Abstract
Congenital dyserythropoietic anemias (CDAs) constitute a rare group of inherited red-blood-cell disorders associated with dysplastic changes in late erythroid precursors. CDA type I (CDAI [MIM 224120], gene symbol CDAN1) is characterized by erythroid pathological features such as internuclear chromatin bridges, spongy heterochromatin, and invagination of the nuclear membrane, carrying cytoplasmic organelles into the nucleus. A cluster of 45 highly inbred Israeli Bedouin with CDAI enabled the mapping of the CDAN1 disease gene to a 2-Mb interval, now refined to 1.2 Mb, containing 15 candidate genes on human chromosome 15q15 (Tamary et al. 1998). After the characterization and exclusion of 13 of these genes, we identified the CDAN1 gene through 12 different mutations in 9 families with CDAI. This 28-exon gene, which is transcribed ubiquitously into 4738 nt mRNA, was reconstructed on the basis of gene prediction and homology searches. It encodes codanin-1, a putative o-glycosylated protein of 1,226 amino acids, with no obvious transmembrane domains. Codanin-1 has a 150-residue amino-terminal domain with sequence similarity to collagens and two shorter segments that show weak similarities to the microtubule-associated proteins, MAP1B (neuraxin) and synapsin. These findings, and the cellular phenotype, suggest that codanin-1 may be involved in nuclear envelope integrity, conceivably related to microtubule attachments. The specific mechanisms by which codanin-1 underlies normal erythropoiesis remain to be elucidated.
Congenital dyserythropoietic anemias (CDAs), a group of inherited disorders associated with morphological and functional abnormalities of erythropoiesis, have been classified into three types (I–III), with some patients still unassigned (Wickramasinghe 1997; Delaunay and Iolascon 1999). The autosomal recessive CDA type II (CDAII [MIM 224100]), with more than 250 cases described to date (Iolascon et al. 2001), is the most common form. The disease gene maps to 20q11.2 in most studied families (Gasparini et al. 1997). The least common of the CDAs, the autosomal dominant CDA type III (CDAIII [MIM 105600]), was localized to chromosome 15q22 upon linkage analysis of a large Swedish family (Lind et al. 1995).
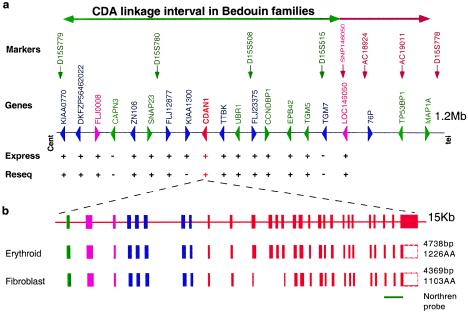
CDA type I (CDAI [MIM 224120]) is an autosomal recessive disease. Patients with CDAI present with moderate-to-severe macrocytic anemia. Bone marrow aspirates reveal binuclear intermediate- and late-erythroid precursors as well as internuclear chromatin bridges. Ultrastructural erythroid features include spongy heterochromatin and invagination of the nuclear membrane, carrying cytoplasm and cytoplasmic organelles into the nucleus. Dysmorphic features, mainly syndactyly and the absence or hypoplasia of phalanges and nails, have also been observed in several patients (Wickramasinghe 1997). Arrest of DNA synthesis (Wickramasinghe 1997) and apoptotic features in erythroid precursors have been described (Tamary et al. 1996). Interferon α2 was incidentally found to ameliorate the anemia and to partially reverse the morphological abnormalities of the erythroid precursors by an unknown mechanism (Lavabre-Bertrand et al. 1995; Wickramasinghe et al. 1997; Parez et al. 2000). A cluster of 45 Israeli Bedouin with CDAI enabled mapping of the disease gene to a region between markers D15S779 and D15S778 (Tamary et al. 1998). All patients showed a similar clinical picture, and in all the subjects, the diagnosis was confirmed by bone marrow electron microscopy. DNA was extracted from whole blood, and RNA was extracted from the diagnostic bone marrow aspiration. All studies were approved by the institutional review board of Rabin Medical Center. Using new polymorphic markers from genomic clones (AC019011, AC018924) and an informative SNP within the defined transcript for the putative transcription factor LOC146050, we further refined the critical CDAI interval to 1.2 Mb (fig. 1a).
Figure 1.
