Abstract
Saccharomyces cerevisiae yeast cells polarize in order to form a single bud in each cell cycle. Distinct patterns of bud-site selection are observed in haploid and diploid cells. Genetic approaches have identified the molecular machinery responsible for positioning the bud site: during bud formation, specific locations are marked with immobile landmark proteins. In the next cell cycle, landmarks act through the Ras-family GTPase Rsr1 to promote local activation of the conserved Rho-family GTPase, Cdc42. Additional Cdc42 accumulates by positive feedback, creating a concentrated patch of GTP-Cdc42, which polarizes the cytoskeleton to promote bud emergence. Using time-lapse imaging and mathematical modelling, we examined the process of bud-site establishment. Imaging reveals unexpected effects of the bud-site-selection system on the dynamics of polarity establishment, raising new questions about how that system may operate. We found that polarity factors sometimes accumulate at more than one site among the landmark-specified locations, and we suggest that competition between clusters of polarity factors determines the final location of the Cdc42 cluster. Modelling indicated that temporally constant landmark-localized Rsr1 would weaken or block competition, yielding more than one polarity site. Instead, we suggest that polarity factors recruit Rsr1, effectively sequestering it from other locations and thereby terminating landmark activity.
Keywords: polarity, Cdc42, bud, yeast, Rsr1
1. Introduction
A polarized cell has a clear axis, with a single ‘front’. Different cells display a variety of polarized morphologies, but a conserved family of polarity GTPases (Cdc42 and Rac in animals and fungi, Rop in plants) appear to control polarization in most eukaryotes [1–3]. The GTPases are anchored to the plasma membrane (and other membranes) by prenylation. A cell's ‘front’ can be defined as the cortical site at which cells accumulate a high concentration of the activated (GTP-bound) form of the GTPase. The active GTPase then organizes cytoskeletal elements through a variety of effectors to yield the polarized morphology appropriate to the cell type.
The direction of polarization can be determined by chemical or physical signals. However, when spatial cues are absent, many cells polarize in a random direction by ‘symmetry breaking’. Theoretical analyses dating back to Turing [4] showed that spontaneous pattern formation from homogeneous starting conditions could occur if autocatalytic biochemical reactions amplified small clusters of ‘morphogens’ (in this context, polarity factors) arising from stochastic fluctuations. During symmetry breaking, some mechanism must ensure that only one among all of the potential sites develops into the cell's front.
The budding yeast Saccharomyces cerevisiae has served as a tractable model to investigate polarity establishment since the pioneering genetic screens by Pringle and co-workers identified the conserved Rho-family GTPase Cdc42 as the master regulator of polarity [5]. Cdc42 localization and activity are affected by GEF (GDP/GTP exchange factor), GAP (GTPase-activating protein) and GDI (guanine nucleotide dissociation inhibitor) regulators. The site of Cdc42 activation is influenced by a ‘bud-site-selection’ system that depends on the Ras-family GTPase Rsr1. Mutants lacking Rsr1 break symmetry and polarize to a single, apparently random, site [6,7].
Symmetry breaking in yeast does not require polymerized actin or tubulin, but (at least in rsr1 cells) it does require the polarity scaffold protein Bem1 [8]. Bem1 acts by forming a complex that links a Cdc42 effector (PAK, p21-activated kinase) to the only Cdc42 GEF, Cdc24 [9–12]. When GTP-Cdc42 binds to a PAK associated with Bem1 and GEF in a complex, the GEF loads GTP on neighbouring Cdc42 (figure 1a). This creates a positive feedback loop that promotes growth of a cortical cluster of GTP-Cdc42. The Bem1 complex diffuses rapidly in the cytoplasm, allowing the complexes to be rapidly recruited to a growing GTP-Cdc42 cluster. By contrast, GTP-Cdc42 diffuses slowly at the membrane, so the cluster does not dissipate too rapidly (see review by Johnson et al. [12]).
Figure 1.
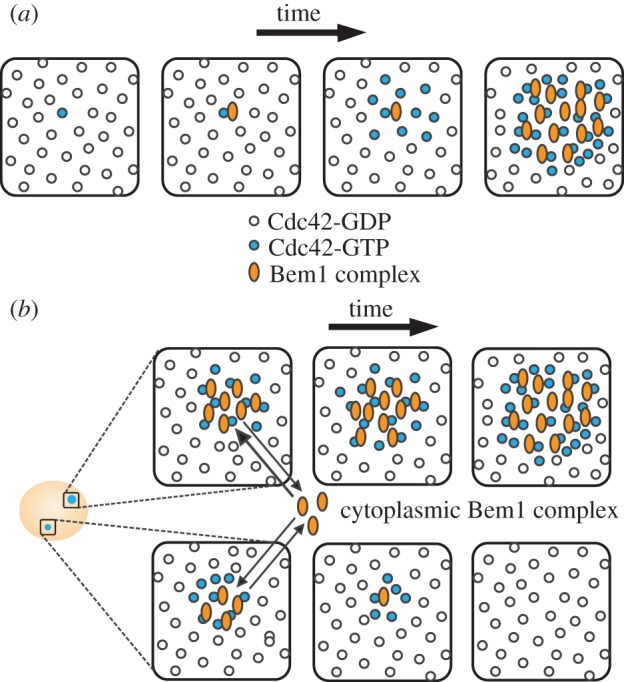
Symmetry-breaking polarization of GTP-Cdc42 by Bem1 complex. (a) A random site on the cell membrane exhibits a stochastic increase in GTP-Cdc42. A Bem1 complex is recruited to this site, which allows the conversion of nearby GDP-Cdc42 to GTP-Cdc42. The additional GTP-Cdc42 recruits more Bem1 complexes in a positive feedback loop, leading to the formation of a polarized cluster of GTP-Cdc42. (b) Stochastic activation of Cdc42 can happen at more than one cortical site and lead to the formation of multiple polarized foci. Competition between foci for cytoplasmic Bem1 complexes ensures that only one focus eventually wins.
Imaging of polarity establishment in rsr1Δ cells revealed that cells frequently ‘grow’ more than one cluster of Cdc42, but that the clusters then compete with each other and a single winner emerges [13] (figure 1b). Computational simulations of the Cdc42/Bem1 system indicated that nascent polarity clusters would compete with each other for cytoplasmic Bem1 complexes and that the largest cluster would eventually win [10]. Competition may be accelerated by the presence of negative feedback in the polarity pathway [13]. Rapid competition could explain why cells only make one bud, and some experimental manipulations intended to slow competition resulted in the simultaneous formation of two buds [11,13].
In wild-type yeast cells, polarization and bud emergence occur at sites influenced by positional markers or ‘landmarks’: transmembrane proteins deposited at specific places during bud formation, anchored to the rigid cell wall, and then inherited by daughter cells [3,14,15]. During cytokinesis, the landmark protein Axl2 is deposited in a ring on either side of the cleavage furrow [16] (figure 2a). The distal (previously the bud tip) and proximal (previously the neck) poles of newborn cells are marked by the landmark proteins Bud8 and Bud9, respectively [15] (figure 2b). Haploid cells use Axl2 to select ‘axial’ sites, in which the new ‘front’ is established adjacent to the immediately preceding cytokinesis site, so that sequential buds emerge next to each other in a chain [17]. Diploid cells use Bud8 and Bud9 to select new sites in a ‘bipolar’ pattern, in which the new ‘front’ is established at one of the two cell poles, and sequential buds may emerge at opposite ends [17]. Additional landmark proteins, Rax1 and Rax2, are deposited in a ring marking each previous bud site, and also contribute to bud-site selection in diploids [18–20].
Figure 2.
