200-nm diameter). In practice, the rigidity of molecules and linkages to the beads are nonlinear and usually require high tensioning to reach stiffness values of ∼1 pN/nm. Moreover, the force exerted by optical tweezers and the resolution of the position detector both decrease with bead size (15), making the use of small beads difficult. Nishiyama et al. (70) developed a dark-field position detection scheme with increased SNR that allowed them to use 200-nm diameter beads in an optical trap. By applying tension to a single kinesin motor proceeding along a microtubule to increase its stiffness, they obtained a response time τ < 20 μs at forces above 3 pN. The high temporal resolution allowed them to detect 4-nm substeps within the 8-nm step of kinesin. Uemura et al. (71) used the same assay to study the stepping of myosin Va at high spatio-temporal resolution, revealing two pathways for the 36-nm steps, one of them composed by 12- and 24-nm substeps. A modified setup was used by Iwaki et al. (72) to tether a single-headed myosin VI to an optically trapped 200-nm bead and rapidly scan the bead along an actin filament. Using this assay, they could observe weak and strong binding of myosin VI heads to actin, and they found that strong binding was greatly enhanced when backward strain was applied.
Trap and molecular stiffness sum up differently in surface-uncoupled geometries such as the two-bead assay depicted in Fig. 2 c and Fig. 4 d. Here, the stiffness of the molecule and the stiffness of one of the two traps act in series, and this combined elastic element acts in parallel with the second trap. As a consequence, even for very rigid molecules, the system stiffness k is always smaller than the combined stiffness of the traps (2ktrap), and the relaxation time is dictated by the trap stiffness.
Thermal noise
Relaxation time constitutes a lower limit to time resolution in optical tweezers, but usually thermal fluctuations pose a more severe limitation on the time resolution achievable in position measurement. In fact, as discussed in the previous section, to distinguish single basepair steps with sufficient SNR, a sufficiently small bandwidth has to be chosen to reduce thermal noise (Eq. 4). A compromise between spatial and temporal resolution is thus necessary. For example, in the conditions reported by Abbondanzieri et al. (40) (r = 350 nm, ktrap = 1.9 pN/nm, Δf = 100 Hz), from Eq. 2 we get a thermal position noise of ∼0.5 Å, close to the ∼1 Å position noise reported. Therefore, the capacity to measure subnanometer displacements from single molecules requires averaging over several tens or hundreds of milliseconds, thus reducing measurement bandwidth and temporal resolution.
Dead time
In general, the above considerations apply when measuring steps of molecular motors and conformational changes or folding trajectories of single molecules. However, additional temporal limitations arise when observing weak molecular bonds or nonprocessive motors, which are characterized by short interaction lifetimes. Nonprocessive motors bind to their track for just one ATP cycle that, under physiological conditions, has a duration on the order of milliseconds. Protein conformational changes or working strokes of motor proteins are usually associated with the formation of molecular bonds and follow binding on the submillisecond timescale. Therefore, an efficient strategy for the detection of brief interaction events and rapid application of loads is necessary to allow the investigation of the kinetics and mechanics of these biological processes.
Methods for the detection of interactions of nonprocessive motors were first developed in studies on muscle myosin in the three-bead geometry (Fig. 2 b). The detection strategy was based on the variation of thermal noise upon binding, which, in turn, depends on the variation of system stiffness upon binding, as described by Eq. 2 in the low-frequency region (f << fc) (21,73,74). When myosin binds to actin, the system stiffness increases and thermal fluctuations at low frequencies consequently decrease (as represented by the turquoise area under the blue and red power spectra in Fig. 3 c). The SNR between thermal noise in the unbound and bound states σxunbound/σxbound = kbound/kunbound can be as high as ∼25 by using weak traps (kunbound ∼ 2ktrap ∼ 0.04 pN/nm) and by high pretensioning of the actin filament (kbound ∼ kmyosin ∼ 1 pN/nm) (22,73). However, if the position variance is measured at f > fc, the SNR decreases with frequency and eventually vanishes (as represented by the pink area in Fig. 3 c, which is the same for the blue and red power spectra). For this reason, position variance in this kind of experiment was calculated using time windows Δt > 5 ms (73), and events with durations below this value could not be detected. This strategy, therefore, sets a temporal limit Δt (the dead time) for detection of molecular interactions.
