Abstract
Objective
For genome-wide association studies (GWAS) in case-control data with stratification, a commonly used association test is the generalized Armitage (GA) trend test implemented in the software EIGENSTRAT. The GA trend test uses principal component analysis to correct for population stratification. It usually assumes an additive disease model and can have high power when the underlying disease model is additive or multiplicative, but may have relatively low power when the underlying disease model is recessive or dominant. The purpose of this paper is to provide a test procedure for GWAS with increased power over the GA trend test under the recessive and dominant models while maintaining the power of the GA trend test under the additive and multiplicative models.
Methods
We extend a Hardy-Weinberg disequilibrium (HWD) trend test for a homogeneous population to account for population stratification, and then propose a robust association test procedure for GWAS that incorporates information from the extended HWD trend test into the GA trend test.
Results and Conclusions
Our simulation studies and application of our method to a GWAS data set indicate that our proposed method can achieve the purpose described above.
Keywords: generalized sequential Bonferroni procedure, genome-wide association studies, Hardy-Weinberg trend test, robust test, recessive model
1. Introduction
In genome-wide association studies with case-control designs, population stratification can happen when the participants are sampled from different subpopulations with significant allele frequency differences [1]. Population stratification can be a confounding factor in the association test and may produce spurious associations if not properly corrected [2, 3, 4]. Many methods have been developed to correct for stratification in association studies [3, 5, 6]. One of the most widely used methods is a generalized Armitage (GA) trend test implemented in the software EIGENSTRAT that uses principal component analysis (PCA) to correct for population stratification [3]. The GA trend test usually assumes that the underlying disease model is additive [3]. In this paper, we use “GA trend test” to denote the GA trend test assuming the additive model. One of the advantages of the GA trend test is that when the underlying disease model is additive or multiplicative, it can have high power in detecting causal variants. However, when the underlying disease model is dominant or recessive, it can have low power.
Some association tests that are robust to different genetic models have been developed for homogenous populations [7, 8, 9, 10]. For example, the MAX3 test statistic [7, 8] uses the maximum of the three Armitage trend test statistics [11] that assume the additive, dominant and recessive model, respectively. MAX3 has been extended to use top principal components (PCs) of genome-wide genotypes as covariates to correct for stratification [12]. This extended MAX3 method is referred to as MAX3-PC in this paper. However, MAX3-PC has two limitations: 1) compared to the GA trend test, when the underlying disease model is additive or multiplicative, MAX3-PC can have relatively lower power; 2) it can be computationally intensive for GWAS, especially for data with more than one million SNPs. Since the additive/multiplicative genetic model is often assumed and fits well for most identified genetic variants [13], in this article, we intend to propose a computationally efficient method for GWAS that can maintain the power of the GA trend test (EIGENSTRAT) under the additive and multiplicative models while it has improved power over the GA trend test under the recessive and dominant models.
To achieve this goal, first, we extend the Hardy-Weinberg disequilibrium (HWD) trend test of Song & Elston [14] for homogenous populations to account for population stratification. The extended HWD trend test can provide useful information under the dominant or recessive models which is different from that provided by the GA trend test. Second, for GWAS we apply the generalized sequential Bonferroni (GSB) procedure to incorporate information from the extended HWD trend test into the GA trend test. Simulation studies indicate that the proposed GSB procedure for GWAS can control the familywise error rate when stratification exists; it has comparable power to that of the GA trend test, when the underlying model is additive or multiplicative, and has higher power than the GA trend test when the underlying model is dominant or recessive. Simulation studies also indicate that MAX3-PC of So and Sham (2011) has relatively lower power than the GA trend test and the proposed GSB procedure under the additive and multiplicative models. Finally we applied our proposed method to four datasets provided by the Wellcome Trust Case-Control Consortium (WTCCC) [13].
2. Methods
In this section, we first extend the HWD trend test of Song & Elston [14] to account for population stratification, and then propose a GSB procedure for GWAS that incorporates information from the extended HWD trend test into the GA trend test. In this study, we assume independence among markers and ignore the LD among markers in the GWAS data.
