Abstract
3-Methylglutaconic aciduria type I is an autosomal recessive disorder clinically characterized by various symptoms ranging from delayed speech development to severe neurological handicap. This disorder is caused by a deficiency of 3-methylglutaconyl-CoA hydratase, one of the key enzymes of leucine degradation. This results in elevated urinary levels of 3-methylglutaconic acid, 3-methylglutaric acid, and 3-hydroxyisovaleric acid. By heterologous expression in Escherichia coli, we show that 3-methylglutaconyl-CoA hydratase is encoded by the AUH gene, whose product had been reported elsewhere as an AU-specific RNA-binding protein. Mutation analysis of AUH in two patients revealed a nonsense mutation (R197X) and a splice-site mutation (IVS8-1G→A), demonstrating that mutations in AUH cause 3-methylglutaconic aciduria type I.
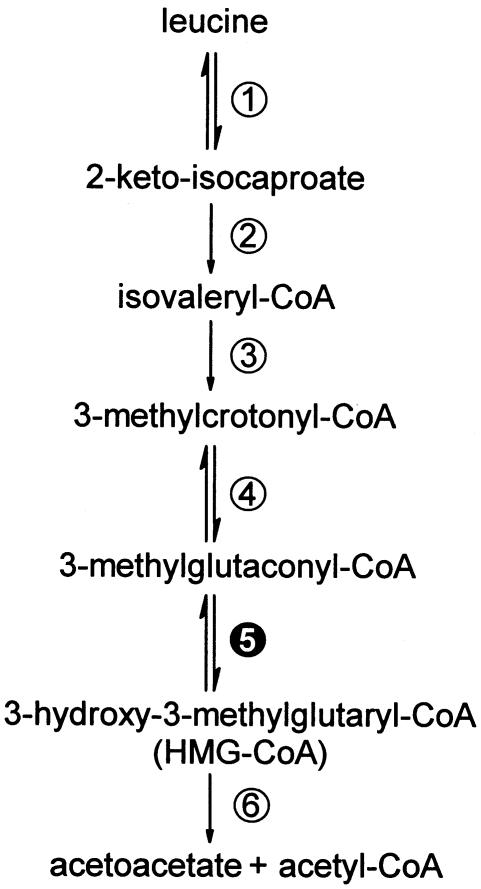
3-Methylglutaconic aciduria is a group of metabolic disorders characterized by increased urinary excretion of 3-methylglutaconic acid and 3-methylglutaric acid. At present, four distinct forms have been recognized. Type I (MIM 250950) represents the isolated 3-methylglutaconyl-CoA hydratase deficiency. This mitochondrial enzyme catalyzes the fifth step in leucine catabolism, as shown in figure 1, which is the conversion of 3-methylglutaconyl-CoA to 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA). Type II (MIM 302060), also referred to as Barth syndrome, is an X-linked mitochondrial cardiomyopathy associated with neutropenia and growth retardation and caused by mutations in the gene encoding tafazzin (TAZ). Type III (MIM 258501) or Costeff syndrome, is an autosomal recessive disorder caused by mutations in the OPA3 gene, which lead to bilateral optic atrophy. Type IV or “unspecified” 3-methylglutaconic aciduria (MIM 250951) comprises a heterogeneous group of patients with progressive neurological symptoms. The hydratase activity in Barth syndrome, Costeff syndrome, and type IV 3-methylglutaconic aciduria is normal, and the excretion of 3-methylglutaconic acid and 3-methylglutaric acid is secondary. Among the four types, patients with type I excrete the highest levels of 3-methylglutaconic acid and 3-methylglutaric acid. Furthermore, these patients additionally excrete increased amounts of 3-hydroxyisovaleric acid; therefore, 3-hydroxyisovaleric acid is an important parameter for differentiation between the isolated 3-methylglutaconyl-CoA hydratase deficiency and the other types. The phenotypic presentation of 3-methylglutaconyl-CoA hydratase deficiency varies from mild, including delayed speech development and hyperchloremic acidosis associated with gastroesophageal reflux, to a much more severe phenotype, including seizures and cerebellar abnormalities. Although the hydratase was purified partially in the late 1950s (Hilz et al. 1958), the gene encoding the protein had not been identified.