Veigel et al. (75) developed a technique to decrease the dead time to ∼1 ms. The idea was to artificially increase the position noise at a high frequency by oscillating one trap at 1 KHz and detecting the amplitude of oscillation at the other bead, which was greatly reduced upon myosin binding. This method enabled them to detect the onset of each binding event within ∼1–2 ms. By monitoring the amplitude of this signal using an analog electronic detection circuit, they were also able to apply a range of loads to a single myosin head with a time delay of ∼3 ms after detection of attachment. They investigated the effects of load on the kinetics of smooth muscle myosin (76) and myosin Va (77). These experiments made a great contribution to contemporary understanding of how load regulates myosin motors function and how it plays a role in coordination of the two motor domains in processive motors. The several-millisecond delay of this technique, however, did not allow the study of load-dependence of faster processes, such as the myosin working stroke. Moreover, techniques capable of clamping force on nonprocessive motors and weak molecular bonds were still lacking.
Ultrafast force-clamp spectroscopy
A different approach, named ultrafast force-clamp spectroscopy, recently allowed application of constant loads between a single intermittently interacting biological polymer and a binding protein with a delay in application of the load of only ∼10 μs (78).
A sketch of the operational principle of the method is shown in Fig. 5 a, in which A is the binding protein, and B is the polymer. A net constant force (Ftot = +ΔF) is applied to the bead-polymer-bead complex (dumbbell) through two feedback systems that clamp the force on the two beads to two different values (−F on the left bead, F+ΔF on the right bead). The dumbbell, thus, moves against viscous drag at constant velocity (v = Ftot/γ) when molecules A and B are not bound. The net force is alternated in direction, so that the dumbbell oscillates in a triangular wave fashion within a limited spatial interval (Fig. 5 b). When A binds to B, the force Ftot is transferred to the surface-coupled molecule A; because the system was designed to maintain a constant force, the dumbbell movement stops, except for possible conformational changes of the molecules after binding.
Figure 5.
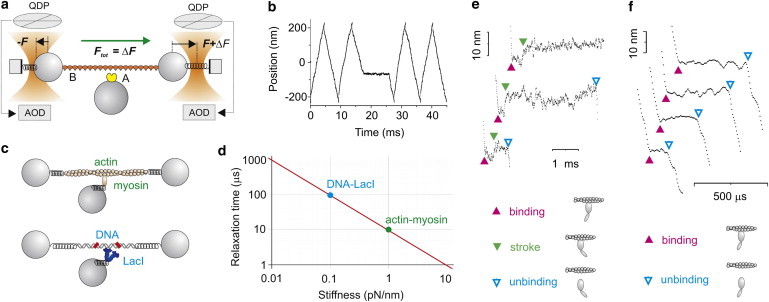
Ultrafast force-clamp spectroscopy. (a) Schematic of the operational principle of the method illustrating constant Ftot = ΔF applied to molecule B through two feedback systems clamping the force on the left to −F and on the right bead to F+ΔF. The force is measured using quadrant detector photodiodes and kept constant by moving the traps via AODs. (b) Position of the dumbbell. The net force is switched between +ΔF and −ΔF to keep the dumbbell within a confined spatial interval (±200 nm). The dumbbell stops when A binds to B. (c) Mechanical model for actin-myosin and DNA-LacI interaction. (d) Relaxation times calculated from the models in panel c using 500-nm diameter beads (78). (e) Actin-myosin interactions longer than 1 ms showed that the myosin working stroke is developed 0.2–1 ms after attachment. (Filled arrowheads pointing down) Myosin working stroke. (f) Submillisecond single actin-myosin interactions detected with ultrafast force-clamp spectroscopy. (Filled arrowheads pointing up) Actin-myosin binding. (Open arrowheads pointing down) Actin-myosin detachment.
The time taken to transfer the force from the viscous solution to molecule A and stop the dumbbell is the relaxation time of the molecular complex (78). When applied to the interaction between fast skeletal muscle myosin and actin using highly pretensioned actin filaments and 500-nm diameter beads, the transfer time was ∼10 μs. For lactose repressor interactions with a weakly tensioned DNA (∼3 pN), the transfer time was ∼100 μs (Fig. 5, c and d). This time is very short compared to the duration of typical protein interactions which, thus, occur under a real force-clamp configuration. Any conformational change occurring after the formation of the molecular bond is also performed under the same constant load (Fig. 5 e). Using this system, Capitanio et al. (78) directly measured load-dependence of the amplitude of myosin working stroke.