Physical map and genomic organization of the CDAN1 locus. a, Relative positions of landmark microsatellite markers are indicated in the CDAI candidate interval defined by markers D15S779 and D15S778. The CDA1 interval is indicated by green and brown arrows. An informative SNP (pink) within the newly defined transcript LOC146050 enabled us to restrict the linkage interval. The genes and transcripts identified are boxed: known genes in green, unknown transcripts in blue, inferred genes in purple, CDAN1 in red. The expression patterns of these genes were tested by RT-PCR on RNA from erythroid precursor cells (Pope et al. 2000) and are marked by a plus sign (+) or a minus sign (−) in the Express lane. Resequenced genes are marked by a plus sign (+) or a minus sign (−) in the Reseq lane. b, Expanded view of the 15-kb segment showing the genomic organization of CDAN1 and of two alternative transcripts isolated and sequenced from erythroid and fibroblast cells. Coding and noncoding exons are depicted as filled and open boxes, respectively. Red and blue exons are based on two partial transcripts, DKFZp434G2127 and BI855138, respectively. Green and pink exons are based on gene prediction and RT-PCR, respectively. The lengths of the corresponding transcripts and inferred proteins are indicated to the right.
In an attempt to identify the underlying mutations, we proceeded with systematic in silico analyses through gene prediction and homology searches of the critical CDAI interval and found 17 transcripts or putative genes. Fifteen genes prevailed as potential candidates, based on their expression as determined by RT-PCR in erythroid cells grown in liquid culture (Pope et al. 2000), including two redefined and two newly characterized transcripts, namely UBR1 (GenBank accession number AF525401), TTBK (GenBank accession number AF525400), FLJ008 (GenBank accession number AF525397) and LOC146050 (GenBank accession number AF525399). The remaining two genes, Calpain3 (CAPN3) and TGM7, were not considered, because they are not expressed in erythroid cells. The predicted exons were amplified from genomic DNA or from mRNA of erythroid precursors of one Bedouin patient with CDAI and of his healthy brother. These were subjected to sequence verification and mutation detection. Such systematic and comprehensive PCR and sequencing analyses were carried out for all coding regions of 14 of the 15 candidate genes (excluding KIAA1300, fig. 1a). All PCR reactions were performed according to standard procedures, and the products were subsequently sequenced using dye terminators of Big-Dyes kits on an ABI 3100 sequencer (Perkin-Elmer/Applied Biosystems). Sequence comparisons were performed using the STADEN package (Bonfield et al. 1995) and Sequencher software (gene codes cooperation).
In the first 13 genes tested, no segregating mutation was identified; however, we eventually identified a homozygous mutation in the Bedouin patient in the fourteenth gene scrutinized (table 1). This gene was reconstructed on the basis of prediction programs and homology searches using two partial transcripts and two EST sequences (fig. 1b). The gene, thus identified as CDAN1, has 28 exons spanning 15 kb of genomic DNA (fig. 1b). It encompasses a putative 4,738-nucleotide mRNA encoding a protein of 1,226 amino acids, which we propose to name Codanin-1, for CDA type I. Subsequently, 11 additional mutations in eight other patient groups with CDAI were identified in this gene (table 1).
Table 1.
CODANIN-I Mutations in Patients with CDAI[Note]
| ID, Family Origin, Genotype, and Exona | NucleotidePositionb | Amino Acid Change | Frequency | Detection Methodc |
| I, Israeli Bedouin, H: | ||||
| 24 | 3238C→T | Arg→Trp (R1040W) | 3/376 | +NcoI |
| II, French Polynesian, H: | ||||
| 26 | 3503C→T | Pro→Leu (P1129L) | 0/142 | −AciI |
| III, European, H: | ||||
| 12 | 1910A→G | Asn→Ser (N598S) | 0/196 | +DdeI |
| IV, European, CH: | ||||
| 14 | 2129C→T | Pro→Leu (P671L) | 1/196 | −MspI |
| 19 | 2716T→A | Phe→Ile (F866I) | 0/170 | MALDI−TOF |
| V, European, CH: | ||||
| 14 | 2158C→T | Arg→stop (R680X) | 0/200 | +BtsI |
| 14 | 2254C→T | Arg→Trp (R712W) | 1/164 | +BsrI |
| VI, European, CH: | ||||
| 23 | 3106C→T | Arg→stop (R996X) | 0/196 | −TaqI |
| 24 | 3242A→T | Asp→Val (D1041V) | 0/180 | MALDI−TOF |
| VII, European, CH: | ||||
| 14 | 2206G→A | Glu→Lys (E696K) | 0/160 | MALDI−TOF |
| VIII, European, CH: | ||||
| 24 | 3221C→A | Ser→Phe (S1034F) | 0/180 | +StyI |
| IX, Arab, CH: | ||||
| 19 | 2719G→A | Val→Met (V867M) | 1/180 | +NlaIII |
Note.— A total of 184 chromosomes from healthy nonrelated Bedouin individuals were analyzed as controls for the Bedouin mutation. For each of the additional mutations, close to 200 chromosomes from unrelated control individuals were analyzed. Three carriers, each for one mutation, were found. This is in accordance with the estimated frequency of CDAI heterozygotes in the European population.