Figure 1.
The metabolic pathway of l-leucine. The different enzymes involved are numbered as follows: 1, Branched-chain amino acid transaminase; 2, Branched-chain 2-keto acid dehydrogenase complex; 3, Isovaleryl-coa dehydrogenase; 4, Methylcrotonyl-CoA carboxylase; 5, 3-Methylglutaconyl-CoA hydratase; and 6, 3-Hydroxy-3-methylglutaryl-CoA lyase.
Several mitochondrial enzymes with enoyl-CoA hydratase activity have been characterized. The first one, called “crotonase,” is most active toward short-chain enoyl-CoAs and participates in the β-oxidation of short-chain fatty acids. Studies performed by Hilz et al. (1958) showed that an enzyme other than crotonase is responsible for the hydratase step in the degradation of leucine. Indeed, our preliminary studies showed that crotonase is not active with HMG-CoA when measured in the reverse direction. A second enzyme harboring hydratase activity is mitochondrial trifunctional protein (MTP), which also catalyzes dehydrogenation and thiolytic cleavage of long-chain fatty acids. Together with very-long-chain acyl-CoA dehydrogenase, this protein is the main enzyme in the long-chain fatty acid β-oxidation. It is unlikely, however, that this enzyme is involved in the degradation of leucine, because patients with MTP deficiency (MIM 143450, MIM 600890) have no abnormal urinary excretion of 3-methylglutaconic acid.
Nakagawa et al. (1995) purified an AU-specific RNA-binding protein, designated “AUH” (GenBank accession numbers NM_001698 and NT_008476), using an affinity column displaying A+U-rich elements. The protein contained a hydratase motif and shared 32% sequence identity with crotonase. The enoyl-CoA hydratase activity with butenoyl-CoA (C4:1-CoA) as substrate was a thousandfold lower when compared with crotonase. The three-dimensional structure of human AUH has been determined at 2.2-Å resolution and predicted a high affinity for short-chain substrates (Kurimoto et al. 2001).
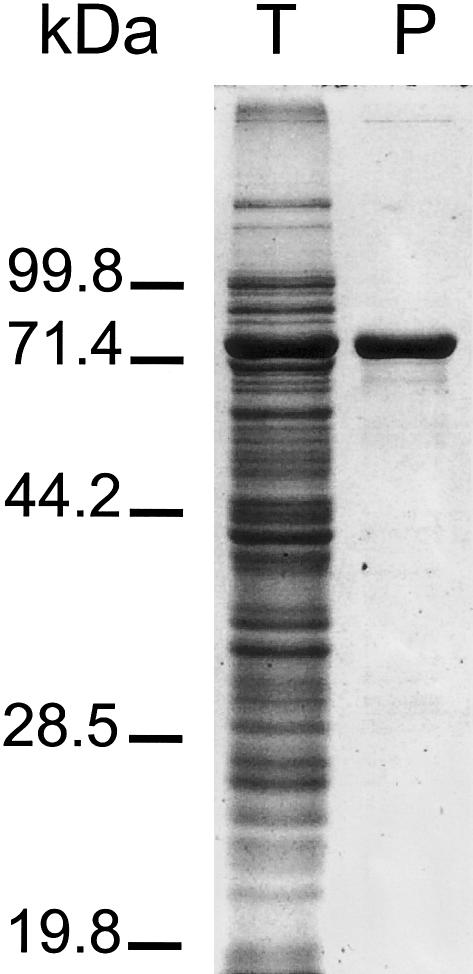
To investigate whether AUH has 3-methylglutaconyl-CoA hydratase activity, we expressed AUH as a fusion protein with maltose-binding protein (MBP). The ORF of AUH was amplified from human cDNA by use of the primers −3AUHfEcoRI (5′-ATA TGA ATT CAA CAT GGC GGC CGC GGT GG-3′) and 1097AUHrXbaI (5′-ATA TTC TAG ACA TAT AGT GGA TCC GAA AGA C-3′), digested with EcoRI and XbaI, and subsequently cloned into the EcoRI and XbaI restriction sites of the pMAL-C2X vector (New England Biolabs). The cloned PCR products were sequenced to assess the integrity of the PCR process. Next, the MBP-AUH was expressed in Escherichia coli and affinity-purified to homogeneity according to the manufacturer’s protocol (fig. 2).