Another advantage of this system is that interaction events are detected through variations in the dumbbell velocity, which display a high SNR. The variation of velocity upon binding grows with the applied force and decreases with the beads size (vunbound = Ftot/γ, vbound = 0). The dead time for the detection of actin-myosin binding was ∼100 μs at ∼5 pN, using 500-nm diameter beads, which allowed the detection of weak binding events (detachment ∼5 × 103 s−1 at 5 pN; Fig. 5 f), and premature detachment of myosin from actin (∼1 × 103 s−1 at 5 pN). On Lac repressor DNA, short-lived interactions (dissociating at ∼1 × 103 s−1 at 4 pN), which are probably involved in a Lac repressor-facilitated diffusion mechanism (79,80), were detected.
Ensemble averages
Ensemble averaging is a useful technique when high spatial and temporal resolution are both required (81,82). This method enabled distinguishing two steps in the working stroke of myosin Va (81), smooth muscle myosin (76), and skeletal muscle myosin (22) and excluded the presence of substeps in kinesin (83). Instead of time-averaging position data of single events to reduce thermal noise, as prescribed by Eq. 2, in ensemble averaging N interaction events are synchronized at the beginning or at the end of the interaction and averaged point-by-point. Position noise thus scales as and can reach the Ångstrom level for N ∼ 1000 (22). Temporal resolution for this kind of average is determined by the accuracy in detecting the time of the beginning or end of the interaction (σA). Such accuracy can be evaluated through simulated data (22,83) or theoretical considerations (78). For the interaction between fast skeletal muscle myosin and actin, σA ∼ 300 μs can be obtained using a classic three-bead assay (22), and in the range 10–50 μs with ultrafast force-clamp spectroscopy (78). Methods have been developed to interpret the kinetics after an ensemble average and extract the rate constants of the different reactions within a biochemical cycle (82).
Conclusions and Perspectives
The spatial and temporal resolution improvements by optical tweezers in recent years have opened new opportunities for investigating the effects of force on biological processes. Many issues about the mechanisms of function and regulation of molecular motors and mechanoenzymes still need to be elucidated (57). The sophisticated technologies that are now available will enable the study of molecular interactions, strength of molecular bonds, and protein and nucleic acid folding with unprecedented resolution. Single-molecule in vitro techniques are just starting to be applied to proteins involved in mechanotransduction, using purely mechanical measurements (23,31,84) or combining mechanical control of a protein and fluorescence detection of its binding partners (85). A growing set of tools for the combined manipulation and localization of single molecules has been developed (64,86–88) and applied mainly to the study of nucleic acid structure (89,90) and DNA binding proteins and processing enzymes (91,92).
Novel tools are moving high-resolution measurements from in-vitro assays to living cells. Among these, molecular tension sensors based on Förster resonance energy transfer (93–95), spectral emission changes due to proximity (96), and fluorescence quenching (97) enabled pN-sensitivity measurements of intramolecular tension. Direct manipulation and force measurement in living cells with optical tweezers have also seen great progress recently, thanks to developments in calibration methods taking into account any variations among cargoes and local viscoelastic properties of the cytoplasm (98–102). All such advancements will be crucial toward providing new insights into the mechanisms of the myriad biological processes that are directly regulated by force or connected to the mechanical conditions in the cell and its surrounding environment.
Acknowledgments
We thank Lucia Gardini for discussion.
This research was funded by the European Union Seventh Framework Programme (No. FP7/2007-2013; grant Nos. B0 211383, B0 228334, and B0 241526), by the Italian Ministry of University and Research (grant No. FIRB 2011 RBAP11X42L006 and Flagship Project NANOMAX).