ID I included five families; all other IDs included one family each. H = homozygote; CH = compound heterozygote.
Nucleotide position in CODANIN-1 cDNA accession number AF525398.
The mutation abolishes (−) or creates (+) a restriction site.
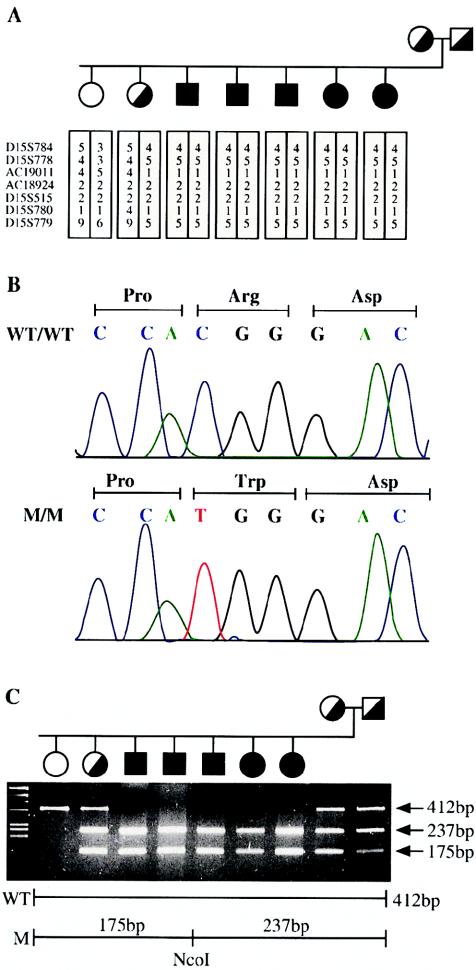
A single homozygous C→T substitution in exon 24, converting arginine to tryptophan at codon 1040 and creating an NcoI restriction site, was found in all 45 analyzed Bedouin patients with CDAI (fig. 2C). One of 184 Israeli Arab and 2 of 192 independent Bedouin control chromosomes were found to be carriers of this mutation (table 1). Subsequently, two other patients with CDAI from two unrelated families were found to be homozygous for two additional CDAN1 missense mutations: a proline→leucine substitution at codon 1129, found in a French Polynesian patient, and an asparagine→serine substitution at codon 598, present in three affected brothers from a family of French origin (table 1). It is noteworthy that the three sibs also exhibit sensorineural deafness and lack of motile sperm cells. These symptoms can be accounted for by a large 70-kb deletion 1 Mb distal to the CDAN1 gene (N. Avidan, H. Tamary, O. Dgany, D. Cattan, A. Pariente, M. Thulliez, N. Borot, L. Moati, A. Barthelme, L. Shalmon, T. Krasnov, E. Ben-Asher, T. Olender, M. Khen, I. Yaniv, R. Zaizov, H. Shalev, J. Delaunay, M. Fellous, D. Lancet, and J. S. Beckmann, unpublished data). Hence, discovery of the A→G substitution in exon 12 indicates that these brothers carry, in addition to the 70-kb deletion, a missense mutation in CDAN1 responsible for the CDAI phenotype. It is surprising to find tightly linked independent mutations on the same haplotype. However, cases of contiguous gene syndromes have been reported (Bitner-Glindzicz et al. 2000; Parvari et al. 2001; Shanske et al. 2001); given the growing number of mapped genetic disorders, such cases may be frequent enough to be encountered.
Figure 2.