2.1 Extended HWD trend test
By assuming that cases and controls are sampled from the same general homogenous population, Song & Elston [14] proposed a HWD trend test by comparing the HWD coefficient in cases Dcase with the HWD coefficient in controls Dcontrol. For a SNP with allele A and B, denote the genotype frequencies of AA, AB, BB in cases by pAA, pAB, pBB and in controls by qAA, qAB, qBB. Then we have Dcase = pBB − {pAB/2 + pBB}2 and Dcontrol = qBB − {qAB/2 + qBB}2. Let Δ = Dcase − Dcontrol. The null hypothesis is H0: Δ = 0, which corresponds to testing whether the HWD coefficient for the test marker in the controls is equivalent to that in cases; if not, the test marker may be associated with the disease. The HWD trend test statistic is given by: where Δ̂ = D̂case − D̂control , and D̂case and D̂control are the maximum likelihood estimates of Dcase and Dcontrol respectively. is the estimate of the variance of Δ̂ under H0 [for details, see 14]. THWDTT asymptotically follows a standard normal distribution and the square of the test follows the chi-square distribution with 1 degree of freedom.
The HWD trend test statistic THWDTT can be used to test association between a marker and a disease under the assumption that Hardy-Weinberg equilibrium holds in the general population from which the cases and controls are sampled. Song & Elston [14] stated that the HWD trend test can automatically control for the genotyping errors, if the cases and controls are genotyped by the same methods at the same time, with the cases and controls randomized. However, a weakness of the HWD trend test is that it can have an inflated type I error rate when population stratification exists (see our simulation results below). Below we extend the HWD trend test to account for population stratification.
First, we assign cases and controls in the data into K groups (or clusters) that correspond to K putative subpopulations. Specifically, we calculate the top principal components (PCs), e.g., the top 10 PCs, of genome-wide genotypes and then apply a clustering algorithm, e.g., the k-means clustering algorithm [15], to the top PCs to partition the individuals into K groups.
In order to determine the number of subpopulations (K), we use the fpc package (http://cran.r-project.org/web/packages/fpc/index.html) that implements the k-means algorithm with options to determine the optimal number (K) of clusters [16]. More specifically, the fpc package uses the Duda-Hart test [17] to test if all individuals in the data are from one cluster. If they are not from one cluster, then the k-means cluster algorithm is executed with the cluster numbers k = 2, 3, …, Kmax (we set Kmax = 4 in our data analysis). The Calinski-Harabasz criterion is used to determine the optimal number of clusters from 2 to Kmax [18]. The Calinski-Harabasz criterion can be calculated as , where SSW is the sum of variances within the clusters, SSB is the sum of variances among the clusters, and N is the number of individuals in the case-control data. The number of subpopulations is selected as .
For the cases and controls assigned to the i-th group where i = 1, 2, …, K, we define an effect size to be the difference (Δi) of HWD coefficients between cases and controls, and denote its maximum likelihood estimation by βi = Δ̂i. Under the null hypothesis H0: Hardy-Weinberg equilibrium holds in the i-th subpopulation (corresponding to the i-th group) and the test marker is not associated with the disease, the variance of the estimated effect VarH0 (Δ̂i) can be estimated (see Song and Elston [14]). If any assigned group only contains cases, then we set the HWD coefficient for the controls to and use the variance , as given in Song and Elston [14]. If any assigned groups only contain controls, this group is removed from the analysis because subpopulations consisting of controls only do not provide information for the association study.
Following the method in meta-analysis using a fixed effect model [19], the combined effect over the K groups (or subpopulations) is given by
where is the inverse of the estimated variance of the estimated effect for the i-th subpopulation. The variance of the combined effect is given by . Under the null hypothesis that Hardy-Weinberg equilibrium holds in each of the subpopulation and the test marker is not associated with a disease, the extended HWD trend test statistic is then defined as E2/V, which has an approximate chi-square distribution with one degree of freedom.