References
- 1.Hoffman B.D., Grashoff C., Schwartz M.A. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wozniak M.A., Chen C.S. Mechanotransduction in development: a growing role for contractility. Nat. Rev. Mol. Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaalouk D.E., Lammerding J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 2009;10:63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cecconi C., Shank E.A., Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 5.Marshall B.T., Long M., Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 6.Rief M., Rock R.S., Spudich J.A. Myosin-V stepping kinetics: a molecular model for processivity. Proc. Natl. Acad. Sci. USA. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reconditi M., Linari M., Lombardi V. The myosin motor in muscle generates a smaller and slower working stroke at higher load. Nature. 2004;428:578–581. doi: 10.1038/nature02380. [DOI] [PubMed] [Google Scholar]
- 8.Laakso J.M., Lewis J.H., Ostap E.M. Myosin I can act as a molecular force sensor. Science. 2008;321:133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howard J. Sinauer Associates; Sunderland, MA: 2001. Mechanics of Motor Proteins and the Cytoskeleton. [Google Scholar]
- 10.Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashkin A., Dziedzic J.M., Chu S. Observation of a single-beam gradient force optical trap for dielectric particles. Opt. Lett. 1986;11:288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 12.Fazal F.M., Block S.M. Optical tweezers study life under tension. Nat. Photonics. 2011;5:318–321. doi: 10.1038/nphoton.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenleaf W.J., Woodside M.T., Block S.M. Passive all-optical force clamp for high-resolution laser trapping. Phys. Rev. Lett. 2005;95:208102. doi: 10.1103/PhysRevLett.95.208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gittes F., Schmidt C.F. Signals and noise in micromechanical measurements. Methods Cell Biol. 1998;55:129–156. doi: 10.1016/s0091-679x(08)60406-9. [DOI] [PubMed] [Google Scholar]
- 15.Capitanio M., Romano G., Finzi L. Calibration of optical tweezers with differential interference contrast signals. Rev. Sci. Instrum. 2002;73:1687–1696. [Google Scholar]
- 16.Neuman K.C., Block S.M. Optical trapping. Rev. Sci. Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svoboda K., Schmidt C.F., Block S.M. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 18.Svoboda K., Block S.M. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 19.Yin H., Wang M.D., Gelles J. Transcription against an applied force. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- 20.Finer J.T., Simmons R.M., Spudich J.A. Single myosin molecule mechanics: picoNewton forces and nanometer steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 21.Molloy J.E., Burns J.E., White D.C.S. Movement and force produced by a single myosin head. Nature. 1995;378:209–212. doi: 10.1038/378209a0. [DOI] [PubMed] [Google Scholar]
- 22.Capitanio M., Canepari M., Bottinelli R. Two independent mechanical events in the interaction cycle of skeletal muscle myosin with actin. Proc. Natl. Acad. Sci. USA. 2006;103:87–92. doi: 10.1073/pnas.0506830102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren Y., Effler J.C., Robinson D.N. Mechanosensing through cooperative interactions between myosin II and the actin crosslinker cortexillin I. Curr. Biol. 2009;19:1421–1428. doi: 10.1016/j.cub.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter W.J., Brenner B., Steffen W. Cytoplasmic dynein is not a conventional processive motor. J. Struct. Biol. 2010;170:266–269. doi: 10.1016/j.jsb.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 25.Walter W.J., Koonce M.P., Steffen W. Two independent switches regulate cytoplasmic dynein’s processivity and directionality. Proc. Natl. Acad. Sci. USA. 2012;109:5289–5293. doi: 10.1073/pnas.1116315109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.deCastro M.J., Fondecave R.M., Stewart R.J. Working strokes by single molecules of the kinesin-related microtubule motor ncd. Nat. Cell Biol. 2000;2:724–729. doi: 10.1038/35036357. [DOI] [PubMed] [Google Scholar]