Figure 2.
Expression and purification of MBP-AUH fusion protein in E. coli. Total E. coli extract (T) and affinity-purified fusion protein (P) were analyzed by 10% SDS-PAGE and stained with Coomassie brilliant blue.
The 3-methylglutaconyl-CoA hydratase activity of the expressed protein was measured in the reverse direction, using HMG-CoA as substrate. The reaction mixture contained 50 mM Tris pH 7.4, 10 mM EDTA, 1 mg/ml BSA, and 0.1 mM HMG-CoA and was started by addition of purified protein. The formation of 3-methylglutaconyl-CoA was followed spectrophotometrically at 260 nm using a molar extinction coefficient of 6200 L/mol/cm. The short-chain enoyl-CoA hydratase activity was measured using butenoyl-CoA as a substrate as described by Wanders et al. (1992). For comparison, both activities were also measured using bovine liver crotonase obtained from Sigma.
As reported above for AUH, we also found very low hydratase activity with C4:1-CoA. In addition, we found that AUH has high 3-methylglutaconyl-CoA hydratase activity. Indeed, the HMG/C4:1 activity ratio was >20. In contrast, crotonase has no detectable 3-methylglutaconyl-CoA hydratase activity (table 1). Therefore, AUH is a strong candidate for the underlying enzymatic defect of 3-methylglutaconic aciduria type I.
Table 1.
Hydratase Activity Measurements of AUH and Crotonase with HMG-CoA and Butenoyl-CoA (C4:1-CoA) as Substrate
| Hydratase Activity | MBP-AUH(nmol/min/mg) | Bovine LiverCrotonase(nmol/min/mg) |
| HMG-CoA | 110 | <1 |
| C4:1-CoA | 4.8 | 8.3 ×105 |
| Ratio HMG-CoA/C4:1-CoA | 23 | <10−6 |
To date, eight patients have been described with isolated 3-methylglutaconyl-CoA hydratase deficiency (Duran et al. 1982; Narisawa et al. 1986; Gibson et al. 1992; Gibson et al. 1998; Shoji et al. 1999; Ensenauer et al. 2000). The hydratase deficiency in these patients has been determined by use of an overall enzyme assay measuring three steps of leucine degradation, from 3-methylcrotonyl-CoA to acetoacetic acid (Narisawa et al. 1989). We reinvestigated fibroblasts from two patients (Duran et al. 1982; Ensenauer et al. 2000) with our novel enzyme assay, using the same conditions as described above, but the reaction was started with 0.3 mg/ml fibroblast homogenate. The reaction mixture was incubated for 30 min at 37°C, stopped with 0.2 M hydrochloric acid, and subsequently neutralized with sodium hydroxide. Formation of 3-methylglutaconyl-CoA was quantified on HPLC using a reverse-phase C18 column (Supelcosil LC-18-DB, 250 mm × 10 mm, 5 μm, Supelco). Separation of the acyl-CoAs was achieved by elution with a linear gradient of methanol (10%–37.6% in 22 min) in potassium phosphate buffer (50 mM, pH 5.3). Both patients showed clear deficiency of 3-methylglutaconyl-CoA hydratase (patients <0.1; control subjects 2.8±0.6 nmol/min/mg protein), whereas the crotonase activity was normal for both patients.