2.2 Generalized Armitage (GA) trend test in EIGENSTRAT
To effectively control false positive rates caused by population stratification, Price et al. [3] proposed the GA trend test, which is implemented in the software EIGENSTRAT. The GA trend test calculates the top principal components (PCs) from genome-wide genotype values for each individual. To test the association between a marker and the disease status, the GA trend test adjusts the individual’s genotype value (assuming an additive model) at the marker by regressing out the top L PCs (e.g., L = 10) in a linear model (see Price et al. 2006) and then adjusts the phenotypic value (0 for a control and 1 for a case) by the same L PCs in the similar way. The GA trend test statistic is defined as (N-L-1)R2, where R2 is the correlation between the ancestry-adjusted genotype and ancestry-adjusted phenotype, and N is the number of individuals in the case-control study. Under the null hypothesis that the test marker is not associated with the disease, the GA trend test statistic asymptotically follows the chi-square distribution with 1 degree of freedom.
2.3 Generalized sequential Bonferroni procedure for multiple testing
For testing multiple hypotheses H1, H2, …, Hm, Holm [20] proposed a generalized sequential Bonferroni (GSB) procedure by assigning different weights to hypotheses of different importance. Let α denote the nominal familywise error rate (FWER), which is defined as the probability of rejecting at least one true null hypothesis. We describe the GSB procedure below.
Let p1, p2, ⋯, pm denote the p-values from m tests corresponding to hypotheses H1, H2, …, Hm. Given weights w1, w2, …, wm for these tests, define B-values as Bi = pi / wi. First we order B-values as B(1) ≤ B(2) ≤… ≤ B(m), so that w(1), w(2), …, w(m) and H(1), H(2), …, H(m) are the corresponding weights and hypotheses of the ordered B-values. Starting from i = 1, given H(1), H(2), …, H(i−1) have been tested and rejected, if , reject H(i); otherwise, accept H(i), H(i+1), …, H(m), and stop the GSB procedure. Holm [20] proved that the GSB procedure controls the FWER at the nominal significance level of α if the condition that the weights are independent of the m tests is satisfied. From the proof of Holm, we can see that the GSB is relatively conservative and that the condition that the weights are independent of the m tests is a sufficient condition but not a necessary condition. We will show by simulation studies (see Appendix) that under some situations, even though the weights are weakly correlated with the m tests, the GSB procedure can still control FWER well. In addition, from the rejection criteria above, in order to reject the hypothesis H(i), the corresponding p-value p(i) must be less than or equal to α, otherwise the GSB procedure will not reject the hypothesis H(i).
We note that when the GSB procedure of Holm is applied to testing association at genome-wide markers, it cannot provide a p-value for each marker, which may be needed when comparing the GSB procedure with other methods in real data analysis. Therefore, we define the adjusted p-value for each test in the GSB procedure following the idea of Westfall and Young [21, p64]: Let p̃(i) be the adjusted p-value corresponding to B(i) and H(i), i = 1, 2, …, m. Then we define . Any adjusted p-value greater than 1.0 should be set to 1.0. The adjusted p-values should be compared to the nominal level α for the entire test procedure: if an adjusted p-value is less than or equal to α, then reject the corresponding hypothesis.
2.4 GSB procedure for GWAS in case-control data with population stratification
Here we describe how to apply the GSB procedure for multiple testing to GWAS data with population stratification. In the original GSB procedure, Holm [20] did not describe how to estimate the weights, although the GSB procedure has the potential to improve the power of multiple hypotheses testing when prior information is available to estimate the weights. Since the extended HWD trend test may provide different information from that provided by the GA trend test, we propose to use the extended HWD trend test to calculate a weight for each marker, and use the GA trend test to calculate a p-value for the marker. Then we use the weight to adjust the GA trend test p-value in the GSB procedure. Specifically, let qi denote the p-value for the i-th marker calculated by the extended HWD trend test. We let the weight wi for the i-th marker be the inverse of the extended HWD trend test p-value qi, i.e., wi = 1 / qi. We then apply the GSB procedure to incorporate the weight into the corresponding GA trend test in GWAS.
