Abstract
We studied 11 new kindreds with primary pigmented nodular adrenocortical disease (PPNAD) or Carney complex (CNC) and found that 82% of the kindreds had PRKAR1A gene defects (including seven novel inactivating mutations), most of which led to nonsense mRNA and, thus, were not expressed in patients’ cells. However, a previously undescribed base substitution in intron 6 (exon 6 IVS +1G→T) led to exon 6 skipping and an expressed shorter PRKAR1A protein. The mutant protein was present in patients’ leukocytes and tumors, and in vitro studies indicated that the mutant PRKAR1A activated cAMP-dependent protein kinase A (PKA) signaling at the nuclear level. This is the first demonstration of an inactivating PRKAR1A mutation being expressed at the protein level and leading to stimulation of the PKA pathway in CNC patients. Along with the lack of allelic loss at the PRKAR1A locus in most of the tumors from this kindred, these data suggest that alteration of PRKAR1A function (not only its complete loss) is sufficient for augmenting PKA activity leading to tumorigenesis in tissues affected by CNC.
Carney complex (CNC [MIM 160980]) is a familial multiple neoplasia syndrome transmitted as an autosomal dominant trait (Carney et al. 1986). CNC was initially described as the association of myxomas, spotty skin pigmentation, and endocrine overactivity (Carney et al. 1985). A variety of endocrine and nonendocrine tumors occur in patients with CNC (Carney 1990; Carney and Toorkey 1991; Carney and Stratakis 1996, 1998; Premkumar et al. 1997; Stratakis et al. 1997; Pack et al. 2000; Raff et al. 2000; Stratakis et al. 2000; Carney et al. 2001). Primary pigmented nodular adrenocortical disease (PPNAD), a rare cause of ACTH-independent Cushing syndrome, is the main endocrine manifestation of CNC. PPNAD is observed in one-fourth of patients with CNC (Stratakis et al. 1999, 2001).
Approximately half of the cases of CNC are familial (Stratakis et al. 2001). Putative genetic loci have been identified by linkage analysis at chromosome 2p16 and 17q22-24 (Stratakis et al. 1996; Casey et al. 1998). Recently, the responsible gene on 17q22-24, PRKAR1A, was identified (Kirschner et al. 2000a); additional mutations in this gene were described later in other kindreds (Casey et al. 2000; Kirschner et al. 2000b). PRKAR1A encodes the type 1α regulatory subunit of cAMP-dependent protein kinase A (PKA). Overall, inactivating mutations of this gene have been observed in ∼41% of CNC kindreds (Kirschner et al. 2000b).
All of the PRKAR1A defects reported so far are functionally null mutations. All sequence changes were predicted to cause premature stop codons, with the exception of one mutation that altered the transcriptional start site (at the ATG codon) (Kirschner et al. 2000b). It was then demonstrated that mutant mRNAs bearing a premature stop codon were unstable, as a result of nonsense-mediated mRNA decay (NMD) (Kirschner et al. 2000b). It is interesting that PRKAR1A seems to function as a classic tumor-suppressor gene in tumors from CNC patients as demonstrated by loss of the normal allele in CNC lesions (Kirschner et al. 2000a). Loss of the normal PRKAR1A protein and NMD of the mutant allele in these studies suggested that oncogenesis in CNC tumors was due to the complete absence of a functional PRKAR1A.
In the present study, 11 kindreds with CNC, in which almost all index cases were referred for Cushing syndrome caused by PPNAD, were investigated for PRKAR1A germline mutations. Family members were evaluated by a thorough history and physical exam. Affection status was determined on the basis of the diagnostic criteria proposed recently (Stratakis et al. 2001) and as reported by Groussin et al. (2002). Clinical data of all the index cases are shown in table 1.
Table 1.
Clinical Manifestations among the Index Cases [Note]
| Family | Lentigines | HeartMyxoma | PPNAD | Endocrine Manifestations | Other |
| CNC07 | + | + | |||
| CNC03 | + | + | |||
| CNC09 | + | + | GH-PRL pituitary-producing adenoma | Ethmoidal osteochondromyxoma; PMS; blue nevus; breast ductal adenoma | |
| CNC04 | + | + | + | Toxic multinodular goiter; ovarian cyst | Benign hepatoma; PMS; breast ductal adenoma; splenic lipoma; skin myxoma; mesenteric myxoma; pelvic leiomyoma; pancreatic acinar adenocarcinoma |
| CNC08 | + | Ovarian cyst | Blue nevus | ||
| CNC06 | + | + | + | ||
| CNC02 | + | + | Calcifications on testicular ultrasonography | Blue nevus | |
| CNC01 | + | + | + | Thyroid tumor Acromegaly | Benign hepatoma |
| CNC05 | + | + | |||
| CNC10 | + | + | |||
| CNC11 | + | Ovarian cyst | PMS; breast ductal adenoma |
Note.— Abbreviations: +=present; GH=growth hormone; PRL=prolactin; PMS=psammomatous melanotic schwannoma.
DNA was extracted as reported elsewhere (Groussin et al. 2002), and the 12 exons and the flanking intronic sequences of the PRKAR1A gene were separately PCR amplified using the primers and the conditions described elsewhere for exons 1A, 1B, 2, and 7 (Kirschner et al. 2000b) and the oligonucleotides listed in table 2 for exons 3, 4A, 4B, 5, 6, 8, 9, and 10. Sequencing was performed as reported elsewhere (Groussin et al. 2002). Ethnically matched controls (N=90) and the CEPH collection of DNA samples (N=100) were tested for PRKAR1A pathogenic disease-causing mutations; none was found, although some of the common polymorphisms of this gene were present (data not shown).