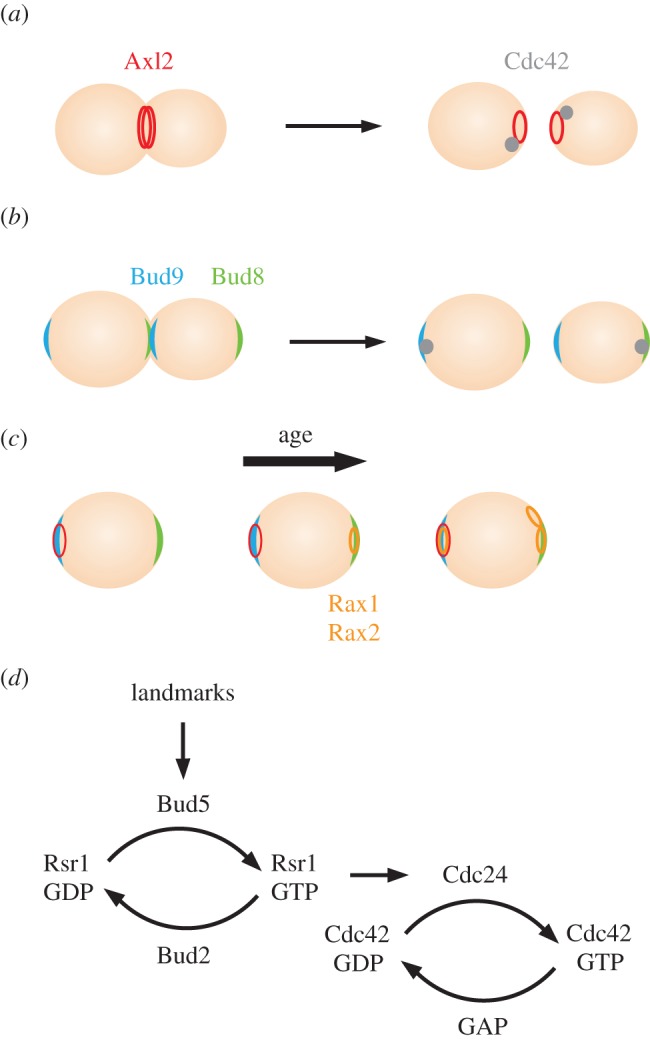
Bud-site selection in yeast. (a) Haploids. (b) Diploids. (c) Additional landmarks deposited as cells replicate. (d) Landmarks influence GTP-loading of Cdc42 via Rsr1 and its regulators.
Although haploids and diploids prefer to use different landmark proteins, all landmarks are deposited in both haploid and diploid cells. Moreover, genetic findings indicate that all landmarks are potentially active: if the preferred landmark-encoding gene is deleted, the other landmarks are used instead. Thus, a first-generation daughter cell is born with three landmarks, and mother cells acquire more marked sites as they age (figure 2c).
The landmark proteins interact with Bud5, a GEF that promotes localized activation of Rsr1 [21–26] (figure 2d). The GAP, Bud2, promotes GTP hydrolysis by Rsr1, thereby restricting Rsr1-GTP accumulation to the vicinity of the landmarks [21,22,27]. Rsr1-GTP binds to the Cdc42-directed GEF, Cdc24, promoting localized GTP-loading of Cdc42 [28,29] (figure 2d). In this way, the pre-localized landmarks influence where Cdc42 GTP-loading takes place, and hence where a new front will form.
As has long been recognized [17], the landmarks define many possible polarization sites: there is an entire ring of potential sites marked by Axl2, and both poles are marked by Bud8 and Bud9. How, then, do cells select a single polarity ‘front’ from these many potential sites?
2. Material and methods
(a). Yeast strains
All yeast strains (listed in table 1) are in the YEF473 background (his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52). We used strains with the polarity marker Bem1-GFP replacing endogenous Bem1, because previous work indicated that Bem1-GFP is functional, whereas GFP-Cdc42 is only partially functional and can have mildly toxic effects [13].
Table 1.
Yeast strains.
| strain | relevant genotype | source |
|---|---|---|
| DLY9069 | a BEM1-GFP:LEU2 | |
| DLY9200 | a/α BEM1-GFP:LEU2/BEM1-GFP:LEU2 rsr1::TRP1/rsr1::TRP1 | [11] |
| DLY9201 | a/α BEM1-GFP:LEU2/BEM1-GFP:LEU2 | this report |
| DLY11780 | a/α BEM1-GFP:LEU2/BEM1-GFP:LEU2 rsr1::TRP1/rsr1::TRP1 SPC42-mCherry::kanR/SPC42 | this report |
| DLY15125 | a/α BEM1-GFP:LEU2/BEM1-GFP:LEU2 rga1::HIS3/rga1::HIS3 | this report |
(b). Live cell microscopy
Cells were grown in synthetic medium (MP Biomedicals) with dextrose. Prior to imaging, cells were arrested with 200 mM hydroxyurea (Sigma) at 30°C for 3 h, washed, released into fresh synthetic medium for 1 h, harvested and mounted on a slab composed of medium solidified with 2% agarose (Denville Scientific Inc.). The slab was put in a temperature-control chamber set to 30°C. In this study, we imaged cells using a spinning disc confocal microscope as detailed below. Comparison of polarization kinetics suggested that imaging in this system was not significantly more phototoxic than with the wide-field system used previously (see electronic supplementary material, figure S1).
Images were acquired with an Andor XD revolution spinning disc confocal microscope (Olympus) with a Yokogawa CSU-X1 5000 r.p.m. disc unit, and a 100×/1.4 UPlanSApo oil-immersion objective controlled by MetaMorph software (Universal Imaging; http:://microscopy.duke.edu). Images (stacks of 30 images taken at 0.24 μm z-steps) were captured by an iXon3 897 EMCCD camera with 1.2× auxiliary magnification (Andor Technology). The fluorescence light source was used at 10% maximal output. An EM-Gain setting of 150 was used for the EMCCD camera. Exposure to the 488 nm diode laser was 150 ms.
For latrunculin treatment, asynchronous log phase cells were harvested, resuspended in synthetic medium containing 200 µM latrunculin A, and mounted on a 2% agarose slab with 200 µM latrunculin A. The cells were on the slab for at least 15 min prior to imaging.
(c). Deconvolution and image analysis
Images were deconvolved using Huygens Essential software (Scientific Volume Imaging). The classic maximum-likelihood estimation and predicted point spread function method with signal-to-noise ratio 3 was used with a constant background across all images from the same day. The output format was 16-bit, unscaled images to enable comparison of pixel values. Comparison of raw and deconvolved images suggested that deconvolution improved signal-to-noise ratio without introducing artefacts (electronic supplementary material, figure S2).
To detect polarity foci in different focal planes, maximum intensity projections were constructed and scored visually for the presence of more than one focus. The coexistence time is the interval between the first frame in which more than one spot was detected and the frame when only one spot was detected. Bem1-GFP intensities were quantified using Volocity (Improvision). A threshold was set that would only select the polarized signal, and the summed, polarized intensity was recorded. Changes in intensity are reported as percent of maximum for that cell. To quantify the percentage of Bem1-GFP in the polarized focus at peak frame, two thresholds were set to separately select the entire cell and the polarized signal. The percentage was determined by these two summed intensities with background fluorescence subtracted. Images were processed for presentation using MetaMorph and Photoshop (Adobe).
(d). Modelling
To ask how the presence landmark-localized Rsr1–GEF complexes would impact the dynamics of polarization, we turned to computational modelling. To provide context for the model, we provide a brief historical synopsis below.
(i). Background on model development
The model used here evolved from one first formulated by Goryachev & Pokhilko [10]. That model used mass action kinetics to describe the known biochemical interactions and activities of three molecular species: Cdc42, a GDI and a Bem1–GEF complex. There was also an implicit GAP activity. The model contained positive feedback but no negative feedback.
Some model parameters (representing Bem1–GEF and implicit GAP activities, as well as interaction rate constants) were subsequently adjusted using in vitro biochemical data as a guide [11]. For example, the total GEF activity measured in yeast lysates was used to constrain product of two model parameters: GEF abundance and GEF specific activity. Thus, the model is based on documented biochemistry, but several individual parameters (as opposed to their product) remain poorly constrained.
That model was then elaborated to include negative feedback by Howell et al. [13], based on the observed oscillatory dynamics of polarization. The mechanism of negative feedback remains to be established. Two hypothetical mechanisms (acting by positive regulation of a GAP or by negative regulation of the Bem1–GEF complex) were considered in that study and yielded qualitatively similar behaviour. Here, we chose to use the simpler of the models, involving Bem1–GEF regulation. We note that because the mechanism remains speculative, the negative feedback parameters are also speculative and not constrained by data.
Two other features of the 2012 model are noteworthy. First, the action of the GDI in the model was simplified. The 2008 model included a GDI able to bind GDP-Cdc42 and extract it reversibly from the membrane to the cytoplasm. In the 2012 model, there is an implicit GDI represented by allowing GDP-Cdc42 to spontaneously exchange between membrane and cytoplasm. Second, the model added a Gaussian noise term to the Bem1–GEF species. Both the implicit GDI and the Bem1–GEF noise were retained in our model.