The mutation in a Bedouin family. A, Pedigree of a Bedouin family, showing the segregation of the founder chromosome with the disease. B, Sequence chromatogram from a wild-type (WT) and diseased (M) genotype showing a C→T substitution in exon 25, converting arginine to tryptophan at codon 863. C, Segregation of the Bedouin mutation, as assayed by NcoI restriction. A 412-bp PCR product is generated between the F and R primers. The mutation results in a gain of restriction site and, therefore, the restriction digest produced fragments of 175 and 237 bp, instead of the 412-bp band observed in healthy individuals.
Three sporadic European patients with CDAI were found to be compound heterozygotes for CDAN1 mutations. One patient has two distinct missense mutations, whereas the other two have one null and one missense mutation each (table 1). Segregation analysis of each mutation in respective families and genotyping of control chromosomes was done by restriction fragment analysis (NEBiolabs) or by mass spectrometry SNP scoring (Sauer et al. 2002) (primers available upon request). Segregation analysis demonstrated that all patients inherited one mutation from each parent, thus confirming that the patients are true compound heterozygotes. In addition, two patients of European descent and one of Arab descent were shown to carry one distinct missense mutation each (table 1). The unidentified mutations may be located in the promoter or introns of CDAN1 or in some as-yet-unidentified exons. It is noteworthy that one additional patient (diagnosed by A.I.), who presented the clinical characteristics of CDAI, was found to be merely a compound heterozygote for two synonymous nucleotide changes (data not shown). For this patient with CDAI, pathogenic mutations remain to be identified either in CDAN1 or in a second locus involved in the etiology of this disease. In any event, our data imply a considerable molecular homogeneity for CDAI.
In total, 12 different mutations in nine families with CDAI were identified: 4 in exon 14, 3 in exon 24, 2 in exon 19, and 1 each in exons 12, 23, and 26. The mutation-clustering pattern suggests either that these exons are more prone to mutation or that they encode essential functional domains (table 1). Furthermore, although nonsense mutations were found in two compound heterozygotes, no homozygote for null-type mutations has been identified. This suggests that codanin-1 may have a unique and essential function, with very little redundancy. Future knockout studies and further screening of patients with CDAI should indicate whether the lack of this protein is lethal. In addition, the diversity of the severity of the disease among homozygous Bedouin patients with CDAI suggests that modifier genes may modulate the phenotype. Hence, at this stage, it is impossible to establish precise phenotype-genotype correlation.
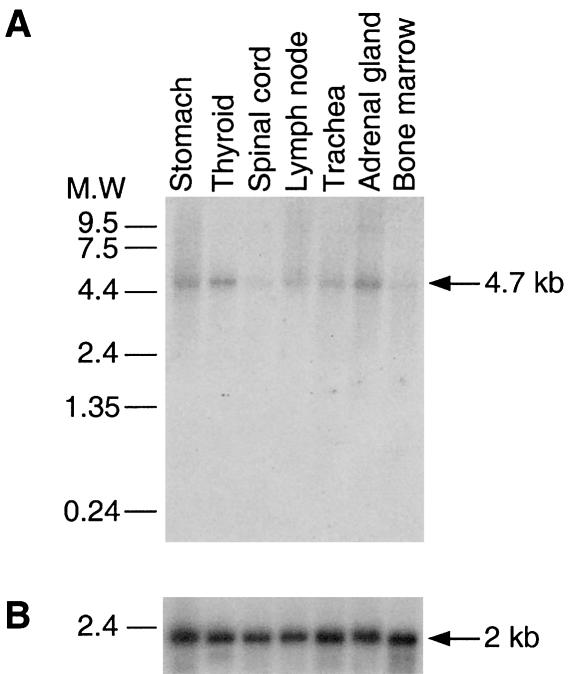
Northern blot analysis was performed with a cDNA probe for exons 26–28 on RNA from eight different human tissues (Clontech). All tissues expressed the same 4.7-kb band (fig. 3A), suggesting that the gene is ubiquitously expressed and that the inferred mRNA is close to full length. A 369-bp shorter, alternatively spliced variant was found upon RT-PCR (data not shown) to be coexpressed in fibroblast cell line cultures from both patients with CDAI and healthy control individuals but not in erythroid cells. It is generated by in-exon splicing of exons 11–14 (fig. 1b), which preserves the original reading frame. This mRNA isoform encodes a putative protein of 1,103 amino acids. This major CDAN1 mRNA appears to be expressed at a level at least tenfold lower than β-actin control as evidenced by their relative exposure times (fig. 3B). This observation is in agreement with the results of expression array experiments (data not shown) and the low number of CDAN1 ESTs present in the databases.