Table 2.
Novel PRKAR1A Oligonucleotide Sequences for Mutation Detection
| Exon and Direction | Sequence |
| Exon 3 | |
| Sense: | 5′- GAATTGGTGTTTTCCTCTTAACTT-3′ |
| Antisense: | 5′-TATGATTCATTCATCAAAGGAGAC-3′ |
| Exon 4A | |
| Sense: | 5'-AATGTTTTTGGTTTATGGAATTGT-3′ |
| Antisense: | 5′-CACACCCTTACTTGAAAAATAGTG-3′ |
| Exon 4B | |
| Sense: | 5′-GACAGTCTGGGGTCTTTAATTCTA-3′ |
| Antisense: | 5′-TCAAAGAGGAAAACAAACTTCAAT-3′ |
| Exon 5 | |
| Sense: | 5′-TTTCTTTAATTTGGAATATGCTTC-3′ |
| Antisense: | 5′-ATCTGACATACAAGGGATGTAATG-3′ |
| Exon 6 | |
| Sense: | 5′-TTTTTAAAACAAAGTTCAGGATTG-3′ |
| Antisense: | 5′-CTAAATCACACTCTCAAACACCAT-3′ |
| Exon 8 | |
| Sense: | 5′-GGCTATTTGGTTGAATCTCTTTAT-3′ |
| Antisense: | 5′-TGAGTTCTTTACCTCTAAAATTCAA-3′ |
| Exon 9 | |
| Sense: | 5′-TTGTTTAGCTTTTTGGTGATTTTA-3′ |
| Antisense: | 5′-GGAGAAGACAAAATTATGGAAGAC-3′ |
| Exon 10 | |
| Sense: | 5′-TATTGTCTTCTTTCTCAGAAGTGC-3′ |
| Antisense: | 5′-GTGCAATAAAAGCAACTTTCAATA-3′ |
To prepare total cellular protein extracts, cultured lymphocytes were harvested and pelleted. Tumor tissues were obtained as reported elsewhere (Groussin et al. 2000). Protein assays were performed using the protein assay kit (Bio-Rad Laboratories). Equivalent protein concentrations were resolved by electrophoresis on 10% SDS-polyacrylamide gel and were transferred to nitrocellulose sheets. Western blotting was performed with primary mouse antibody to the RIα subunit (1/250; Becton Dickinson Transduction Laboratories). For detection of the first antibody with a goat anti-mouse IgG antibody (1/5000; Santa Cruz; sc-2005), chemiluminescence was used.
For loss of heterozygosity (LOH) analysis, DNA from different tumors of the proband from family CNC04 was analyzed along with a paired DNA sample from peripheral blood. Seven microsatellite markers located on 17q22-24 were used: centromere-D17S807, D17S1882, D17S1813, PRKAR1A(CA)n, D17S795, D17S789, and D17S840-telomere. The sequences of the primers and genomic order of their loci were derived from the publicly available genomic databases (Genome Database; Whitehead Institute MIT, Center for Genome Research Web site). Markers were PCR amplified with end labeled 32P-radiolabeled oligonucleotide primers, and 30 cycles were performed (95°C for 30 s, 51°C for 40 s, 72°C for 40 s), followed by a final 5-min extension at 72°C. Aliquots of amplified DNA were mixed with an equal volume of loading buffer, were denatured at 94°C for 5 min, and were electrophoresed on a 6% polyacrylamide gel. Gels were dried and placed on Kodak X-Omat films. Marker PRKAR1A(CA)n PCR fragments were analyzed by electrophoresis through 4% polyacrylamide gel and were visualized with ultraviolet light after staining with ethidium bromide.
A PCR-cloning method was used to construct both wild-type and exon 6–skipping mutant expression constructs. Total RNA was extracted from peripheral blood leukocytes of family CNC04 proband using RNABle (Eurobio). cDNA was synthesized by Moloney murine leukemia virus-reverse transcriptase (Invitrogen). RIα cDNA from lymphocytes was amplified using the primers 5′-GAG CAA AGC GCT GAG GGA GCT C-3′ (sense) and 5′-AAG CAT GGA TTG GGG AGA GGA G-3′ (antisense). The reaction performed with Expand Long Template PCR System (Roche) consisted of 2 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 58°C, and 2 min at 72°C. The PCR fragments corresponding to the full-length wild-type RIα cDNA and the natural mutant with the exon 6 skipping were electrophoresed on agarose gel and were purified by using a QIA quick gel extraction kit (Qiagen). These were introduced into the pGEM-T easy vector by using a TA cloning kit (Promega). The two constructs were verified by sequencing before a second PCR amplification was performed using Pwo DNA Polymerase (Roche). Each sample was incubated successively at 94°C for 15 s, 63°C for 30 s, 72°C for 1 min, for a total of 30 cycles, followed by a final extension at 72°C for 7 min. The following specific oligonucleotide primers were used: HA-RIα sense primer containing a HindIII site with the hemagglutinin HA sequence (underlined): 5′-CCT CCA AGC TTG CCA CCA TGG CTT ACC CAT ACG ACG TCC CAG ACT ACG CTG AGT CTG GCA GTA CCG CCG CC-3′ or nonHA-RIα sense primer: 5′-CCT CCA AGC TTG AGA ACC ATG GAG TCT GGC-3′; a common antisense primer containing a Xho site: 5′-CCG TTC TCG AGT CAG ACA GAC AGT GAC ACA AAA CT-3′. The four products were purified by gel electrophoresis, were digested with HindIII and Xho, and were cloned into the HindIII/Xho sites of the pREP4 expression vector (Invitrogen) to create HA-RIα-WT, HA-RIα-Δ184–236, RIα-WT, and RIα-Δ184–236. All constructs were sequenced prior to their use in expression studies. Lymphocytic cell culture and cycloheximide treatment were performed as reported elsewhere (Kirschner et al. 2000b).