To determine whether 3-methylglutaconic aciduria type I is caused by mutations in AUH, the ORF of AUH was amplified from human cDNA in two overlapping fragments, using the following M13-tagged primers: fragment A: −28AUHf-21M13 (5′-tgt aaa acg acg gcc agt TCG CAG GTC CAC GCC GTA AAC-3′) and 680AUHrM13 (5′-cag gaa aca gct atg acc CGC GTG GCA ATC GCT GTG TC-3′); fragment B: 505AUHf-21M13 (5′-tgt aaa acg acg gcc agt GCT ATT CTT CCA GTG CCA AC-3′) and 1097AUHrM13 (5′-cag gaa aca gct atg acc CAT ATA GTG GAT CCG AAA GAC-3′). PCR fragments obtained from patients and two control subjects were sequenced with Big Dye Terminators and were analyzed on an Applied Biosystems 377A automated DNA sequencer, following the manufacturer’s protocols (PE Applied Biosystems).
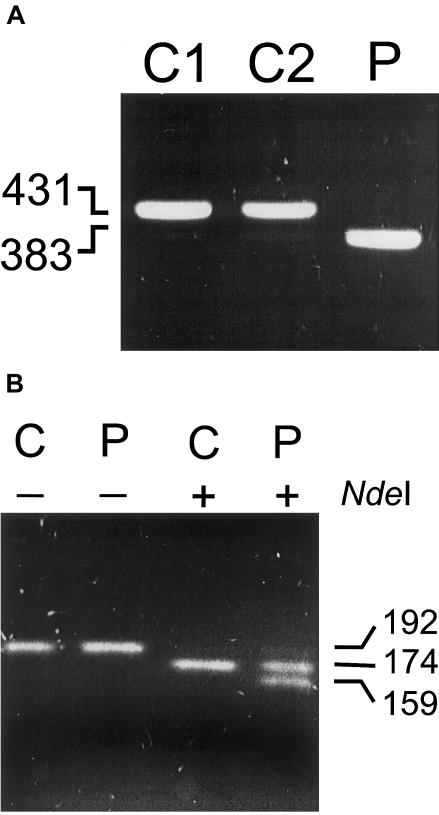
The first patient, the third son of consanguineous parents, of Afghan origin, had retardation in speech development. A delayed motor development became evident in retrospect (Ensenauer et al. 2000). The amplification product of fragment B was ∼50 bp smaller than that found in controls (fig. 3A). Sequence analysis revealed that this was caused by skipping of exon 9. The splice acceptor and donor site neighboring exon 9 conform to the AG/GT rule. To identify the underlying mutation causing missplicing, we amplified splice sites flanking exon 9 from genomic DNA, using the following primers: In8AUHf-21M13 (5′-tgt aaa acg acg gcc agt GAC ATG CCT CTT TGA AGC AC-3′) and In9AUHrM13 (5′- cag gaa aca gct atg acc GCA AGG GTA ATC TTG CTC AG-3′). In patient 1, we identified a homozygous IVS8-1G→A mutation, which disrupts the consensus sequence of the splice acceptor site. Exon 9 encodes α-helix H9 (Kurimoto et al. 2001), which crystal structure analysis suggests to be a constituent of the active-site pocket and is also necessary for subunit interaction and trimer stability.
Figure 3.
Mutation analysis of AUH in two patients. A, PCR fragment B amplified from cDNA of two control subjects (C1 and C2) and patient 1 (P) showing the 42-bp deletion (exon 9) as a consequence of a IVS8-1G→A mutation. B, PCR-RFLP analysis of the 589C→T mutation. Part of intron 5 and exon 5 containing the c.589C→T mutation was amplified using a mismatch primer (558AUHfNdeI 5′-TGG CTT TAG CCT GTG ACA TA-3′) creating an NdeI restriction site in the 589T allele. For convenient control of the restriction, the reverse primer (In5AUHrM13NdeI 5′-cag gaa aca gct atg acc CAT ATG ACC ATT AGG ACC AAC AAG TG-3′) contained an M13 extension and a second NdeI restriction site. PCR products from genomic DNA of a control subject (C) and patient 2 (P) were loaded directly (−) or digested with NdeI (+) before electrophoresis.
The second patient is the younger of two affected brothers of healthy nonconsanguineous Moroccan parents. He had no physical abnormalities; only his speech development was retarded (Duran et al. 1982). Sequence analysis at the cDNA level showed an apparently homozygous nonsense mutation, 589C→T. PCR-RFLP analysis on genomic DNA showed heterozygosity for the 589C→T mutation (fig. 3B), indicating that the other allele is a null allele or produces an unstable mRNA. The R197X nonsense mutation causes the translation to be terminated before glutamate 209, which together with glutamate 189 forms the catalytic group of the active-site pocket. In conclusion, our data show that mutations in AUH cause 3-methylglutaconic aciduria type I.