A subsequent study obtained a better-constrained value for the abundance of Cdc42 (1 µM) [30], which we have adopted in the current model. In addition, we manually tuned some of the other parameters so that the model would reproduce three features extracted from imaging data: (i) the measured shape of the Cdc42 peak (peak width at half height, 1.9 μm [31]); (ii) Cdc42 dynamics in the peak (FRAP recovery half-time, 3.5 s [32]) and our estimate of the amount of Cdc42 in the peak (proportion of the total Cdc42, 4.6%). All parameter values are listed in the electronic supplementary material, table S2.
In our model, we also made the GAP explicit, rather than implicit. The parameter-adjusted model was used to represent rsr1 mutant cells, in which landmark proteins do not affect Cdc42 behaviour. Unique to this study is the addition of the Rsr1–GEF and the Rga1–GAP.
Previous work indicated that the Cdc42-directed GEF, Cdc24, was present in both cytoplasmic and local cortical pools that exchanged rapidly [33]. The cytoplasmic pool was in significant excess compared with the localized pool, which presumably represents the sum of Bem1-bound and Rsr1-bound GEF. Because the GEF is in excess, we assume here that Rsr1- and Bem1-bound pools of GEF are not in competition with each other. This allowed us to simplify the model, using only two GEF species (Rsr1–GEF and Bem1–GEF) and ignoring the excess cytoplasmic GEF. Bem1–GEF behaves as in the previous models. The new Rsr1–GEF is represented as an immobile GEF located at the sites demarcated by landmarks (a ring in haploids and two circular patches at the cell poles in diploids).
The localization of Rga1 at a circular patch at the cytokinesis site was determined experimentally [34]. Thus, we modelled Rga1–GAP as an immobile GAP located at that site. The Rsr1–GEF and Rga1–GAP activities were set as described below: there are no available data to constrain these values, except that the Rga1–GAP must be strong enough to exclude polarization within the previous division site [34].
The full model is described in the electronic supplementary material.
3. Results
Imaging of rsr1Δ/rsr1Δ cells breaking symmetry revealed key features of polarity establishment [13]; here, we report similar studies with wild-type (RSR1/RSR1) yeast cells with intact bud-site selection. One could imagine that the symmetry-breaking process is a backup pathway that is only used when the normal cues are absent. In this view, an intact bud-site-selection system actually chooses the future polarity site prior to activating Cdc42. If that is correct, then unlike in rsr1Δ/rsr1Δ cells, all RSR1/RSR1cells would form only one polarity cluster. Alternatively, the bud-site-selection system may simply activate a little Cdc42 at several landmark-designated ‘permitted’ sites for polarity. In that scenario, we might see more than one initial polarity cluster in RSR1/RSR1 cells, with the final polarity site determined by competition between clusters as seen in rsr1Δ/rsr1Δ cells.
(a). Imaging Bem1-GFP in RSR1 cells
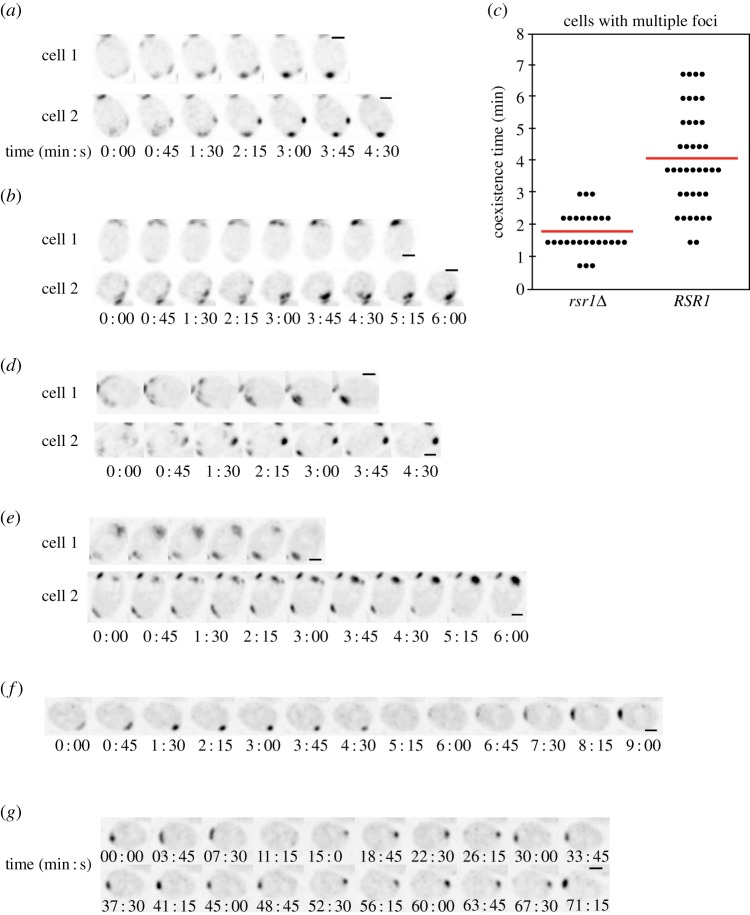
We imaged polarity establishment using the functional polarity marker Bem1-GFP [11]. To enrich the imaged population for cells about to polarize, we used hydroxyurea arrest-release synchronization as previously described [13]. This has the added benefit of reducing phototoxicity, allowing higher temporal resolution. In both haploid (RSR1) and diploid (RSR1/RSR1) cells, polarity sites were established in the expected locations (adjacent to the division site in haploids, at one or the other pole in diploids). Many cells (40 of 65 diploids imaged) initially developed more than one polarity cluster, and the multi-cluster intermediates resolved to a single site in a manner suggestive of competition (figure 3a,b). This observation suggests that polarity establishment in RSR1 cells proceeds via the same basic positive feedback and competition process (figure 1) as in cells breaking symmetry, with the exception that Rsr1 biases the location of initial polarity cluster growth.
Figure 3.
Competition among polarity clusters in wild-type cells. (a, b) Growth of multiple Bem1-GFP clusters and resolution to a single cluster in wild-type diploid (a, DLY9201) and haploid cells (b, DLY9069). Cells were synchronized by hydroxyurea arrest-release prior to imaging. A residual Bem1-GFP signal at the previous cytokinesis site is also visible at the top (a) or bottom (b) of the cropped frames. t = 0 indicates first detection of multiple polarized signals. (c) Coexistence time of multiple clusters: time between the detection of more than one cluster and the first time frame with only one cluster in diploid rsr1Δ/rsr1Δ (n = 27) and RSR1/RSR1 cells (n = 39). Each dot represents one cell; the line is the average coexistence time. (d) Competition between Bem1-GFP clusters is also observed in unsynchronized RSR1/RSR1 cells. (e) Competition between Bem1-GFP clusters is also observed in latrunculin A-treated RSR1/RSR1 cells lacking F-actin. (f) Latrunculin A-treated RSR1/RSR1 cells sometimes display ‘relocation’ of the Bem1-GFP cluster from one pole to the other. (g) In extreme cases, the Bem1-GFP cluster in latrunculin A-treated cells relocates back and forth repeatedly between the two poles. Scale bars, 2 µm. (Online version in colour.)
Quantification of the time taken to resolve multi-cluster intermediates indicated that competition was slower in RSR1/RSR1 cells than in rsr1Δ/rsr1Δ cells (figure 3c). Potential reasons for this difference are considered in the Discussion.
It was conceivable that the synchrony protocol we used might alter the polarization process. However, we detected competition between polarity clusters even in unsynchronized proliferating cells (15 of 40 diploids imaged; figure 3d).
To assess how polarization dynamics might be affected by actin-mediated processes like vesicle trafficking, we imaged unsynchronized cells treated with latrunculin A to depolymerize F-actin. As before, several cells (eight of 32 diploids imaged) displayed competition between polarity clusters (figure 3e). However, whereas in untreated cells the polarity site remained stably located until bud emergence, a subset of latrunculin-treated cells (nine of 32 diploids imaged) displayed polarity-site ‘relocation’ (figure 3f). In these cases, the Bem1 cluster disappeared from one pole and appeared at the other, sometimes repeatedly (a ‘ping-pong’ pattern: figure 3g). This phenotype suggests that in the absence of F-actin, landmarks at both poles (in diploids) continue to compete for polarity proteins even after a large polarity cluster has formed. Thus, F-actin may be needed to ‘lock in’ the initially chosen polarity cluster.