COS7 cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% FCS, glutamine, and gentamycine. The cells were plated in six-well dishes and were transfected 24 h later by using Lipofectamine Plus (Invitrogen) following the protocol provided. We used the luciferase reporter pSS-CRE-LUC containing a sequence of the rat somatostatin gene −71–53 (including the CRE site) inserted in the 5′-region of the luciferase gene. The reporter gene RSV-βGal (Bertherat et al. 1995) was used as an internal control for transfection efficiency. Cells were cotransfected with 0.2 μg of pSS-CRE-LUC, 0.25 μg of RSV-βGal, and 1.5 μg of plasmid expressing HA-RIα-WT or molar equivalent of HA-RIα-Δ184–236 and empty pREP4 plasmids. Six hours prior to harvesting, half of the transfected dishes were incubated in a mixture of 10−5 M forskolin (Sigma) and 0.5 mM 3-isobutyl-1-methylxanthine (Sigma). Cells were harvested 24 h after transfection and were subjected to lysis; luciferase activity was then assayed for and was normalized to β-galactosidase activity. All transfection experiments were performed in triplicate and were repeated five times; the results are expressed as the means. Statistical significance was assessed by a Student's t test (StatView 5.0., SAS Institute). Control immunoblotting analysis was performed to confirm that HA-RIα constructs had equivalent levels of expression. Total lysates from empty vector, HA-RIα-WT, or HA-RIα-Δ184–236 COS transfected cells were used in Western blotting, which then was evaluated by chemiluminescence using anti-HA antibody (Santa Cruz, sc-805).
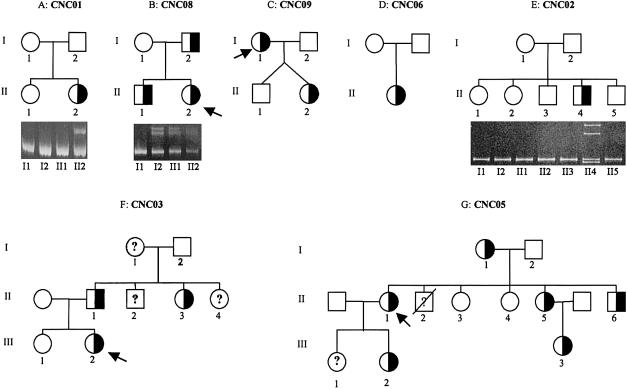
Sequencing of the 12 coding exons of the genomic DNA from the index cases revealed a PRKAR1A mutation in the heterozygotic state in 9 of the 11 kindreds (details are given in table 3 and figs. 1, 2, and 3). Seven of these mutations have not been previously reported. Four of them occurred de novo (fig. 1). The 578 delTG and the exon 8 IVS + 3 A→G mutations were reported elsewhere (Kirshner et al. 2000b). The 578 delTG mutation is the most frequent CNC mutation found to occur de novo in more than eight kindreds so far; founder effect has been excluded in most of these kindreds by extensive genotyping of chromosome 17 markers when relatives were available (data not shown).
Table 3.
PRKAR1A Mutations in Nine Families with CNC
| Family | Mutation | Effect on RIα |
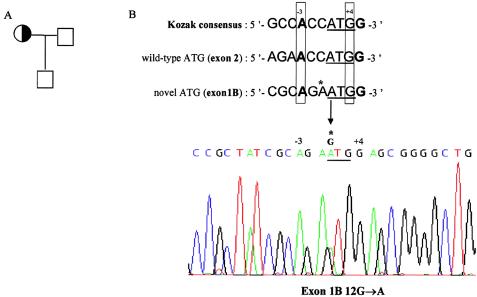
| CNC07 | Exon 1B 12G→A | Additional out-of-frame ATG initiation codon within a consensus sequence for translation initiation; could abolish translation of the wild-type protein |
| CNC03 | Exon 4B IVS +1G→A | Exon skipping |
| CNC09 | Exon 4B 578 delTG | Frameshift after codon 163; stop codon after 4 missense residues |
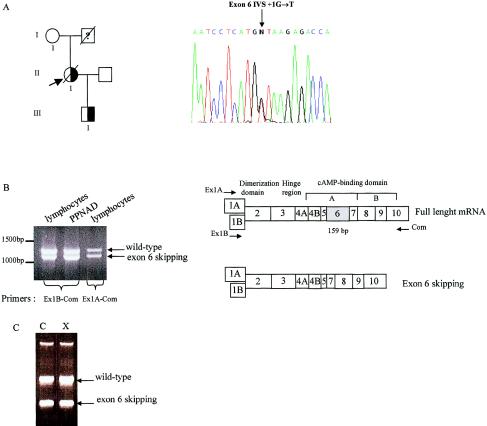
| CNC04 | Exon 6 IVS +1G→T | Exon skipping |
| CNC08 | Exon 7 IVS del (−7→−2) | Exon skipping |
| CNC06 | 810 ins A (exon 7) | Frameshift after codon 241; stop codon after 6 missense residues |
| CNC02 | 815 del 13 bp (exon 7) | Frameshift after codon 243; stop codon after 9 missense residues |
| CNC01 | 850 del AT (exon 7) | Frameshift after codon 255; stop codon after 13 missense residues |
| CNC05 | Exon 8 IVS +3A→G | Activation of a cryptic splice site |
Figure 1.