Figure 3.
Northern blot analysis. Panels A and B show CDAN1 and β-actin mRNA exposed for 1 week and overnight, respectively. The membrane was purchased from Clontech and was hybridized according to the manufacturer’s instructions. The membrane was first probed with codanin-1 (exons 26–28 [fig. 1]) and subsequently reprobed with β-actin cDNA as an internal control.
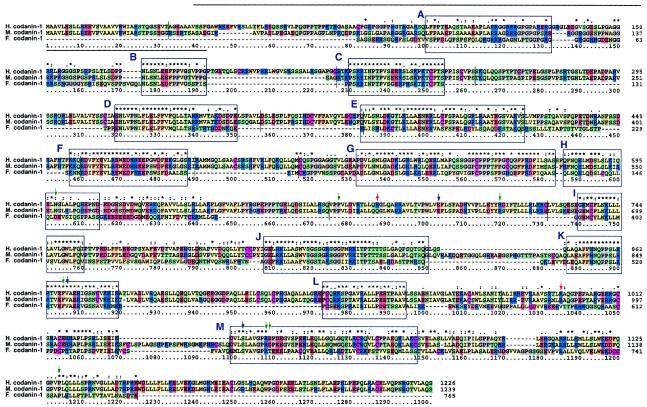
BLAST homology searches (Altschul et al. 1990) against several genomic sequence databases showed no obvious human codanin-1 paralog but revealed two putative orthologs: one in the mouse syntenic region (84% identity) and one in Fugu rubripes (44% identity) (fig. 4). We also identified a putative ortholog in Drosophila melanogaster (GenBank accession number AF487678S2), which shares 23% identity with codanin-1. This protein, vanaso, has been proposed to be involved in the fly’s olfactory behavior and is unlikely to be a functional CDAN1 ortholog. Codanin-1 has no clear intracellular location-specific domains, such as a transmembrane or a signal peptide segments; it contains, however, numerous o-glycosylation consensus sites. In addition, the amino-terminal domain shares a significant homology with fibrillar collagens as revealed by FASTA (Pearson and Lipman 1988) (fig. 4). A multiple alignment was performed using the CLUSTALX program with the default parameters (Higgins et al. 1996). Further domain analysis was performed by BLOCKS (Henikoff et al. 2000), defining 13 interspecies sequence conservation regions (fig. 4). Subsequently, these BLOCKs were compared with all BLOCK database motifs, using the Local Alignment of Multiple Alignment method (Pietrokovski 1996). Similarities to two domains of the microtubule-associated protein MAP1B (Noble et al. 1989) (fig. 4, green shaded blocks C and E) and to two domains of fibrillar collagen triple-helix repeats (Silver et al. 2002) (fig. 4, pink shaded blocks A and G) were unveiled. Using CYRCA, more remote, indirect similarities were further detected between the triple-helix repeats blocks and synapsins blocks, a protein family known to be involved in binding diverse substrates including actin, spectrin, and microtubules (Angers et al. 2002). CYRCA is a method developed to identify blocks of potentially similar function and structure appearing in different contexts (Kunin et al. 2001). On the basis of the disease phenotype, which includes spongy heterochromatin and invagination of the nuclear membrane, and the protein similarity results, we speculate that codanin-1 might be involved in nuclear membrane integrity by connecting the nuclear membrane and microtubules (Dreger et al. 2001; Lippincott-Schwartz 2002).
Figure 4.
Multiple alignment of human codanin-1. Multiple alignment with its putative mouse and fugu orthologs. Identical and similar residues in the multiple-alignment chart are represented by asterisks (*) and colons (:), respectively; weak conservation is represented by periods (.). Conserved BLOCKS are confined within blue boxes. Shaded boxes are used for known block similarity: green, fibrillar collagens (IPB00885) (A, G); pink box, MAP1B (IPB000102) (C, E). Arrows indicate positions of mutated residues; green and blue arrows specify missense mutations within families I–VI and VII–IX, respectively, and red arrows specify null mutations. Note the cluster of mutations around aa 1034–41 and 866–867. The thick black arrow indicates the region with shared similarity to fibrillar collagens. The thin black line represents a split between exons.