As described in the previous section, to reject the null hypothesis that a marker is not associated with the disease, the p-value from the GA trend test at that marker must be less than or equal to the nominal FWER α. This property can prevent that a null marker, which usually has a p-value from the GA trend test greater than α (such as 0.05), to be called significant because of the extremely high weight that is calculated from the extended HWD trend test due to genotyping errors.
The extended HWD trend test can have high power under both the recessive and dominant models, but it has no (or almost no) power under the multiplicative (or additive) model. Therefore the weights calculated from the extended HWD trend test only generate noise when the underlying disease model is multiplicative or additive. This results in that under these two models, the GSB procedure (using p-values from the GA trend test) has relatively lower power than the GA trend test (with Bonferroni correction) in GWAS. On the other hand, when the underlying disease models are recessive or dominant, weights calculated from the extended HWD trend test can provide useful information, and the GSB procedure can have much higher power than the GA trend test (see our simulation results below).
Since the additive model is often assumed in association studies in the literature when the true underlying genetic models are unknown, we hope to modify the GSB procedure for GWAS so that it can maintain the power of the GA trend test under the additive and multiplicative models, but still have increased power under the dominant and recessive models. To achieve this goal, we employ a smoothing method of Roeder [22] to calculate smoothed weights. Suppose wi is the original weight for the i-th marker, w̄ is the average value of the weights for all markers. We calculate a smoothed weight for the i-th marker as , where λ is a parameter and 0 ≤ λ ≤ 1. We call the GSB procedure using the smoothed weights smooth-GSB procedure. When λ is close to 1, then the smoothed weight is close to the constant w̄, and the GSB procedure has similar results as the GA trend test. On the other hand, when λ is close to 0, the smoothed weight is close to the original weight wi, and the smooth-GSB procedure has similar results as the GSB procedure using the original weight wi. Since we hope that the smooth-GSB procedure has power close to that of the GA trend test in GWAS under the additive and multiplicative models, the value of λ should be close to 1. On the other hand, if λ is very close to 1, only a little information from the original weight (i.e. from the extended HWD trend test) can be used, this will result in that the smooth-GSB procedure does not have much increased power compared to the GA trend test under the recessive and dominant models. As a tradeoff, we would suggest the value of λ should be greater than 0.5 but less than 0.9.
In the GSB procedure for GWAS, if the weight calculated from the extended HWD trend test is (asymptotically) independent of the GA trend test at each marker, then the GSB procedure for GWAS can control FWER. Joo et al. [23] proved that the HWD trend test statistic and the Cochran Armitage trend test statistic assuming the additive model are asymptotically independent in homogeneous populations. We expect that, as the extension of these two test statistics to correct for stratification, the extended HWD trend test statistic and the GA trend test statistic are approximately independent or at most are very weakly correlated. However, it is difficult to prove the independence theoretically. Below we will show by simulation studies that at a marker, the correlation between the extended HWD trend test statistic and the GA trend test statistic is negligible. We also show that in simulated case-control GWAS data, the GSB and smooth-GSB procedures can control FWER.