Pedigrees of seven families with a heterozygous mutation of PRKARIA gene. Half-filled squares and circles represent heterozygous mutated male and female patients, respectively. Open squares and circles represent unaffected patients. The individuals who were studied in each generation are numbered. Question marks (?) represent individuals of unknown affection status. The proband is indicated by the arrow. The results of nondenaturing polyacrylamide gel electrophoretic analysis of mutated exons are shown below the three pedigrees. The presence of heteroduplexes (upper bands), which are formed by the combination of mismatched alleles, confirms heterozygosity for mutated patients. Migration of the PCR products of family CNC02 shows abnormal bands only for proband with a heterozygous 13-bp deletion in exon 7. The upper bands correspond to heteroduplexes, the lower bands to normal and mutant alleles.
Figure 2.
A heterozygous G→A transversion in exon 1B gives rise to a novel ATG translation initiation codon. A, Pedigree of family CNC07. The proband is indicated by the arrow. B, Sequencing of PRKAR1A exon 1B revealed a heterozygous mutation that creates an upstream, out-of-frame ATG codon in the RIα1B mRNA within a consensus sequence for translation initiation. This AUG codon is classified as a strong start site on the basis of the A and G, respectively, at position −3 and +4 (Kozak 1997). According to the scanning model of eukaryotic translation, the novel ATG should initiate translation of a truncated protein and decrease translation from the wild-type start codon (Kozak 1999), as administrated for CDKN2A gene (Liu et al. 1999). The location of mutant ATG codon is shown with a horizontal band. The mutation is indicated by an arrow.
Figure 3.
An exon 6 splice-site mutation leads to an exon 6 skipping in family CNC04. A, Pedigree of family CNC04. The subjects who were studied in each generation are numbered. The question mark (?) represents an individual of unknown affection status. The proband (II.1) is indicated by the arrow. She presented with a severe form of Carney complex and died of a pancreatic adenocarcinoma with rapidly growing liver metastasis. Her unaffected mother was also investigated for the presence of the mutation. Only the proband and her affected son were heterozygous for the intron 6 splice mutation. The proband’s sequence trace of the sense strand showed a G→T transversion in the splice donor site of intron 6. B, Migration, on a 1% agarose gel, of RT-PCR products from transformed lymphocytes and PPNAD from family CNC04’s proband. After gel extraction and purification, RT-PCR products were directly sequenced. Top, Schematic representation of the gene organization of coding exons giving rise to the mature RIα mRNA. Regions that encode functional domains are denoted. The localization of the primers used for the RT-PCR is indicated with arrows. Bottom, Schematic representation of exon 6 skipping mRNA. C, RT-PCR products from transformed lymphocytes from patient carrying the exon 6 IVS del(−9→−2) mutation. No change is obvious after 4 h of treatment with 100 μg/ml cycloheximide (X) compared to the vehicle (C).
The mutation found in one kindred (CNC04) was a G→T transversion in the 5′ splice-donor site of intron 6. The proband had died of a severe form of CNC; her severely affected son also had the mutation, but her unaffected mother did not (fig. 3A). This unique mutation was predicted to lead to exon skipping; however, the sequence change was in frame, because exon 6 contains 53 triplet codons, making it unlikely that NMD was ongoing (fig. 3B). In fact, the mutant mRNA was present in the proband’s peripheral lymphocytes before and after treatment with cycloheximide (fig. 3C), consistent with an exon 6 skipping mutation, exon 6 IVS del(−9→−2), shown elsewhere (Kirschner et al. 2000b).
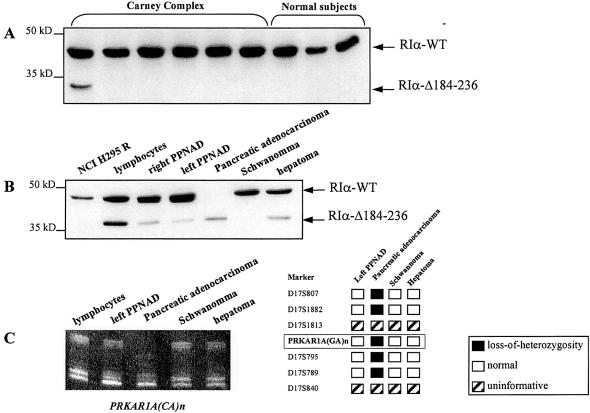
Exon 6 is located in the region encoding for cAMP-binding domain A and gives rise to 53 amino acids (184–236) of the human RIα subunit. To detect shortened RIα forms (RIα-Δ184–236), we performed western-blot analysis of cell lysates from peripheral lymphocytes from six CNC patients with known mutations of the PRKAR1A gene, including the proband of family CNC04 and three control subjects (fig. 4A). Only the patient with the exon 6 mRNA skipping mutation showed the shortened RIα form. We then studied five different tumors from this patient (adrenal nodules [left and right PPNAD], a pancreatic adenocarcinoma, schwannoma, and hepatoma); the deleted form of RIα was found in all tumors (fig. 4B). It is interesting that only the pancreatic adenocarcinoma demonstrated loss of expression of the RIα wild-type protein. To understand the mechanism underlying this observation, we performed LOH analysis using peripheral blood DNA and DNA from the tumors examined above. Seven markers located around the PRKAR1A gene were studied, including the intragenic dinucleotide repeat. Consistent with the western blotting data, only the malignant tumor showed LOH for all informative markers, including the intragenic one (fig. 4C).