Acknowledgment
We thank H. R. Waterham for critically reading this manuscript.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for AUH [accession numbers NM_001698 and NT_008476]
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for 3-methylglutaconic aciduria type I [MIM 250950], Barth syndrome [MIM 302060], Costeff syndrome [MIM 258501], 3-methylglutaconic aciduria type IV [MIM 250951], and MTP deficiency [MIM 143450, MIM 600890]
References
- Duran M, Beemer FA, Tibosch AS, Bruinvis L, Ketting D, Wadman SK (1982) Inherited 3-methylglutaconic aciduria in two brothers: another defect of leucine metabolism. J Pediatr 101:551–554 [DOI] [PubMed] [Google Scholar]
- Ensenauer R, Muller CB, Schwab KO, Gibson KM, Brandis M, Lehnert W (2000) 3-Methylglutaconyl-CoA hydratase deficiency: a new patient with speech retardation as the leading sign. J Inherit Metab Dis 23:341–344 [DOI] [PubMed] [Google Scholar]
- Gibson KM, Lee CF, Wappner RS (1992) 3-Methylglutaconyl-coenzyme—a hydratase deficiency: a new case. J Inherit Metab Dis 15:363–366 [DOI] [PubMed] [Google Scholar]
- Gibson KM, Wappner RS, Jooste S, Erasmus E, Mienie LJ, Gerlo E, Desprechins B, De Meirleir L (1998) Variable clinical presentation in three patients with 3-methylglutaconyl-coenzyme A hydratase deficiency. J Inherit Metab Dis 21:631–638 [DOI] [PubMed] [Google Scholar]
- Hilz H, Knappe J, Ringelmann E, Lynen F (1958) Methylglutaconase, eine neue hydratase, die am stoffwechsel verzweigter carbonsäuren beteiligt ist. Biochem Z 329:476–489 [PubMed] [Google Scholar]
- Kurimoto K, Fukai S, Nureki O, Muto Y, Yokoyama S (2001) Crystal structure of human AUH protein, a single-stranded RNA binding homolog of enoyl-CoA hydratase. Structure 9:1253–1263 [DOI] [PubMed] [Google Scholar]
- Nakagawa J, Waldner H, Meyer-Monard S, Hofsteenge J, Jeno P, Moroni C (1995) AUH, a gene encoding an AU-specific RNA binding protein with intrinsic enoyl-CoA hydratase activity. Proc Natl Acad Sci USA 92:2051–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narisawa K, Gibson KM, Sweetman L, Nyhan WL (1986) Deficiency of 3-methylglutaconyl-coenzyme A hydratase in two siblings with 3-methylglutaconic aciduria. J Clin Invest 77:1148–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narisawa K, Gibson KM, Sweetman L, Nyhan WL (1989) 3-Methylglutaconyl-CoA hydratase, 3-methylcrotonyl-CoA carboxylase and 3-hydroxy3-methylglutaryl-CoA lyase deficiencies: a coupled enzyme assay useful for their detection. Clin Chim Acta 184:57–64 [DOI] [PubMed] [Google Scholar]
- Shoji Y, Takahashi T, Sawaishi Y, Ishida A, Matsumori M, Shoji Y, Enoki M, Watanabe H, Takada G (1999) 3-Methylglutaconic aciduria type I: clinical heterogeneity as a neurometabolic disease. J Inherit Metab Dis 22:1–8 [DOI] [PubMed] [Google Scholar]
- Wanders RJA, IJlst L, Poggi F, Bonnefont JP, Munnich A, Brivet M, Rabier D, Saudubray JM (1992) Human trifunctional protein deficiency: a new disorder of mitochondrial fatty acid oxidation. Biochem Biophys Res Commun 188:1139–1145 [DOI] [PubMed] [Google Scholar]