Further analysis of codanin-1, using antibodies and animal models, should help to unravel the mechanism by which this protein ensures nuclear integrity during erythropoiesis, to clarify the pathogenesis of the disease, and possibly to facilitate the development of novel strategies for its management. Furthermore, identification of the gene involved in CDAI may facilitate the cloning of the genes involved in other types of CDA.
Acknowledgments
This work was supported in part by the Israel Ministry of Health Chief Scientist Grant (to H.T.), the Arc-en-Ceil/Keshet exchange program (to H.T. and J.D.), and by an Israel Ministry of Science Culture and Sports grant for the National Laboratory for Genome Infrastructure (to D.L.). It was also supported by the Crown Human Genome Center, the Alfred Krupp Foundation, the German-Israeli Foundation for Scientific Research and Development, and the Weizmann Institute Glasberg, Levy, Nathan Brunschwig and Levine funds (all to D.L.). R.Z. is incumbent of the J. Maus and G. Ceaseman-Maus Chair in Pediatric Hematology; D.L. holds the Ralph and Lois Silver Chair in Human Genomics; and J.S.B. is a recipient of the Hermann-Mayer chair. We also thank Profs. J.-P. Dommergues and G. Tchernia and Drs. F. Bernaudin, L. Roda, and H. Chambost for referring their patients to us; and Dr. M. Meunier-Rotival and Ms. C. Driancourt and M. A. Proust for their invaluable help.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ (for codanin-1 homologs and orthologs, as well as genetic markers)
- Blocks, http://bioinformatics.weizmann.ac.il/blocks/ (for biological sequence analysis) (Henikoff et al. 2000)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for KIAA0770 [AB018313], DKFZP564G2022 [NM_015497], ZFP106 [NM_022473], SNAP23 [NM_003825], FLJ10460 [NM_018097], FLJ23375 [NM_024956], CCNDBP1 [NM_012142], EPB42 [NM_000119], TGM5 [NM_004245], UBR1 [AF525401], TTBK [AF525400], FLJ008 [AF525397], CDANI [AF525398], LOC146050 [AF525399], and vanaso [AF487678S2])
- Local Alignment of Multiple Alignment (LAMA), http://bioinformatics.weizmann.ac.il/blocks-bin/LAMA_search.sh (for sequence motifs search) (Pietrokovski 1996)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CDAI [MIM 224120], CDAII [MIM 224100], and CDAIII [MIM 105600])
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- Angers A, Fioravante D, Chin J, Cleary LJ, Bean AJ, Byrne JH (2002) Serotonin stimulates phosphorylation of aplysia synapsin and alters its subcellular distribution in sensory neurons. J Neurosci 22:5412–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitner-Glindzicz M, Lindley KJ, Rutland P, Blaydon D, Smith VV, Milla PJ, Hussain K, Furth-Lavi J, Cosgrove KE, Shepherd RM, Barnes PD, O'Brien RE, Farndon PA, Sowden J, Liu XZ, Scanlan MJ, Malcolm S, Dunne MJ, Aynsley-Green A, Glaser B (2000) A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat Genet 26:56–60 [DOI] [PubMed] [Google Scholar]
- Bonfield JK, Smith K, Staden R (1995) A new DNA sequence assembly program. Nucleic Acids Res 23:4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay J, Iolascon A (1999) The congenital dyserythropoietic anaemias. Baillieres Best Pract Res Clin Haematol 12:691–705 [DOI] [PubMed] [Google Scholar]
- Dreger M, Bengtsson L, Schoneberg T, Otto H, Hucho F (2001) Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc Natl Acad Sci USA 98:11943–11948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini P, del Giudice EM, Delaunay J, Totaro A, Granatiero M, Melchionda S, Zelante L, Iolascon A (1997) Localization of the congenital dyserythropoietic anemia II locus to chromosome 20q11.2 by genomewide search. Am J Hum Genet 61:1112–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff J, Greene E, Pietrokovski S, Henikof S (2000) Increased coverage of protein families with the blocks database servers. Nucleic Acids Res 28:228–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266:383–402 [DOI] [PubMed] [Google Scholar]