Figure 4.
Identification of a deleted form of RIα subunit in proband of family CNC04 and absence of LOH except for the pancreatic adenocarcinoma. A, Western blotting of RIα subunit in 5 μg protein lysates from transformed lymphocytes of Carney complex patients with mutation in PRKAR1A gene and three control subjects. Only the CNC patient with an exon 6 skipping mRNA presents a shortened form of the protein. Size markers are indicated at left. B, Western blotting analysis of 20 μg total cellular proteins prepared from lymphocytes and five different tumors of family CNC04’s proband. Two bands are present in all tissues of the patient except for the pancreatic adenocarcinoma where the wild-type RIα is lost. In the schwannoma, RIα-Δ184–236 was present after longer exposure. The human adrenal cell line H295R is shown as a control for RIα expression. Size markers are indicated at left. C, LOH analysis was performed using DNA samples from peripheral blood and four tumors from the same patient. Seven markers located around PRKAR1A were used, including PRKAR1A(CA)n which was located within the 5′ region of the gene. The five informative loci demonstrated LOH only in the tumor DNA of the pancreatic adenocarcinoma (a finding consistent with Western blotting data).
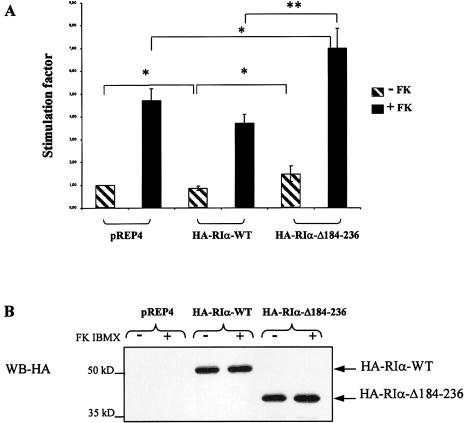
To investigate the consequences of the deleted form of the RIα protein at the transcriptional level, transient transfections were performed under the assumption that changes in expression of cAMP-responsive genes reflect PKA activity, as shown elsewhere (Gonzalez and Montminy 1989). We created HA-expression vectors for the wild-type RIα protein (RIα-WT) and the deleted mutant (RIα-Δ184–236). Forskolin, an adenylyl-cyclase activator and IBMX, a phosphodiesterase inhibitor, were used as stimulants of the cAMP signaling pathway. We then examined the effects of the wild-type RIα and RIα-Δ184–236 on the activity of a luciferase reporter gene under the control of the somatostatin promoter containing a cAMP-responsive element (CRE), a binding site for ATF/CREB transcription factors. As shown in figure 5A, RIα-Δ184–236 showed higher transcriptional activity of the CRE-somatostatin gene than empty vector or wild-type RIα transfected cells. This effect was present at baseline but was augmented after exposure to forskolin and IBMX. As shown in figure 5B, this is not explained by difference of expression between the two HA-tagged proteins. We obtained similar results with the non–HA-expression vectors (data not shown).
Figure 5.
Transcriptional activity of the RIα-WT and RIα-Δ184–236 on the somatostatin CRE promoter stimulated by forskolin. A, COS cells were transiently cotransfected with a reporter plasmid expressing luciferase under the control of the somatostatin CRE sequence and equivalent molar quantity of pREP4 expression vectors for HA-RIα-WT and HA-RIα-Δ184–236. Cells were left untreated or were stimulated in the presence of forskolin and IBMX. Experiments were repeated five times with triplicate samples. The figure shows the mean of the five experiments. All error bars represent standard error of the mean; * denotes significance at P<.05; ** denotes significance at P<.01. The stimulation factor is reported to the transcription level observed in the cells without transfected RIα. B, Control immunoblotting analysis of whole-cell extracts of HA-RIα expression. Anti-HA antibodies were used in Western blotting with total lysates from empty vector, HA-RIα-WT, or HA-RIα-Δ184–236 COS transfected cells.
In the present study of five sporadic and six familial cases of PPNAD or CNC, we found nine PRKAR1A mutations. In four kindreds, the mutation was de novo, as proven by the investigation of parental DNA (fig. 1), suggesting a high rate of spontaneously occurring mutations in this gene, as suggested by Kirschner et al. (2000a). With the exception of two mutations, all others were unique inactivating heterozygous mutations that are predicted to lead to a premature termination codon. This is in accordance with the mutations described to date, which have been demonstrated to be functionally null. Mutant mRNAs are degraded by NMD, and predicted truncated RIα forms are not detected (Kirschner et al. 2000b). Accordingly, we were not able to detect truncated protein or mutant mRNA from lymphocytes or tumors from patients harboring any of the novel PRKAR1A mutations, with one notable exception (see below).