- 27.Smith S.B., Cui Y., Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 28.Liphardt J., Onoa B., Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 29.Gebhardt J.C., Bornschlögl T., Rief M. Full distance-resolved folding energy landscape of one single protein molecule. Proc. Natl. Acad. Sci. USA. 2010;107:2013–2018. doi: 10.1073/pnas.0909854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stigler J., Ziegler F., Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 31.Rognoni L., Stigler J., Rief M. Dynamic force sensing of filamin revealed in single-molecule experiments. Proc. Natl. Acad. Sci. USA. 2012;109:19679–19684. doi: 10.1073/pnas.1211274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visscher K., Schnitzer M.J., Block S.M. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 33.Rock R.S., Rice S.E., Sweeney H.L. Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci. USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang M.D., Schnitzer M.J., Block S.M. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 35.Bianco P., Bongini L., Lombardi V. PicoNewton-millisecond force steps reveal the transition kinetics and mechanism of the double-stranded DNA elongation. Biophys. J. 2011;101:866–874. doi: 10.1016/j.bpj.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lang M.J., Asbury C.L., Block S.M. An automated two-dimensional optical force clamp for single molecule studies. Biophys. J. 2002;83:491–501. doi: 10.1016/S0006-3495(02)75185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valentine M.T., Guydosh N.R., Block S.M. Precision steering of an optical trap by electro-optic deflection. Opt. Lett. 2008;33:599–601. doi: 10.1364/ol.33.000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elms P.J., Chodera J.D., Marqusee S. Limitations of constant-force-feedback experiments. Biophys. J. 2012;103:1490–1499. doi: 10.1016/j.bpj.2012.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nambiar R., Gajraj A., Meiners J.C. All-optical constant-force laser tweezers. Biophys. J. 2004;87:1972–1980. doi: 10.1529/biophysj.103.037697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abbondanzieri E.A., Greenleaf W.J., Block S.M. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodside M.T., Anthony P.C., Block S.M. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenleaf W.J., Frieda K.L., Block S.M. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takagi Y., Homsher E.E., Shuman H. Force generation in single conventional actomyosin complexes under high dynamic load. Biophys. J. 2006;90:1295–1307. doi: 10.1529/biophysj.105.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piazzesi G., Lucii L., Lombardi V. The size and the speed of the working stroke of muscle myosin and its dependence on the force. J. Physiol. 2002;545:145–151. doi: 10.1113/jphysiol.2002.028969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laakso J.M., Lewis J.H., Ostap E.M. Control of myosin-I force sensing by alternative splicing. Proc. Natl. Acad. Sci. USA. 2010;107:698–702. doi: 10.1073/pnas.0911426107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greenberg M.J., Lin T., Ostap E.M. Myosin IC generates power over a range of loads via a new tension-sensing mechanism. Proc. Natl. Acad. Sci. USA. 2012;109:E2433–E2440. doi: 10.1073/pnas.1207811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans E. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 48.Guo B., Guilford W.H. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc. Natl. Acad. Sci. USA. 2006;103:9844–9849. doi: 10.1073/pnas.0601255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewalle A., Steffen W., Sleep J. Single-molecule measurement of the stiffness of the rigor myosin head. Biophys. J. 2008;94:2160–2169. doi: 10.1529/biophysj.107.119396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arya M., Kolomeisky A.B., Anvari B. Dynamic force spectroscopy of glycoprotein Ib-IX and von Willebrand factor. Biophys. J. 2005;88:4391–4401. doi: 10.1529/biophysj.104.046318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dame R.T., Noom M.C., Wuite G.J.L. Bacterial chromatin organization by H-NS protein unraveled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 52.Bianco P., Nagy A., Kellermayer M.S. Interaction forces between F-actin and titin PEVK domain measured with optical tweezers. Biophys. J. 2007;93:2102–2109. doi: 10.1529/biophysj.107.106153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pyrpassopoulos S., Shuman H., Ostap E.M. Single-molecule adhesion forces and attachment lifetimes of myosin-I phosphoinositide interactions. Biophys. J. 2010;99:3916–3922. doi: 10.1016/j.bpj.2010.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Messieres M., Chang J.C., La Porta A. Single-molecule study of G-quadruplex disruption using dynamic force spectroscopy. Phys. Rev. Lett. 2012;109:058101. doi: 10.1103/PhysRevLett.109.058101. [DOI] [PubMed] [Google Scholar]