- Iolascon A, Delaunay J, Wickramasinghe SN, Perrotta S, Gigante M, Camaschella C (2001) Natural history of congenital dyserythropoietic anemia type II. Blood 98:1258–1260 [DOI] [PubMed] [Google Scholar]
- Kunin V, Chan B, Sitbon E, Lithwick G, Pietrokovski S (2001) Consistency analysis of similarity between multiple alignments: prediction of protein function and fold structure from analysis of local sequence motifs. J Mol Biol 307:939–949 [DOI] [PubMed] [Google Scholar]
- Lavabre-Bertrand T, Blanc P, Navarro R, Saghroun M, Vannereau H, Braun M, Wagner A, Taib J, Lavabre-Bertrand C, Navarro M (1995) Alpha-interferon therapy for congenital dyserythropoiesis type I. Br J Haematol 89:929–932 [DOI] [PubMed] [Google Scholar]
- Lind L, Sandstrom H, Wahlin A, Eriksson M, Nilsson-Sojka B, Sikstrom C, Holmgren G (1995) Localization of the gene for congenital dyserythropoietic anemia type III, CDAN3, to chromosome 15q21-q25. Hum Mol Genet 4:109–112 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J (2002) Cell biology: ripping up the nuclear envelope. Nature 416:31–32 [DOI] [PubMed] [Google Scholar]
- Noble M, Lewis SA, Cowan NJ (1989) The microtubule binding domain of microtubule-associated protein MAP1B contains a repeated sequence motif unrelated to that of MAP2 and tau. J Cell Biol 109:3367–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parez N, Dommergues M, Zupan V, Chambost H, Fieschi JB, Delaunay J, Miélot F, Cramer EM, Dommergues JP, Wickramasinghe SN, Tchernia G (2000) Severe congenital dyserythropoietic anaemia type 1: prenatal management, transfusion support and alpha-interferon therapy. Br J Haematol 110:420–423 [DOI] [PubMed] [Google Scholar]
- Parvari R, Brodyansky I, Elpeleg O, Moses S, Landau D, Hershkovitz E (2001) A recessive contiguous gene deletion of chromosome 2p16 associated with cystinuria and a mitochondrial disease. Am J Hum Genet 69:869–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrokovski S (1996) Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res 24:3836–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope SH, Fibach E, Sun J, Chin K, Rodgers GP (2000) Two-phase liquid culture system models normal human adult erythropoiesis at the molecular level. Eur J Haematol 64:292–303 [DOI] [PubMed] [Google Scholar]
- Sauer S, Gelfand DH, Boussicault F, Bauer K, Reichert F, Gut IG (2002) Facile method for automated genotyping of single nucleotide polymorphisms by mass spectrometry. Nucleic Acids Res 30:e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanske A, Ferreira JC, Leonard JC, Fuller P, Marion RW (2001) Hirschsprung disease in an infant with a contiguous gene syndrome of chromosome 13. Am J Med Genet 102:231–236 [DOI] [PubMed] [Google Scholar]
- Silver F, Horvath I, Foran D (2002) Mechanical implications of the domain structure of fiber-forming collagens: comparison of the molecular and fibrillar flexibilities of the alpha1-chains found in types I–III collagen. J Theor Biol 216:243–254 [DOI] [PubMed] [Google Scholar]
- Tamary H, Shalev H, Luria D, Shaft D, Zoldan M, Shalmon L, Gruinspan A, Stark B, Chaison M, Shinar E, Resnitzky P, Zaizov R (1996) Clinical features and studies of erythropoiesis in Israeli Bedouins with congenital dyserythropoietic anemia type I. Blood 87:1763–1770 [PubMed] [Google Scholar]
- Tamary H, Shalmon L, Shalev H, Halil A, Dobrushin D, Ashkenazi N, Zoldan M, Resnitzky P, Korostishevsky M, Bonne-Tamir B, Zaizov R (1998) Localization of the gene for congenital dyserythropoietic anemia type I to a <1-cM interval on chromosome 15q15.1-15.3. Am J Hum Genet 62:1062–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe SN (1997) Dyserythropoiesis and congenital dyserythropoietic anemias. Br J Haematol 98:785–797 [DOI] [PubMed] [Google Scholar]
- Wickramasinghe SN, Hasan R, Smythe J (1997) Reduced interferon-alpha production by Epstein-Barr virus transformed B-lymphoblastoid cell lines and lectin-stimulated lymphocytes in congenital dyserythropoietic anaemia type I. Br J Haematol 98:295–298 [DOI] [PubMed] [Google Scholar]