PRKAR1A gene defects were identified in ∼41% of 52 CNC kindreds (Kirschner et al. 2000b). In this series, 81% (9/11) of our kindreds had mutations. This difference, albeit not statistically significant, may be real; it may be attributed to the different population that we studied, since our referral basis is Cushing syndrome caused by PPNAD: indeed, 10 of our 11 probands and 15 of 20 patients with a PRKAR1A mutation had PPNAD. Moreover, most of our patients were adults. It was reported recently that pediatric patients with PPNAD represent a different group of CNC patients (Sandrini et al. 2002). Taken together, these data suggest that adult CNC patients presenting with PPNAD and Cushing syndrome are perhaps more likely to have an inactivating PRKAR1A. This is also supported by the presence of frequent germline PRKAR1A mutations in patients with isolated, sporadic PPNAD who do not have any other signs of CNC (Groussin et al. 2002).
Among the PRKAR1A new mutations described in this study, two deserve a special note. First, a 5′ UTR point mutation in exon 1B created an additional out-of-frame ATG initiation codon within which a strong consensus sequence for translation initiation was found. This is the first described mutation of exon 1B. A similar alteration has been reported for the CDKN2A gene in melanoma-prone families (Liu et al. 1999) and for the POMC gene in severe early-onset obesity, adrenal insufficiency, and red hair pigmentation (Krude et al. 1998). This mutation involved only one of the two different RIα mRNAs originating from the alternative splicing of the two distinct leader exons, 1A and 1B. But it seems that the RIα 1B mRNA isoform is particularly important in mediating tissue-specific regulation of RIα expression (Dahle et al. 2001).
The second novel mutation that is of particular interest is the one that creates an alternatively spliced variant of RIα, RIα Δ184–236; it is interesting that this mutation was identified in a kindred with a severe form of CNC. This is the first example of abnormal RIα subunit mRNA and protein being present in cells from CNC patients: the exon 6 donor splice-site mutation leads to an exon 6 skipping that creates an in-frame, shorter PRKAR1A mRNA that does not undergo NMD. Accordingly, the predicted truncated PRKAR1A protein products could be detected in both peripheral lymphocytes and tumors from this kindred. The identification of this mutation allowed us to examine, for the first time for a CNC-causing mutation, the in vivo effects that a shorter RIα has on the PKA signaling system.
The R subunits of PKA are modular proteins composed of several distinct, well-defined, and stable domains (fig. 3B) (McKnight et al. 1988). Two tandem gene-duplicated cAMP-binding domains are present at the C terminus; each contains ∼120 amino acids. cAMP-binding domain A interacts directly with the C subunit and is essential for high-affinity binding to C; two structurally and functionally distinct subsites were characterized, one that binds cAMP and another that binds to the C subunit (Huang and Taylor 1998). The mutant RIα Δ184–236 has a 53-aa deletion located in the region of cAMP-binding domain A, which is known to contain Glu202 (E202) and Arg211 (R211) cAMP-binding sites. Cotransfection studies using a cell line expressing both the mutant and the wild-type RIα subunits (to mimic the in vivo state) showed loss of any inhibitory effects on CRE-dependent transcription of the somatostatin promoter when RIα Δ184–236 was overexpressed. Accordingly, measurements of nonstimulated and stimulated luciferase activity were significantly higher when RIα Δ184–236 was overexpressed. This difference was likely to be related to increased PKA activity, since CRE-dependent transcription reflects nuclear translocation of the catalytic subunit (Gonzalez and Montminy 1989). The most likely mechanism by which a truncated RIα could lead to cAMP signaling alterations is a dominant negative effect on RIα-mediated inhibition of the PKA catalytic subunit (Ogreid and Taylor 1990).
To date, PRKAR1A is considered a tumor-suppressor gene because of the constitutional loss of one allele (functionally null germ line mutations) associated with loss of the normal allele in CNC tumors. We have studied several different tumors of the patient with the abnormal subunit RIα Δ184–236, including an unusual pancreatic adenocarcinoma (arising from the acinar cells) to which the patient succumbed after widespread metastasis. It should be noted that CNC appears to predispose to this unusual tumor, since two other patients with CNC have been reported with similar diagnosis and clinical course (Stratakis et al. 2001). In our study, complete LOH was detected only in the pancreatic adenocarcinoma, suggesting that loss of the normal allele is not necessary for abnormal growth and/or proliferation of cells in CNC-affected tissues, unless these results are confounded by “contamination” with normal cells. The latter was unlikely, given the careful dissection of the lesions from this patient. Thus, we speculate that the dominant negative effect of RIα Δ184–236 is responsible for RIα downregulation in the initial stages of this tumor, where LOH was not present. Progression of the tumor was, perhaps, further favored by loss of the normal allele, as demonstrated by LOH of the malignant components of the resected lesion.
In conclusion, we described a high frequency of PRKAR1A gene mutation in 11 CNC kindreds. Analysis of the phenotypes may suggest that adult patients with CNC presenting with Cushing syndrome caused by PPNAD are more likely to have a PRKAR1A gene defect. Perhaps the most important finding in our study is that a defective PRKAR1A protein without concurrent loss of the normal allele can cause CNC and is associated with in vitro evidence of increased PKA activity. The dominant effect of this unique mutant suggests that perturbations of PKA may be enough to cause pathology in CNC-affected cells and has wide implications for the understanding of the cAMP signaling in tumorigenesis.