(b). Dynamics of polarization in RSR1 cells
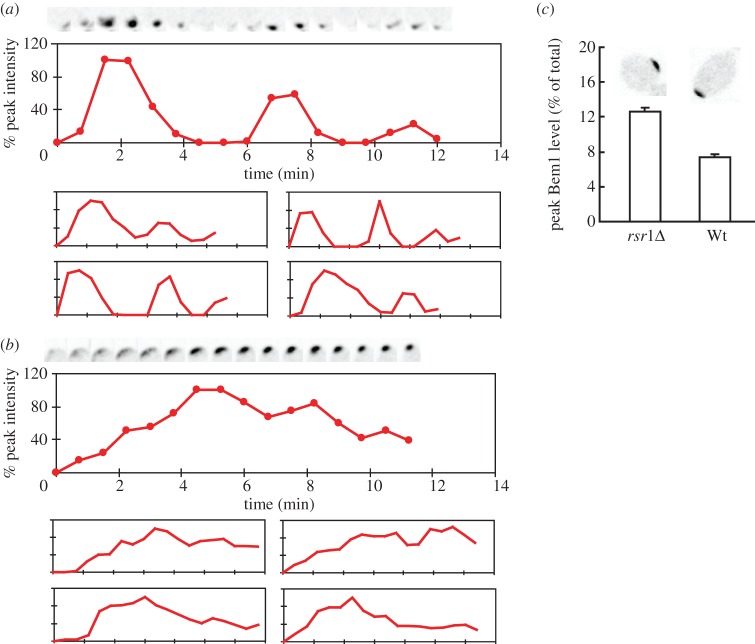
In rsr1Δ/rsr1Δ cells, initial clustering of polarity factors is followed by dispersal and re-clustering in an oscillatory manner, presumably as a result of negative feedback in the polarity circuit [13] (figure 4a). Comparison of RSR1/RSR1 and rsr1Δ/rsr1Δ cells revealed significant differences in the dynamics of polarity clusters: the initial clustering was slower in RSR1/RSR1 cells, and subsequent dispersal was also slower (figure 4b). As a result, Bem1 levels did not oscillate with high amplitude in RSR1/RSR1 cells (because haploid cells polarize near the previous cytokinesis site and Bem1 is localized to both the cytokinesis site and the polarity site, the dynamics of polarization are harder to distinguish in haploids so this analysis focused on diploid cells). Peak levels of Bem1 at the polarity site were lower in RSR1/RSR1 cells than they were in rsr1Δ/rsr1Δ cells (figure 4c and the electronic supplementary material, movie S1).
Figure 4.
Dynamics of Bem1-GFP polarization in diploids. (a,b) Clustering of Bem1-GFP in rsr1Δ/rsr1Δ (a, DLY9200) and RSR1/RSR1 (b, DLY9201) cells. Top panels are cropped images of polarization sites at 45 s intervals. t = 0 is 45 s before the first detection of polarized signal. Middle panels show the intensity changes of Bem1-GFP in the cluster. The trace ends at bud emergence. Bottom panels are traces of four other cells. (c) The fraction of Bem1-GFP that is polarized at the peak frame (mean ± s.e.m.) in rsr1Δ/rsr1Δ (DLY11780) and RSR1/RSR1 (Wt; DLY9201) cells (n = 10). Representative images are shown at top. (Online version in colour.)
In addition to the differences discussed above, a subset of RSR1 and RSR1/RSR1 cells (17 of 51 haploids and 12 of 65 diploids) exhibited a behaviour not seen in rsr1Δ/rsr1Δ mutants: Bem1 accumulated to intermediate levels at one pole but then appeared to fluctuate rapidly at that pole for 10–20 min before strengthening and coalescing to a tighter spot prior to bud emergence (figure 5). This subset of cells appears to polarize by a two-step process in which an initial stage (not obvious in the majority of cells imaged) involves low-level noisy recruitment of Bem1 to Rsr1-demarcated sites. Possible interpretations of this result are considered in the Discussion.
Figure 5.
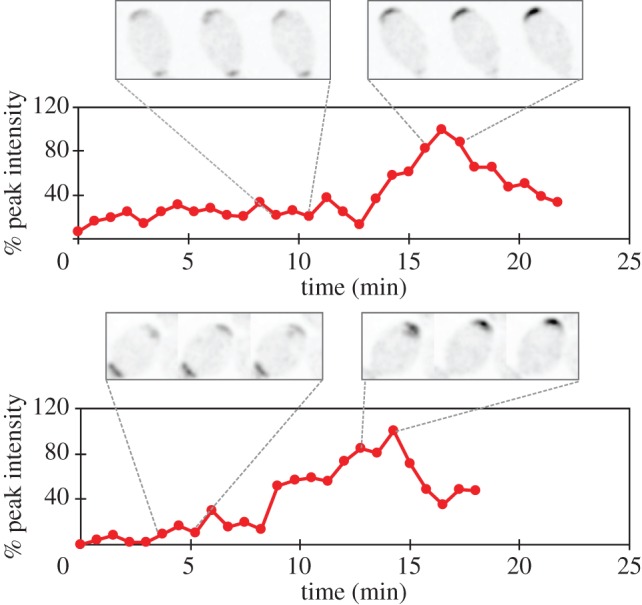
Fluctuating Bem1-GFP clusters. A subset of RSR1/RSR1 cells (DLY9201) initially display a low-level recruitment of Bem1-GFP, which then coalesces to a tighter cluster before bud emergence. Bottom panels: quantification of localized Bem1-GFP signal. Representative frames of cropped images show the fluctuating (top left) and coalescing stages (top right) in two individual cells. Residual Bem1-GFP signal at the previous cytokinesis site is also visible at the bottom of the cropped frames. (Online version in colour.)
(c). Modelling bud-site selection
As discussed above, bud-site-selection landmark proteins are thought to promote local accumulation of Rsr1-GTP, which recruits Cdc24 to sites specified by the landmarks (see Introduction). This would generate a ring of Cdc24 GEF in haploids, and two patches of GEF at opposite poles in diploids. To ask whether such GEF patterns would, in combination with the known symmetry-breaking mechanism, lead to the polarity protein localization observed in RSR1 cells, we turned to computational modelling.
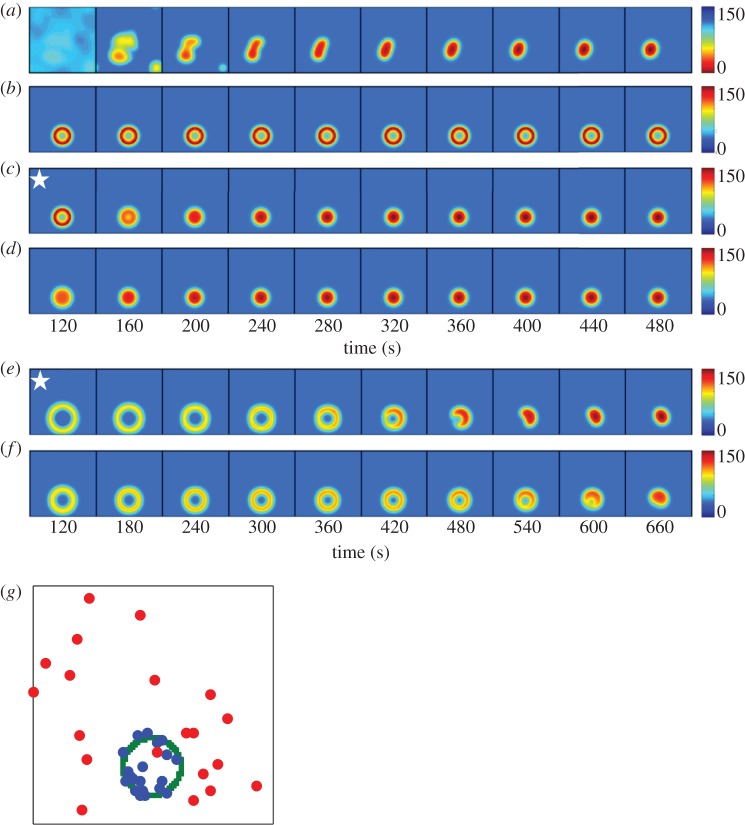
We adapted a model that was originally developed to describe symmetry-breaking polarization in yeast (see Methods) [10,11,13]. The model contains positive feedback owing to the Bem1 complex (figure 1) as well as negative feedback via modification of the Bem1 complex to an inert cytoplasmic state (electronic supplementary material). Stochastic noise was added to the least abundant species (Bem1 complex) as described [13]. However, the model does not incorporate downstream cytoskeletal polarization, and therefore lacks F-actin, which has been suggested to either reinforce [32,33,35,36] or perturb [31,37–39] polarity. The model exhibits the formation of multiple Cdc42 clusters that compete, resulting in a single final polarity cluster (figure 6a). With our model parameters (electronic supplementary material, table S2), the simulations evolve to a single polarity peak in 3–7 min, which is a bit slower than the approximately 2 min it takes, on average, in vivo (figure 3c).