3. Simulation studies
3.1 Simulating case-control data with stratification
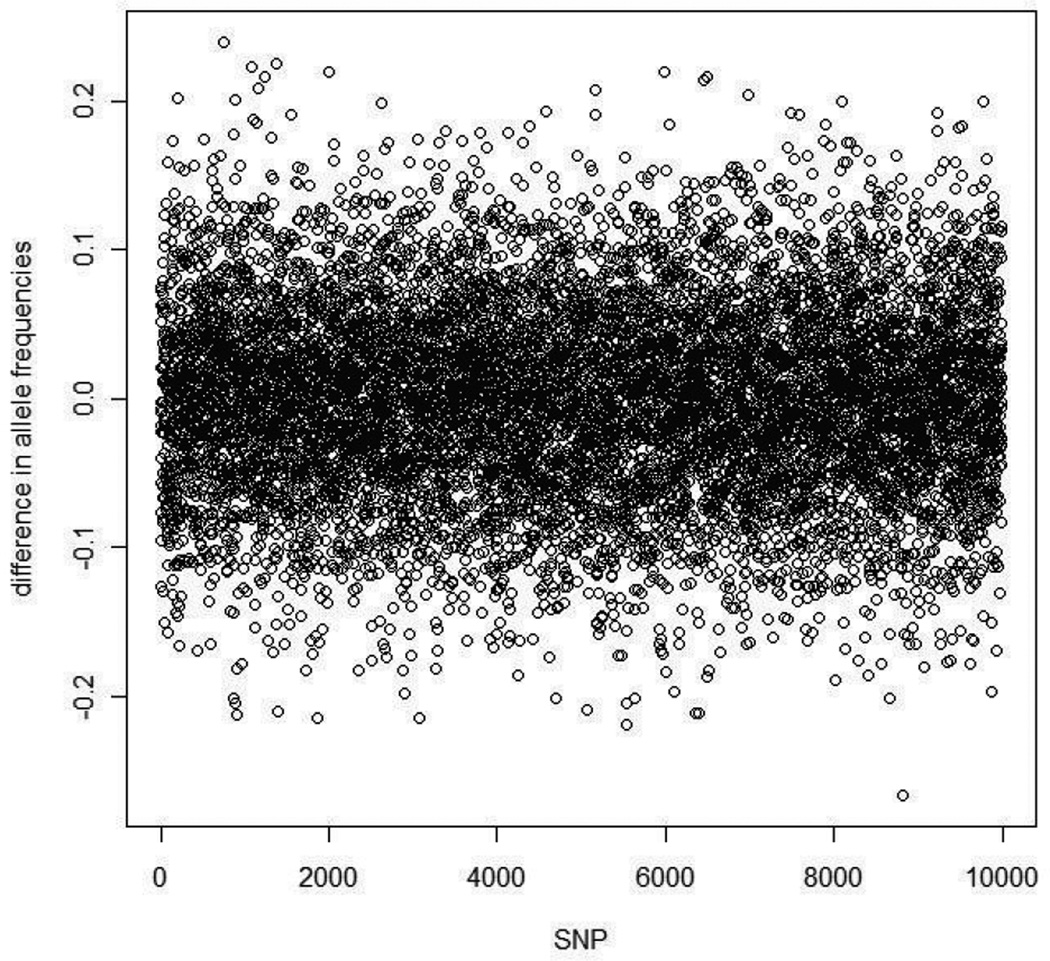
To evaluate the false positive rates and power of the extended HWD trend test and the proposed smooth-GSB procedure in case-control data with population stratification, we simulated data sets in a similar way to that in [3]. Each data set was simulated to include 2000 cases and 2000 controls with 10,000 random single nucleotide polymorphisms (SNPs). To simulate stratification, each individual was sampled from one of two populations. In cases, 40% were sampled from population 1 and 60% were sampled from population 2. In controls, 20% were sampled from population 1 and 80% were sampled from population 2. Allele frequencies for population 1 and population 2 at each of the 10,000 random SNPs were generated using the Balding-Nichols model with FST = 0.01. Specifically, at each random SNP, the allele frequencies for populations 1 and 2 were each drawn from a beta distribution with parameters p(1 − FST)/FST and (1 − p)(1 − FST)/FST, where the ancestral population allele frequency p was sampled from the uniform distribution in the interval [0.1, 0.9]. Usually the allele frequency differences between populations 1 and 2 are less than 0.1 [3]. In addition to the 10,000 random SNPs, we simulated 10 SNPs with differential frequencies in populations 1 and 2. For each of these 10 SNPs, the allele frequency difference between populations 1 and 2 was greater than 0.1, to approximate the effect of selection. In our simulation, we set the allele frequency to 0.2 in population 1 and 0.8 in population 2 as a demonstration of strong selection. As an example, Figure 1 shows the allele frequency differences at the 10,000 random SNPs between population 1 and population 2 in one replicate of our simulated data set. We can see that most of the allele frequency differences between the two populations are less than 0.1.