Acknowledgments
We thank F. René-Corail, for excellent technical assistance; the staff of the Banque de Cellules (Prof. M Delpech, CHU Cochin), for lymphocyte collection; the surgeons (Profs. Y. Chapuis and B. Dousset) and the medical and paramedical staff at the Surgery and the Endocrine Departments of the Hôpital Cochin, who managed the patients; Drs. A. Louvel and F. Tissier, for the pathological examinations; and Dr. A. Dugue and the staff of the Laudat Hormone Laboratory, for hormone assays. We thank also Prof. C. Beldjord, Dr. B. Violet (Institut Cochin), Drs. Papageorgiou and Sandrini (both at NICHD, NIH), and other members of our laboratories for helpful discussions. We thank Profs. J. C. Piette and M. Abid and Drs. Z. Amoura and N. Chabbert-Buffet for referring their patients for endocrine or genetic investigations. This work was supported in part by the Plan Hospitalier de Recherche Clinique (grant AOM 95201 to the COMETE Network, coordinated by Prof. P. F. Plouin and dedicated to the study of adrenal tumors) and the Association pour la Recherche sur le Cancer (grant ARC 4225). L.G. was the recipient of a fellowship from the Association pour la Recherche sur le Cancer.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Genome Database, http://gdbwww.gdb.org/ (for sequence information)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim/ (for CNC [MIM 160980]) [PubMed]
- Whitehead Institute/MIT, Center for Genome Research,http://www.genome.wi.mit.edu/ (for sequence information)
References
- Bertherat J, Chanson P, Montminy M (1995) The cyclic adenosine 3′,5′-monophosphate-responsive factor CREB is constitutively activated in human somatotroph adenomas. Mol Endocrinol 9:777–783 [DOI] [PubMed] [Google Scholar]
- Carney JA (1990) Psammomatous melanotic schwannoma: a distinctive, heritable tumor with special associations, including cardiac myxoma and the Cushing syndrome. Am J Surg Pathol 14:206–222 [PubMed] [Google Scholar]
- Carney JA, Boccon-Gibod L, Jarka DE, Tanaka Y, Swee RG, Unni KK, Stratakis CA (2001) Osteochondromyxoma of bone: a congenital tumor associated with lentigines and other unusual disorders. Am J Surg Pathol 25:164–176 [DOI] [PubMed] [Google Scholar]
- Carney JA, Gordon H, Carpenter PC, Shenoy BV, Go VL (1985) The complex of myxomas, spotty pigmentation, and endocrine overactivity. Medicine 64:270–283 [DOI] [PubMed] [Google Scholar]
- Carney JA, Hruska LS, Beauchamp GD, Gordon H (1986) Dominant inheritance of the complex of myxomas, spotty pigmentation, and endocrine overactivity. Mayo Clin Proc 61:165–172 [DOI] [PubMed] [Google Scholar]
- Carney JA, Stratakis CA (1996) Ductal adenoma of the breast and the Carney complex. Am J Surg Pathol 20:1154–1155 [DOI] [PubMed] [Google Scholar]
- Carney JA, Stratakis CA (1998) Epithelioid blue nevus and psammomatous melanotic schwannoma: the unusual pigmented skin tumors of the Carney complex. Semin Diagn Pathol 15:216–224 [PubMed] [Google Scholar]
- Carney JA, Toorkey BC (1991) Ductal adenoma of the breast with tubular features: a probable component of the complex of myxomas, spotty pigmentation, endocrine overactivity, and schwannomas. Am J Surg Pathol 15:722–731 [DOI] [PubMed] [Google Scholar]
- Casey M, Mah C, Merliss AD, Kirschner LS, Taymans SE, Denio AE, Korf B, Irvine AD, Hughes A, Carney JA, Stratakis CA, Basson CT (1998) Identification of a novel genetic locus for familial cardiac myxomas and Carney complex. Circulation 98:2560–2566 [DOI] [PubMed] [Google Scholar]
- Casey M, Vaughan CJ, He J, Hatcher CJ, Winter JM, Weremowicz S, Montgomery K, Kucherlapati R, Morton CC, Basson CT (2000) Mutations in the protein kinase A R1α regulatory subunit cause familial cardiac myxomas and Carney complex. J Clin Invest 106:R31–R38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahle MK, Knutsen HK, Tasken KA, Pilz R, Tasken K (2001) Cyclic AMP regulates expression of the RIα subunit of cAMP-dependent protein kinase through an alternatively spliced 5′ UTR. Eur J Biochem 268:5920–5929 [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR (1989) Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59:675–680 [DOI] [PubMed] [Google Scholar]
- Groussin L, Jullian E, Perlemoine K, Leheup B, Luton JP, Bertagna X, Bertherat J (2002) Mutations of the PRKAR1A gene in Cushing’s syndrome due to sporadic primary pigmented nodular adrenocortical disease (PPNAD). J Clin Endocrinol Metab 87:4324–4329 [DOI] [PubMed] [Google Scholar]
- Groussin L, Massias J, Bertagna X, Bertherat J (2000) Loss of expression of the ubiquitous transcription factor cAMP response element-binding protein (CREB) and compensatory overexpression of the activator CREMτ in the human adrenocortical cancer cell line H295R. J Clin Endocrinol Metab 85:345–354 [DOI] [PubMed] [Google Scholar]