- 55.Dudko O.K., Hummer G., Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc. Natl. Acad. Sci. USA. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim J., Zhang C.Z., Springer T.A. A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature. 2010;466:992–995. doi: 10.1038/nature09295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veigel C., Schmidt C.F. Moving into the cell: single-molecule studies of molecular motors in complex environments. Nat. Rev. Mol. Cell Biol. 2011;12:163–176. doi: 10.1038/nrm3062. [DOI] [PubMed] [Google Scholar]
- 58.Greenleaf W.J., Woodside M.T., Block S.M. High-resolution, single-molecule measurements of biomolecular motion. Annu. Rev. Biophys. Biomol. Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allersma M.W., Gittes F., Schmidt C.F. Two-dimensional tracking of ncd motility by back focal plane interferometry. Biophys. J. 1998;74:1074–1085. doi: 10.1016/S0006-3495(98)74031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gittes F., Schmidt C.F. Interference model for back-focal-plane displacement detection in optical tweezers. Opt. Lett. 1998;23:7–9. doi: 10.1364/ol.23.000007. [DOI] [PubMed] [Google Scholar]
- 61.Gittes F., Schmidt C.F. Thermal noise limitations on micromechanical experiments. Eur. Biophys. J. Biophys. 1998;27:75–81. [Google Scholar]
- 62.Moffitt J.R., Chemla Y.R., Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc. Natl. Acad. Sci. USA. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Capitanio M., Cicchi R., Pavone F.S. Position control and optical manipulation for nanotechnology applications. Eur. Phys. J. B. 2005;46:1–8. [Google Scholar]
- 64.Capitanio M., Maggi D., Pavone F. Fiona in the trap: the advantages of combining optical tweezers and fluorescence. J. Opt. A, Pure Appl. Opt. 2007;9:S157. [Google Scholar]
- 65.Steffen W., Smith D., Sleep J. Mapping the actin filament with myosin. Proc. Natl. Acad. Sci. USA. 2001;98:14949–14954. doi: 10.1073/pnas.261560698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carter A.R., King G.M., Perkins T.T. Stabilization of an optical microscope to 0.1 nm in three dimensions. Appl. Opt. 2007;46:421–427. doi: 10.1364/ao.46.000421. [DOI] [PubMed] [Google Scholar]
- 67.Carter A.R., Seol Y., Perkins T.T. Precision surface-coupled optical-trapping assay with one-basepair resolution. Biophys. J. 2009;96:2926–2934. doi: 10.1016/j.bpj.2008.12.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng W., Arunajadai S.G., Bustamante C. Single-base pair unwinding and asynchronous RNA release by the hepatitis C virus NS3 helicase. Science. 2011;333:1746–1749. doi: 10.1126/science.1206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng W., Hou X., Ye F. Use of tapered amplifier diode laser for biological-friendly high-resolution optical trapping. Opt. Lett. 2010;35:2988–2990. doi: 10.1364/OL.35.002988. [DOI] [PubMed] [Google Scholar]
- 70.Nishiyama M., Muto E., Higuchi H. Substeps within the 8-nm step of the ATPase cycle of single kinesin molecules. Nat. Cell Biol. 2001;3:425–428. doi: 10.1038/35070116. [DOI] [PubMed] [Google Scholar]
- 71.Uemura S., Higuchi H., Ishiwata S. Mechanochemical coupling of two substeps in a single myosin V motor. Nat. Struct. Mol. Biol. 2004;11:877–883. doi: 10.1038/nsmb806. [DOI] [PubMed] [Google Scholar]
- 72.Iwaki M., Iwane A.H., Yanagida T. Brownian search-and-catch mechanism for myosin-VI steps. Nat. Chem. Biol. 2009;5:403–405. doi: 10.1038/nchembio.171. [DOI] [PubMed] [Google Scholar]
- 73.Smith D.A., Steffen W., Sleep J. Hidden-Markov methods for the analysis of single-molecule actomyosin displacement data: the variance-Hidden-Markov method. Biophys. J. 2001;81:2795–2816. doi: 10.1016/S0006-3495(01)75922-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Knight A.E., Veigel C., Molloy J.E. Analysis of single-molecule mechanical recordings: application to acto-myosin interactions. Prog. Biophys. Mol. Biol. 2001;77:45–72. doi: 10.1016/s0079-6107(01)00010-4. [DOI] [PubMed] [Google Scholar]
- 75.Veigel C., Coluccio L.M., Molloy J.E. The motor protein myosin-I produces its working stroke in two steps. Nature. 1999;398:530–533. doi: 10.1038/19104. [DOI] [PubMed] [Google Scholar]
- 76.Veigel C., Molloy J.E., Kendrick-Jones J. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat. Cell Biol. 2003;5:980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- 77.Veigel C., Schmitz S., Sellers J.R. Load-dependent kinetics of myosin-V can explain its high processivity. Nat. Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 78.Capitanio M., Canepari M., Pavone F.S. Ultrafast force-clamp spectroscopy of single molecules reveals load dependence of myosin working stroke. Nat. Methods. 2012;9:1013–1019. doi: 10.1038/nmeth.2152. [DOI] [PubMed] [Google Scholar]