Figure 1.
The allele frequency differences between the two populations in one simulated data set.
3.2 Type I error evaluation of the original HWD trend test and the extended HWD trend test
To compare the type I errors of the original HWD trend test of song and Elston [14] and the extended HWD trend test, we chose a null SNP, i.e., non-disease-causing SNP, from the simulated 10,000+10 SNPs and evaluated the type I errors on this SNP. The null SNP was chosen in two ways: 1) from the 10,000 random SNPs and 2) from the 10 SNPs with differential frequencies, where we set the allele frequency as 0.2 in subpopulation 1 and 0.8 in subpopulation 2. We simulated 10,000 replicated data sets to estimate the type I error and the results are showed in Table I. We can see that with the random null SNP, both the original HWD trend test and the extended HWD trend test could control the type I error well. However, at the SNP with highly differentiated allele frequencies in the two subpopulations, the original HWD trend test had obviously inflated type I error and the extended HWD trend test could still control the type I error.
Table I.
Empirical type I error of the original HWD trend test (HWDTT) and the extended HWDTT (E-HWDTT) based on 10,000 replicates with significance level of 0.05.
The allele frequency differences of the candidate null SNP between the two populations were randomly simulated based on the Fst = 0.01
The allele frequency differences of the candidate null SNP between the two populations were set to 0.2 and 0.8 respectively.
3.3 Evaluation of correlation between the GA trend test and the extended HWD trend test
We estimated the correlation coefficients between the GA trend test and the extended HWD trend test under the null hypothesis that no markers are associated with the disease and that Hardy-Weinberg equilibrium holds in each subpopulation. Under this null hypothesis we generated 10,000 replicates of the case-control data with 10,000+10 SNPs (see section 3.1). For each SNP, we calculated the GA trend test statistic and the extended HWD trend test statistic. The sample Spearman’s rank correlation coefficient between the GA trend test and the extended HWD trend test was calculated for each SNP based on the 10,000 replicates. The mean and variance of the absolute values of the Spearman’s rank correlation coefficients for the 10,000+10 markers were 0.00799 and 0.0060, respectively. These values indicate that the correlation between the two statistics is negligible.
3.4 FWER evaluation
To evaluate the FWERs of our proposed GSB procedures, we generated 100,000 replicates of the case-control data (with 10,000+10 SNPs) under the null hypothesis that no SNPs were associated with the disease (see the section on simulating case-control data above). The FWER was estimated as the proportion of replicates with at least one false discovery. We also estimated the FWERs of the extended HWD trend test and the GA trend test. For these individual SNP based tests, the significance level for individual SNPs was set using Bonferroni correction, i.e., the nominal FWER divided by the number of SNPs. For the smooth-GSB procedure, different values from 0.55 to 0.9 for λ were evaluated. The estimated FWERs were reported in Table II. We can see that all methods controlled the FWER well. We did not estimate the FWER of the MAX3-PC because of the high computational cost. The evaluation of the type I error of MAX3-PC can be found in So and Sham’s paper [12].
Table II.
Empirical FWER based on 100,000 replicates with nominal FWER = 0.01, 0.05 and 0.1*.
| FWER | E- HWDTTa |
GA trendb |
GSBc | S-GSBd |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| λ=.55 | λ=.6 | λ=.65 | λ=.7 | λ=.75 | λ=.8 | λ=.85 | λ=.9 | ||||
| .01 | .0088 | .0095 | .0092 | .0093 | .0095 | .0094 | .0094 | .0093 | .0093 | .0094 | .0095 |
| .05 | .0417 | .0478 | .0487 | .0478 | .0477 | .0476 | .0475 | .0473 | .0473 | .0473 | .0475 |
| .10 | .0827 | .0927 | .0944 | .0942 | .0942 | .0939 | .0937 | .0937 | .0933 | .0931 | .0927 |
For the individual SNP based tests (E-HWDTT and the GA trend test), the significance level for individual SNPs was set using Bonferroni correction, i.e., the nominal FWER divided by the number of SNPs
E-HWDTT: the extended HWD trend test
GA trend = the GA trend test
GSB = generalized sequential Bonferroni procedure
S-GSB: the smooth-GSB procedure with different λ from 0.55 to 0.9.