- Huang LJ, Taylor SS (1998) Dissecting cAMP binding domain A in the RIα subunit of cAMP-dependent protein kinase: distinct subsites for recognition of cAMP and the catalytic subunit. J Biol Chem 273:26739–26746 [DOI] [PubMed] [Google Scholar]
- Kirschner LS, Carney JA, Pack SD, Taymans SE, Giatzakis C, Cho YS, Cho-Chung YS, Stratakis CA (2000a) Mutations of the gene encoding the protein kinase A type I-α regulatory subunit in patients with the Carney complex. Nat Genet 26:89–92 [DOI] [PubMed] [Google Scholar]
- Kirschner LS, Sandrini F, Monbo J, Lin JP, Carney JA, Stratakis CA (2000b) Genetic heterogeneity and spectrum of mutations of the PRKAR1A gene in patients with the Carney complex. Hum Mol Genet 9:3037–3046 [DOI] [PubMed] [Google Scholar]
- Kozak M (1997) Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. Embo J 16:2482–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1999) Initiation of translation in prokaryotes and eukaryotes. Gene 234:187–208 [DOI] [PubMed] [Google Scholar]
- Krude H, Biebermann H, Luck W, Horn R, Brabant G, Gruters A (1998) Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat Genet 19:155–157 [DOI] [PubMed] [Google Scholar]
- Liu L, Dilworth D, Gao L, Monzon J, Summers A, Lassam N, Hogg D (1999) Mutation of the CDKN2A 5′ UTR creates an aberrant initiation codon and predisposes to melanoma. Nat Genet 21:128–132 [DOI] [PubMed] [Google Scholar]
- McKnight GS, Clegg CH, Uhler MD, Chrivia JC, Cadd GG, Correll LA, Otten AD (1988) Analysis of the cAMP-dependent protein kinase system using molecular genetic approaches. Recent Prog Horm Res 44:307–335 [DOI] [PubMed] [Google Scholar]
- Ogreid D, Taylor SS (1990) Expression and characterization of a truncated form of regulatory subunit of cAMP-dependent protein kinase I lacking binding domain A. Adv Second Messenger Phosphoprotein Res 24:196–201 [PubMed] [Google Scholar]
- Pack SD, Kirschner LS, Pak E, Zhuang Z, Carney JA, Stratakis CA (2000) Genetic and histologic studies of somatomammotropic pituitary tumors in patients with the “complex of spotty skin pigmentation, myxomas, endocrine overactivity and schwannomas” (Carney complex). J Clin Endocrinol Metab 85:3860–3865 [DOI] [PubMed] [Google Scholar]
- Premkumar A, Stratakis CA, Shawker TH, Papanicolaou DA, Chrousos GP (1997) Testicular ultrasound in Carney complex: report of three cases. J Clin Ultrasound 25:211–214 [DOI] [PubMed] [Google Scholar]
- Raff SB, Carney JA, Krugman D, Doppman JL, Stratakis CA (2000) Prolactin secretion abnormalities in patients with the “syndrome of spotty skin pigmentation, myxomas, endocrine overactivity and schwannomas” (Carney complex). J Pediatr Endocrinol Metab 13:373–379 [PubMed] [Google Scholar]
- Sandrini F, Matyakhina L, Bourdeau I, Farmakidis C, Keil M, Kirschner LS, Stratakis CA (2002) PRKAR1A, a regulator of protein kinase A, is mutated in children with Carney complex and isolated micronodular adrenal hyperplasia. Abstract presented at the Society for Pediatric Research (SPR) Annual Meeting, Baltimore, May 6 [Google Scholar]
- Stratakis CA, Carney JA, Lin JP, Papanicolaou DA, Karl M, Kastner DL, Pras E, Chrousos GP (1996) Carney complex, a familial multiple neoplasia and lentiginosis syndrome: analysis of 11 kindreds and linkage to the short arm of chromosome 2. J Clin Invest 97:699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratakis CA, Courcoutsakis NA, Abati A, Filie A, Doppman JL, Carney JA, Shawker T (1997) Thyroid gland abnormalities in patients with the syndrome of spotty skin pigmentation, myxomas, endocrine overactivity, and schwannomas (Carney complex). J Clin Endocrinol Metab 82:2037–2043 [DOI] [PubMed] [Google Scholar]
- Stratakis CA, Kirschner LS, Carney JA (2001) Clinical and molecular features of the Carney complex: diagnostic criteria and recommendations for patient evaluation. J Clin Endocrinol Metab 86:4041–4046 [DOI] [PubMed] [Google Scholar]
- Stratakis CA, Papageorgiou T, Premkumar A, Pack S, Kirschner LS, Taymans SE, Zhuang Z, Oelkers WH, Carney JA (2000) Ovarian lesions in Carney complex: clinical genetics and possible predisposition to malignancy. J Clin Endocrinol Metab 85:4359–4366 [DOI] [PubMed] [Google Scholar]
- Stratakis CA, Sarlis N, Kirschner LS, Carney JA, Doppman JL, Nieman LK, Chrousos GP, Papanicolaou DA (1999) Paradoxical response to dexamethasone in the diagnosis of primary pigmented nodular adrenocortical disease. Ann Intern Med 131:585–591 [DOI] [PubMed] [Google Scholar]