Figure 6.
Modelling polarity establishment in haploids. Each panel represents the concentration of Cdc42 (colour) at the plasma membrane (square) at a particular time (indicated below) after initiating a simulation. (a) Positive and negative feedback loops with noise in the Bem1 complex, no Rsr1–GEF or Rga1–GAP. Because polarity is initiated by noise in this case, each simulation evolves differently. (b) Rsr1–GEF ring added. Rsr1–GEF at 2.5% of total Bem1–GEF. (c) Rsr1–GEF turned off at 120 s (indicated by star). (d) Persistent Rsr1–GEF at 0.025% of total Bem1–GEF. (e) Rga1–GAP plug in the centre of a transient Rsr1–GEF ring. Rsr1–GEF at 2.5% of total Bem1–GEF, turned off after 15 s. Rga1–GAP turned off after 120 s (star). (f) Rga1–GAP plug in the centre of a weak Rsr1–GEF ring. Persistent Rsr1–GEF at 0.025% of total Bem1–GEF. Note longer timescales for (e,f): whereas simulations in (a–d) change little after the last panel, (e,f) continue to evolve towards a single peak. (g) Locations of the Cdc42 peak for 20 simulations of the type shown in (a) (red dots: no Rsr1–GEF) or (f) (blue dots: Rsr1–GEF ring depicted in green). The dots indicate positions with maximum Cdc42 concentration after 1000 s.
To this symmetry-breaking model, we added spatially patterned GEF activity to represent the Cdc24 recruited by Rsr1 (hereafter ‘Rsr1–GEF’). Haploid RSR1 cells were modelled with a ring of Rsr1–GEF activity surrounding the cytokinesis site. The total Rsr1–GEF activity was initially set equal to 2.5% of the Bem1-associated GEF in the model. Under these conditions, Cdc42 accumulated in a ring and remained there for more than 1000 s without evolving to a single peak at the periphery of the ring (figure 6b). As we never observed a ring of Cdc42 in cells, this model fails to recapitulate the effect of Rsr1 on polarity establishment.
Our simulations indicate that even a modest amount of spatially patterned GEF can have a powerful influence on the symmetry-breaking system, overriding its ability to generate a single peak. We considered two possible adjustments that might limit this effect. First, we eliminated the Rsr1–GEF after initiating the simulation. As shown in figure 6c, shutting Rsr1–GEF off did allow the system to evolve to a single peak, but the peak was always centred in the middle of the ring. Second, we tested whether a weaker Rsr1-associated GEF might bias (rather than override) the Bem1 system. With a persistent Rsr1–GEF at 0.025% of the Bem1–GEF, the initial ring also evolved to a single Cdc42 peak centred in the middle of the ring (figure 6d).
In cells, polarization in the centre of the ring would lead to formation of a bud within the previous division site. Although this does not occur in wild-type cells, it does occur at high frequency in rga1Δ mutants [34]. Rga1 is a Cdc42-directed GAP that accumulates at the division site in cells undergoing cytokinesis. It is thought to remain after cytokinesis, creating a ‘plug’ within the Rsr1–GEF ring that discourages polarization within the ring [34].
To take into account the localized GAP activity provided by Rga1, we raised the model GAP activity within the ring. We then simulated the two scenarios above: transient Rsr1–GEF or weak Rsr1–GEF. In the transient Rsr1–GEF simulation, we also made the Rga1–GAP plug transient, as Rga1 relocalizes to the polarity site following polarization [40]. If both Rsr1–GEF and Rga1–GAP were turned off simultaneously, simulations again evolved to a peak in the centre of the ring (not shown). But if the Rga1–GAP was switched off more than 45 s after the Rsr1–GEF, then the initial ring gradually evolved to a Cdc42 peak at the ring periphery (figure 6e). With a weak persistent Rsr1–GEF, the system also evolved to a single Cdc42 peak at the periphery of the ring (figure 6f). The final position of the peak varied from simulation to simulation as a result of noise, but in contrast to the random placement observed without Rsr1–GEF, most peaks were near the ring (figure 6g). Thus, an immobile ring of Rsr1–GEF with an internal plug of Rga1–GAP can recapitulate the final Cdc42 distribution attained in RSR1 haploids, but only if the Rsr1–GEF is either weak or transient.
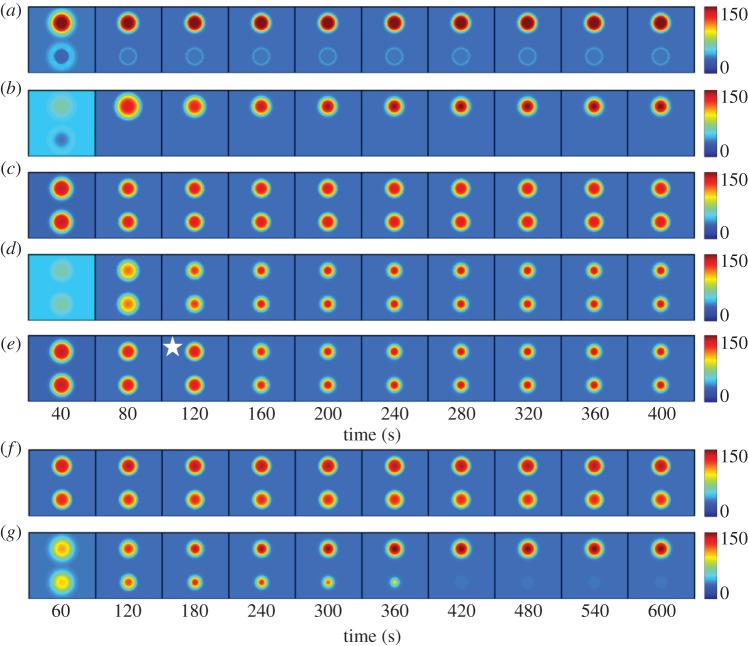
Diploid RSR1 cells were modelled by assuming that they have two patches of Rsr1–GEF at the proximal and distal poles. An Rga1–GAP plug was added at the proximal pole, as described above. In this scenario, simulations rapidly developed a single peak of Cdc42 at the distal pole (figure 7a). Similar behaviour was observed for weak Rsr1–GEF (0.025% of Bem1–GEF: figure 7b). Polarization was more rapid than it was in simulations lacking the Rsr1–GEF (figure 6a), and the RSR1 diploid simulations never developed more than one polarity cluster. Thus, a little Rsr1-localized GEF is sufficient to bias the location of polarization.
Figure 7.
Modelling polarity establishment in diploids. (a) An Rsr1–GEF patch was placed at either end of the cell, with an Rga1–GAP plug in the lower patch. Rsr1–GEF at 2.5% of Bem1–GEF. (b) Rsr1–GEF patches with an Rga1–GAP plug in the lower patch. Rsr1–GEF at only 0.025% of Bem1–GEF. (c) Rsr1–GEF patches with no Rga1–GAP plug. Rsr1–GEF at 2.5% of Bem1–GEF. (d) Rsr1–GEF patches with no Rga1–GAP plug. Rsr1–GEF at only 0.025% of Bem1–GEF. (e) Rsr1–GEF patches with no Rga1–GAP plug. Rsr1–GEF at 2.5% of Bem1–GEF but Rsr1–GEF was turned off at 120 s (indicated by star). (f) Uneven Rsr1–GEF patches (55 : 45 ratio), with no Rga1–GAP. Rsr1–GEF at 2.5% of Bem1–GEF. (g) Uneven Rsr1–GEF patches (55 : 45 ratio), with no Rga1–GAP. Rsr1–GEF at only 0.025% of Bem1–GEF.