- 79.Berg O.G., Winter R.B., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 80.Monico C., Capitanio M., Pavone F.S. Optical methods to study protein-DNA interactions in vitro and in living cells at the single-molecule level. Int. J. Mol. Sci. 2013;14:3961–3992. doi: 10.3390/ijms14023961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Veigel C., Wang F., Molloy J.E. The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 82.Chen C., Greenberg M.J., Shuman H. Kinetic schemes for post-synchronized single molecule dynamics. Biophys. J. 2012;102:L23–L25. doi: 10.1016/j.bpj.2012.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carter N.J., Cross R.A. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 84.Ferrer J.M., Lee H., Lang M.J. Measuring molecular rupture forces between single actin filaments and actin-binding proteins. Proc. Natl. Acad. Sci. USA. 2008;105:9221–9226. doi: 10.1073/pnas.0706124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.del Rio A., Perez-Jimenez R., Sheetz M.P. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ishijima A., Kojima H., Yanagida T. Simultaneous observation of individual ATPase and mechanical events by a single myosin molecule during interaction with actin. Cell. 1998;92:161–171. doi: 10.1016/s0092-8674(00)80911-3. [DOI] [PubMed] [Google Scholar]
- 87.Lang M.J., Fordyce P.M., Block S.M. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat. Methods. 2004;1:133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Comstock M.J., Ha T., Chemla Y.R. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat. Methods. 2011;8:335–340. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Mameren J., Gross P., Peterman E.J. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc. Natl. Acad. Sci. USA. 2009;106:18231–18236. doi: 10.1073/pnas.0904322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hohng S., Zhou R., Ha T. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the Holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Candelli A., Wuite G.J., Peterman E.J. Combining optical trapping, fluorescence microscopy and micro-fluidics for single molecule studies of DNA-protein interactions. Phys. Chem. Chem. Phys. 2011;13:7263–7272. doi: 10.1039/c0cp02844d. [DOI] [PubMed] [Google Scholar]
- 92.Zhou R., Kozlov A.G., Ha T. SSB functions as a sliding platform that migrates on DNA via reptation. Cell. 2011;146:222–232. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meng F., Suchyna T.M., Sachs F. A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ. FEBS J. 2008;275:3072–3087. doi: 10.1111/j.1742-4658.2008.06461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grashoff C., Hoffman B.D., Schwartz M.A. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Borghi N., Sorokina M., Dunn A.R. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc. Natl. Acad. Sci. USA. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iwai S., Uyeda T.Q. Visualizing myosin-actin interaction with a genetically-encoded fluorescent strain sensor. Proc. Natl. Acad. Sci. USA. 2008;105:16882–16887. doi: 10.1073/pnas.0805513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stabley D.R., Jurchenko C., Salaita K.S. Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nat. Methods. 2012;9:64–67. doi: 10.1038/nmeth.1747. [DOI] [PubMed] [Google Scholar]
- 98.Fischer M., Berg-Sorensen K. Calibration of trapping force and response function of optical tweezers in viscoelastic media. J. Opt. A, Pure Appl. Opt. 2007;9:S239–S250. [Google Scholar]
- 99.Fischer M., Richardson A.C., Berg-Sørensen K. Active-passive calibration of optical tweezers in viscoelastic media. Rev. Sci. Instrum. 2010;81:015103. doi: 10.1063/1.3280222. [DOI] [PubMed] [Google Scholar]
- 100.Rai A.K., Rai A., Mallik R. Molecular adaptations allow dynein to generate large collective forces inside cells. Cell. 2013;152:172–182. doi: 10.1016/j.cell.2012.11.044. [DOI] [PubMed] [Google Scholar]
- 101.Hendricks A.G., Holzbaur E.L., Goldman Y.E. Force measurements on cargoes in living cells reveal collective dynamics of microtubule motors. Proc. Natl. Acad. Sci. USA. 2012;109:18447–18452. doi: 10.1073/pnas.1215462109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blehm B.H., Schroer T.A., Selvin P.R. In vivo optical trapping indicates kinesin’s stall force is reduced by dynein during intracellular transport. Proc. Natl. Acad. Sci. USA. 2013;110:3381–3386. doi: 10.1073/pnas.1219961110. [DOI] [PMC free article] [PubMed] [Google Scholar]