We also simulated a diploid lacking the Rga1–GAP, and in this case, the simulations developed two peaks of Cdc42, one at each pole (figure 7c). Even with weak Rsr1–GEF (0.025% of Bem1–GEF: figure 7d), or transient Rsr1–GEF (figure 7e), we did not observe competition between the peaks.
The ineffective competition in these simulations could result from the development of comparably sized Cdc42 peaks. During competition, the relative advantage of the winning peak depends on the magnitude of the difference between the peaks [11]. Thus, close-to-equal peaks take a long time to develop asymmetry. Consistent with this hypothesis, simulations lacking the Rga1–GAP in which the Rsr1–GEF was uneven (so that more Cdc42 was recruited to one pole than the other) led to more effective competition between the two peaks (figure 7f,g). Competition was considerably slower with high Rsr1–GEF (2.5% of Bem–GEF: figure 7f) than with low Rsr1–GEF (0.025% of Bem–GEF: figure 7g), indicating that the continuing presence of the Rsr1–GEF antagonizes competition.
(d). Imaging Bem1-GFP in rga1Δ/rga1Δ diploid cells
To ask whether diploids lacking Rga1 would develop two polarity peaks at opposite poles, we imaged polarity establishment in rga1Δ/rga1Δ homozygous mutants. Most cells developed a single peak (figure 8a; electronic supplementary material, movie S2). Some cells (four of 21 diploids imaged) did develop two polarity clusters, but these intermediates were rapidly resolved to a single peak (figure 8b). Under our imaging conditions, RSR1/RSR1 diploids had a marked preference to polarize at the pole opposite the neck (62 of 65 cells). This preference was less strong in the rga1Δ/rga1Δ mutants (16 of 21 cells), suggesting that the Rga1 GAP accounts for part, but not all, of the bias.
Figure 8.
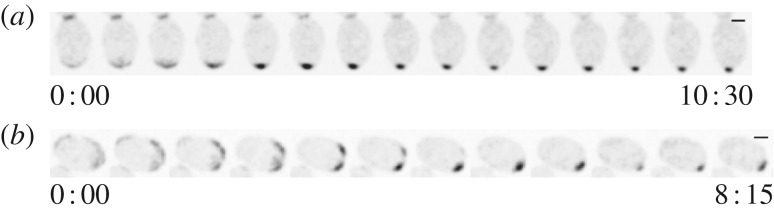
Polarization of rga1Δ/rga1Δ cells. Diploid rga1Δ/rga1Δ cells (DLY15125) polarizing with (a) single or (b) multiple clusters. Scale bar, 2 µm.
4. Discussion
(a). Wild-type cells exhibit competition between polarity clusters
The bud-site-selection system in yeast is often said to dictate the position at which the next bud will emerge. However, it has long been recognized that bud-site-selection landmarks actually specify a restricted subset of preferred positions, rather than dictating a single site [17]; the basis for selecting the final bud-site position (and the reason why there is only one) has been mysterious. One possibility is that Rsr1, which has the capacity to oligomerize [41], picks one among the permitted positions and then recruits Cdc42 and other polarity factors to the chosen site. In this view, the yeast cell's symmetry-breaking capability might be considered a backup or failsafe system, not normally called upon to participate in wild-type cells.
Arguing against the idea that Rsr1 picks a unique site, we found that RSR1/RSR1 cells often developed more than one cluster of polarity factors before evolving to a single polarity peak. This behaviour suggests that competition between the clusters is important in selecting the single winning polarity site in wild-type cells. It could be that Rsr1 simply biases the symmetry-breaking process to initiate polarity clusters at any of the permitted sites. In this view, polarity establishment occurs in much the same way with or without bud-site selection, and the uniqueness of the final polarity site is attained by competition (between distant clusters) and/or merging (of nearby clusters).
(b). Effect of the bud-site-selection system on the dynamics of polarization
The dynamics of polarity establishment, as revealed by imaging Bem1-GFP, displayed surprising differences between RSR1/RSR1 and rsr1Δ/rsr1Δ cells. As reported previously [13], in rsr1Δ/rsr1Δ cells Bem1 clusters formed rapidly, and then dispersed and reformed in an oscillatory manner. By comparison, in RSR1/RSR1 cells the Bem1 clusters grew more slowly, peaked at a lower intensity and then approached an intermediate intensity without marked oscillation (figure 4). These features may all be linked: given a system with both positive feedback and delayed negative feedback, the dynamics of the system will depend on the relative timeframe with which the feedback loops take effect. If positive feedback is fast, then the system will tend to polarize a lot of Bem1 before the negative feedback kicks in, after which the strongly activated negative feedback would disperse much of the polarized protein. But if the initial positive feedback is slow, then the negative feedback will ‘catch up’ as the system polarizes, reducing the peak polarity and dampening oscillations. Given these considerations, we interpret the observed differences in polarity dynamics to stem from a primary difference in the rate at which positive feedback builds the polarity cluster. The unexpected conclusion is that the presence of Rsr1 somehow slows the initial growth of the polarity cluster.
How might Rsr1 slow initial polarization? It seems possible that at early stages during polarity establishment, the Rsr1–GEF distributed over a relatively large area leads to prolonged competition between the allowed sites. This scenario might explain another behaviour we observed in a subset of both haploid RSR1 and diploid RSR1/RSR1 cells (figure 5): a faint and fluctuating Bem1-GFP signal was observed for several minutes in the areas expected to harbour active Rsr1 (the division site in haploids and the distal tip in diploids) prior to development of a single strong polarity site. Perhaps this reflects prolonged and ineffective competition, as seen in our simulations containing an Rsr1–GEF ring. However, our simulations of wild-type diploids did not capture this effect: with a circular patch (rather than a ring) of Rsr1–GEF, the simulations rapidly polarized towards the centre of the patch (figure 7a). Thus, it remains unclear why RSR1/RSR1 cells would polarize more slowly than mutants lacking Rsr1.
(c). Modelling bud-site selection
Previous studies showed that GTP-Rsr1 binds to Cdc24 and that this interaction is required for bud-site selection [28,29]. Thus, the simplest hypothesis to explain how Rsr1 biases polarization is that it recruits Cdc24 from the cytoplasm to all cortical sites containing landmark proteins. Using a computational model previously developed to simulate symmetry-breaking polarization, we explored whether such a localized Rsr1–GEF would suffice to yield the polarity protein behaviour observed in cells. With no Rsr1–GEF, this model initiates Cdc42 clusters owing to molecular noise, and as the clusters grow, they compete or merge with each other to yield a final strong polarity peak (figure 6a).
We added a localized Rsr1–GEF either as a ring, representing Axl2/Rsr1-mediated Cdc24 recruitment in haploids, or as two patches, representing Bud8/Rsr1- and Bud9/Rsr1-mediated Cdc24 recruitment in diploids. We also added a central ‘plug’ with higher GAP activity in the middle of the ring (or one of the patches) to simulate the reported exclusion zone enforced by the centrally located GAP, Rga1 [34]. Our simulations indicated that this system could yield a single polarity site at an appropriate location. However, to obtain that result it was necessary to make the Rsr1–GEF either very weak or transient.
(d). Localized Rsr1–GEF could interfere with competition
Models containing an Rsr1–GEF ring totalling 2.5% of the Bem1–GEF available for positive feedback recruited polarity factors to a stable ring, which did not resolve to a single peak of Cdc42 (figure 6b: this was not affected by the presence or absence of a GAP plug in the centre of the ring). Thus, the presence of even a relatively weak Rsr1–GEF was sufficient to suppress the competition process that yields a single peak. This effect could be overcome either by greatly reducing the amount of Rsr1–GEF or by making the Rsr1–GEF transient. The ability of Rsr1–GEF to interfere with competition may explain the unexpected finding that RSR1/RSR1 cells took longer than rsr1Δ/rsr1Δ cells to transition from a two-polarity-cluster intermediate to the final single-polarity-site state (figure 3c).
Models containing two patches of Rsr1–GEF but lacking a GAP plug (simulating rga1Δ/rga1Δ mutant diploids) developed two polarity peaks that did not resolve to a single peak in a relevant (approx. 10 min) timeframe, even with a very weak or transient Rsr1–GEF (figure 7). By contrast, rga1Δ/rga1Δ mutant cells displayed rapid competition and no defect in developing a single polarity site (figure 8). Why is competition so much more powerful in cells than it is in our model simulations? In the model, competition builds on initial differences between peaks that develop owing to molecular noise. In cells, it may be that noise stemming from vesicle traffic (not present in our model) is more powerful in generating these differences, promoting competition.
It is also possible that competition is enabled by some other factor that discourages polarization at the previous division site. Although haploid rga1Δ cells polarize preferentially at the division site [34], we found that diploid rga1Δ/rga1Δ mutants retained a bias to polarize away from the division site. A very recent study identified a new Cdc42 antagonist present at the division site that collaborates with Rga1 to prevent budding at that site [42]. Modelling a situation with two uneven patches, competition was more effective and our simulations did evolve towards a single polarity site. However, even in this case, a persistent Rsr1–GEF impaired competition (figure 7). Thus, as for the haploid ring simulations, effective polarization would occur only if the Rsr1-recruited GEF were very weak or transient.
(e). Does the Rsr1–GEF play additional roles beyond bud-site selection?
The idea that Rsr1 might interfere with effective polarization (as it does in the simplified model) is at odds with a growing body of evidence that Rsr1 actually promotes polarization, beyond just bud-site selection. In the filamentous fungi Ashbya gossypii and Candida albicans, Rsr1 is important for keeping the polarity site from wandering away from the hyphal tip [43,44]. Moreover, in Candida Bud5 is localized to hyphal tips [45]. Similarly, in S. cerevisiae, Rsr1 and Bud5 become concentrated at the polarity site during bud emergence [22,23,25]. In addition, RSR1 overexpression was found to partly suppress the temperature sensitivity of specific cdc42 alleles [46], suggesting that Rsr1 can assist in polarity establishment per se, in addition to influencing the position of the polarity site.
These studies are all consistent with the hypothesis that once established, a polarity site somehow recruits Bud5, which could then activate Rsr1 to recruit more Cdc24. This would reinforce polarity by providing a Cdc42–Bud5–Rsr1–Cdc24–Cdc42 positive feedback loop acting in parallel to the loop provided by Bem1. Such a parallel feedback loop might help to explain why bem1Δ mutants can polarize at all.
To reconcile our observations with the data discussed above, we suggest the following scenario. At early stages of polarity establishment, the presence of GTP-Rsr1 at multiple landmark-designated sites on the cortex biases the location of polarity cluster initiation but then slows the growth of the cluster and competition between clusters. At later stages, the landmark-localized pool is somehow shut off, and Bud5 is instead recruited to the chosen polarity site, reinforcing Cdc24 recruitment. The most parsimonious idea would be that the loss of Bud5/Rsr1 from landmark sites is simply a consequence of the recruitment of Bud5/Rsr1 to the polarity site. Alternatively, Bud5 might be inactivated at the previous landmark sites by some other mechanism. Thus, we speculate that Rsr1 plays two separate roles: in early G1 it helps to mark the sites that initiate polarity establishment, and in late G1 it participates in a positive feedback that reinforces polarization. The basis for the switch between these roles remains to be determined.
(f). The role of actin
We found that in diploids treated with latrunculin to depolymerize F-actin, the winning polarity site sometimes relocated from one landmark-designated position to another. This observation suggests that F-actin may be required to stop the ‘losing’ landmarks from continuing to attract polarity factors once a large polarity cluster is formed. In light of the hypothesized Cdc42–Bud5–Rsr1–Cdc24–Cdc42 positive feedback loop discussed above, one possibility would be that Cdc42-oriented F-actin mediates delivery of a polarity-site ‘landmark’ that then draws Bud5 to the new polarity site, away from the immobile bud-site-selection landmarks.
A role for actin-mediated vesicle traffic in providing positive feedback for the polarity site has been vigorously advocated by previous studies [32,33,35]. In those studies, it was assumed that actin reinforced polarization by delivering Cdc42 itself to the polarity site, whereas here we suggest that it may deliver a landmark that serves to activate Rsr1 at that site. Consistent with a role for actin and Rsr1 in the same pathway, both deletion of Rsr1 [8] and depolymerization of F-actin [33] can block polarization when cells lack Bem1. Testing the hypothesis will require identification of the proposed landmark(s).
In a few of the latrunculin-treated cells, the polarity site repeatedly switched from one end of the cell to the other in a ‘ping-pong’ manner. This behaviour is strikingly reminiscent of the oscillatory relocation of Cdc42 between the two cell ends that was recently reported to occur during bipolar growth of the cylindrical fission yeast, Schizosaccharomyces pombe [47]. However, whereas in S. cerevisiae the behaviour is only seen in cells with depolymerized actin, in S. pombe it requires F-actin, so the role of actin must be different in the two systems. Three factors are likely to underlie oscillatory relocation in both systems: (i) the two cell ends contain tip proteins that can attract polarity factors to those sites; (ii) competition for limiting factors may explain why only one site at a time can accumulate large amounts of Cdc42; and (iii) negative feedback loops may explain why the winning site then disperses Cdc42, allowing the other tip to gain the upper hand [13,47,48].
5. Conclusions
We suggest that by providing a spatially restricted Cdc24 GEF activity at landmark-designated sites, the bud-site-selection system biases where the symmetry-breaking positive feedback will begin to concentrate polarity factors. Once polarity sites develop, they compete or merge until there is a single winner. Modelling predicts that the continued action of landmark-localized GEF would interfere with competition and merging, and we suggest that polarity factors recruit Rsr1 and its regulators away from the landmarks in order to reduce such interference. Rsr1 may then provide a parallel positive feedback pathway to assist polarization. A surprising observation, which remains to be explained, is that the presence of Rsr1 slows the initial clustering of Bem1.
Acknowledgement
We thank Jayme Dyer, Allie McClure, Ben Woods and Reichen Kuo for comments on the manuscript.
Funding statement
This work was supported by NIH/NIGMS grant no. GM62300 to D.J.L.
References
- 1.Etienne-Manneville S. 2004. Cdc42: the centre of polarity. J. Cell Sci. 117, 1291–1300 (doi:10.1242/jcs.01115) [DOI] [PubMed] [Google Scholar]
- 2.Yang Z, Lavagi I. 2012. Spatial control of plasma membrane domains: ROP GTPase-based symmetry breaking. Curr. Opin. Plant Biol. 15, 601–607 (doi:10.1016/j.pbi.2012.10.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi E, Park HO. 2012. Cell polarization and cytokinesis in budding yeast. Genetics 191, 347–387 (doi:10.1534/genetics.111.132886) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turing A. 1952. The chemical basis of morphogenesis. Phil. Trans. R. Soc. Lond. B 237, 37–72 (doi:10.1098/rstb.1952.0012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams AEM, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. 1990. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111, 131–142 (doi:10.1083/jcb.111.1.131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender A, Pringle JR. 1989. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the Ras-related gene RSR1. Proc. Natl Acad. Sci. USA 86, 9976–9980 (doi:10.1073/pnas.86.24.9976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chant J, Herskowitz I. 1991. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65, 1203–1212 (doi:10.1016/0092-8674(91)90015-Q) [DOI] [PubMed] [Google Scholar]
- 8.Irazoqui JE, Gladfelter AS, Lew DJ. 2003. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 5, 1062–1070 (doi:10.1038/ncb1068) [DOI] [PubMed] [Google Scholar]
- 9.Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ. 2008. Symmetry-breaking polarization driven by a Cdc42p GEF–PAK complex. Curr. Biol. 18, 1719–1726 (doi:10.1016/j.cub.2008.09.060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goryachev AB, Pokhilko AV. 2008. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 582, 1437–1443 (doi:10.1016/j.febslet.2008.03.029) [DOI] [PubMed] [Google Scholar]
- 11.Howell AS, et al. 2009. Singularity in polarization: rewiring yeast cells to make two buds. Cell 139, 731–743 (doi:10.1016/j.cell.2009.10.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JM, Jin M, Lew DJ. 2011. Symmetry breaking and the establishment of cell polarity in budding yeast. Curr. Opin. Genet. Dev. 21, 740–746 (doi:10.1016/j.gde.2011.09.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howell AS, Jin M, Wu CF, Zyla TR, Elston TC, Lew DJ. 2012. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell 149, 322–333 (doi:10.1016/j.cell.2012.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chant J. 1999. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol 15, 365–391 (doi:10.1146/annurev.cellbio.15.1.365) [DOI] [PubMed] [Google Scholar]
- 15.Harkins HA, Page N, Schenkman LR, De Virgilio C, Shaw S, Bussey H, Pringle JR. 2001. Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast. Mol. Biol. Cell 12, 2497–2518 (doi:10.1091/mbc.12.8.2497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao XD, Sperber LM, Kane SA, Tong Z, Tong AH, Boone C, Bi E. 2007. Sequential and distinct roles of the cadherin domain-containing protein Axl2p in cell polarization in yeast cell cycle. Mol. Biol. Cell 18, 2542–2560 (doi:10.1091/mbc.E06-09-0822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chant J, Pringle JR. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129, 751–765 (doi:10.1083/jcb.129.3.751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen T, Hiroko T, Chaudhuri A, Inose F, Lord M, Tanaka S, Chant J, Fujita A. 2000. Multigenerational cortical inheritance of the Rax2 protein in orienting polarity and division in yeast. Science 290, 1975–1978 (doi:10.1126/science.290.5498.1975) [DOI] [PubMed] [Google Scholar]
- 19.Fujita A, Lord M, Hiroko T, Hiroko F, Chen T, Oka C, Misumi Y, Chant J. 2004. Rax1, a protein required for the establishment of the bipolar budding pattern in yeast. Gene 327, 161–169 (doi:10.1016/j.gene.2003.11.021) [DOI] [PubMed] [Google Scholar]
- 20.Kang PJ, Angerman E, Nakashima K, Pringle JR, Park HO. 2004. Interactions among Rax1p, Rax2p, Bud8p, and Bud9p in marking cortical sites for bipolar bud-site selection in yeast. Mol. Biol. Cell 15, 5145–5157 (doi:10.1091/mbc.E04-07-0600) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bender A. 1993. Genetic evidence for the roles of the bud-site-selection genes BUD5 and BUD2 in control of the Rsr1p (Bud1p) GTPase in yeast. Proc. Natl Acad. Sci. USA 90, 9926–9929 (doi:10.1073/pnas.90.21.9926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park HO, Sanson A, Herskowitz I. 1999. Localization of Bud2p, a GTPase-activating protein necessary for programming cell polarity in yeast to the presumptive bud site. Genes Dev. 13, 1912–1917 (doi:10.1101/gad.13.15.1912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang PJ, Sanson A, Lee B, Park HO. 2001. A GDP/GTP exchange factor involved in linking a spatial landmark to cell polarity. Science 292, 1376–1378 (doi:10.1126/science.1060360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marston AL, Chen T, Yang MC, Belhumeur P, Chant J. 2001. A localized GTPase exchange factor, Bud5, determines the orientation of division axes in yeast. Curr. Biol. 11, 803–807 (doi:10.1016/S0960-9822(01)00230-5) [DOI] [PubMed] [Google Scholar]
- 25.Park HO, Kang PJ, Rachfal AW. 2002. Localization of the Rsr1/Bud1 GTPase involved in selection of a proper growth site in yeast. J. Biol. Chem. 277, 26 721–26 724 (doi:10.1074/jbc.C200245200) [DOI] [PubMed] [Google Scholar]
- 26.Zahner JE, Harkins HA, Pringle JR. 1996. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 16, 1857–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park HO, Chant J, Herskowitz I. 1993. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature 365, 269–274 (doi:10.1038/365269a0) [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y, Bender A, Cerione RA. 1995. Interactions among proteins involved in bud-site selection and bud-site assembly in Saccharomyces cerevisiae. J. Biol. Chem. 270, 626–630 (doi:10.1074/jbc.270.2.626) [DOI] [PubMed] [Google Scholar]
- 29.Shimada Y, Wiget P, Gulli MP, Bi E, Peter M. 2004. The nucleotide exchange factor Cdc24p may be regulated by auto-inhibition. EMBO J. 23, 1051–1062 (doi:10.1038/sj.emboj.7600124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das A, Slaughter BD, Unruh JR, Bradford WD, Alexander R, Rubinstein B, Li R. 2012. Flippase-mediated phospholipid asymmetry promotes fast Cdc42 recycling in dynamic maintenance of cell polarity. Nat. Cell Biol. 14, 304–310 (doi:10.1038/ncb2444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Layton AT, Savage NS, Howell AS, Carroll SY, Drubin DG, Lew DJ. 2011. Modeling vesicle traffic reveals unexpected consequences for Cdc42p-mediated polarity establishment. Curr. Biol. 21, 184–194 (doi:10.1016/j.cub.2011.01.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slaughter BD, Das A, Schwartz JW, Rubinstein B, Li R. 2009. Dual modes of Cdc42 recycling fine-tune polarized morphogenesis. Dev. Cell 17, 823–835 (doi:10.1016/j.devcel.2009.10.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wedlich-Soldner R, Wai SC, Schmidt T, Li R. 2004. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J. Cell Biol. 166, 889–900 (doi:10.1083/jcb.200405061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tong Z, Gao XD, Howell AS, Bose I, Lew DJ, Bi E. 2007. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J. Cell Biol. 179, 1375–1384 (doi:10.1083/jcb.200705160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wedlich-Soldner R, Altschuler S, Wu L, Li R. 2003. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299, 1231–1235 (doi:10.1126/science.1080944) [DOI] [PubMed] [Google Scholar]
- 36.Orlando K, Sun X, Zhang J, Lu T, Yokomizo L, Wang P, Guo W. 2011. Exo-endocytic trafficking and the septin-based diffusion barrier are required for the maintenance of Cdc42p polarization during budding yeast asymmetric growth. Mol. Biol. Cell 22, 624–633 (doi:10.1091/mbc.E10-06-0484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozbudak EM, Becskei A, van Oudenaarden A. 2005. A system of counteracting feedback loops regulates Cdc42p activity during spontaneous cell polarization. Dev. Cell 9, 565–571 (doi:10.1016/j.devcel.2005.08.014) [DOI] [PubMed] [Google Scholar]
- 38.Savage NS, Layton AT, Lew DJ. 2012. Mechanistic mathematical model of polarity in yeast. Mol. Biol. Cell 23, 1998–2013 (doi:10.1091/mbc.E11-10-0837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dyer JM, Savage NS, Jin M, Zyla TR, Elston TC, Lew DJ. 2013. Tracking shallow chemical gradients by actin-driven wandering of the polarization site. Curr. Biol. 23, 32–41 (doi:10.1016/j.cub.2012.11.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caviston JP, Longtine M, Pringle JR, Bi E. 2003. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14, 4051–4066 (doi:10.1091/mbc.E03-04-0247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang PJ, Beven L, Hariharan S, Park HO. 2010. The Rsr1/Bud1 GTPase interacts with itself and the Cdc42 GTPase during bud-site selection and polarity establishment in budding yeast. Mol. Biol. Cell 21, 3007–3016 (doi:10.1091/mbc.E10-03-0232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meitinger F, Richter H, Heisel S, Hub B, Seufert W, Pereira G. 2013. A safeguard mechanism regulates rho GTPases to coordinate cytokinesis with the establishment of cell polarity. PLoS Biol. 11, e1001495 (doi:10.1371/journal.pbio.1001495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauer Y, Knechtle P, Wendland J, Helfer H, Philippsen P. 2004. A Ras-like GTPase is involved in hyphal growth guidance in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 15, 4622–4632 (doi:10.1091/mbc.E04-02-0104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hausauer DL, Gerami-Nejad M, Kistler-Anderson C, Gale CA. 2005. Hyphal guidance and invasive growth in Candida albicans require the Ras-like GTPase Rsr1p and its GTPase-activating protein Bud2p. Eukaryot. Cell 4, 1273–1286 (doi:10.1128/EC.4.7.1273-1286.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pulver R, Heisel T, Gonia S, Robins R, Norton J, Haynes P, Gale CA. 2013. Rsr1 focuses Cdc42 activity at hyphal tips and promotes maintenance of hyphal development in Candida albicans. Eukaryot. Cell 12, 482–495 (doi:10.1128/EC.00294-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozminski KG, Beven L, Angerman E, Tong AH, Boone C, Park HO. 2003. Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Mol. Biol. Cell 14, 4958–4970 (doi:10.1091/mbc.E03-06-0426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das M, Drake T, Wiley DJ, Buchwald P, Vavylonis D, Verde F. 2012. Oscillatory dynamics of Cdc42 GTPase in the control of polarized growth. Science 337, 239–243 (doi:10.1126/science.1218377) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu CF, Lew DJ. In press Beyond symmetry-breaking: competition and negative feedback in GTPase regulation. Trends Cell Biol. (doi:10.1016/j.tcb.2013.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]