3.5 Power evaluation
To evaluate the power of the proposed methods, we simulated a causal SNP besides the 10,000+10 non-causal SNPs as follows. At the causal SNP with allele A and B, let B denote the risk allele. We assume Hardy-Weinberg equilibrium in the subpopulations 1 and 2. Suppose pk (k = 1, 2) is the frequency of allele B in population k. The genotype frequencies of (AA, AB, BB) for controls (population control) of population k are set to {(1 − pk)2, 2pk(1−pk), pk2}. Let the genetic relative risks be R1 = Pr (affected | AB) / Pr (affected | AA), R2 = Pr (affected | BB) / Pr (affected | AA). When the genetic model is additive, multiplicative, dominant or recessive, we have R2 = 2 R1 − 1, R2 = R12, R1 = R2, or R1 = 1, respectively. The genotype frequencies of (AA, AB, BB) in cases is set to {(1 − pk)2/c, 2pk(1−pk)R1/c, pk2R2/c }, where c = (1 − pk)2 + 2pk(1−pk)R1 + pk2R2. With the specified genotype frequencies, cases and controls were sampled from the corresponding trinomial distribution with parameters {(1 − pk)2/c, 2pk(1−pk)R1/c, pk2R2/c } and {(1 − pk)2, 2pk(1−pk), pk2}, respectively. We compared the power of five different methods: 1) the extended HWD trend test, 2) the GA trend test as implemented in EIGENSTRAT, 3) the GSB procedure, 4) the smooth-GSB procedure, and 5) MAX3-PC. The top 10 PCs of the genome-wide genotypes were used as covariates in the GA trend test and MAX3-PC. We simulated data under the four genetic disease models: the additive, multiplicative, dominant and recessive models, and considered different genotype relative risks. For each scenario, we simulated 10,000 replicates of the case-control data. The nominal FWER was set to be 0.05. For the individual SNP based tests (the extended HWD trend test, the GA trend test, and MAX3-PC), the corresponding significance level for each SNP was set to 0.05/m, where m is the number of SNPs. The power was estimated as the proportion of replicates with identified significant SNPs.
To test the influence of different values of λ on the smooth-GSB procedure, we estimated power of the smooth-GSB procedure with different λ values ranging from 0.55 to 0.90 based on 10,000 replicated data sets. Results were reported in Table III. We can see that the power of the smooth-GSB procedure is robust to different values of λ when 0.55 ≤ λ ≤0.9. When λ increased from 0.55 to 0.90, the power of the smooth-GSB procedure slightly increased under the additive and the multiplicative models and slightly decreased under the dominant and recessive models. More specifically as λ increased from 0.60 to 0.85, the power increase of the smooth-GSB procedure under the additive and multiplicative models was usually less than 0.02 and the power decrease under the dominant and recessive models was usually less than 0.03. Given this, we would further suggest setting the value of λ in the smooth-GSB procedure such that 0.60 ≤ λ ≤0.85. For example we can set λ = 0.7. Compared to the GA trend test, our smooth-GSB procedure has much higher power than the GA trend test under both the recessive and dominant models. On the other hand, the smooth-GSB procedure (when 0.60 ≤ λ ≤ 0.85) has power very close to the GA trend test (the difference is often less than 0.02) under both the additive and multiplicative models. For example, under the recessive model with genotypic relative risk R2 = 1.5, the power of the smooth-GSB procedure with λ = 0.6 was 0.531, which was about 10% (in absolute terms) higher than the power 0.438 of the GA trend test. We can also see that the robust test MAX3-PC of So and Sham (2011) had relatively lower power than the GA trend test and our smooth-GSB procedure under the additive and multiplicative models while it had higher power than the other two methods under the dominant and recessive models. For example, under the additive model with genotype relative risk R1 = 1.3, the power of the GA trend test, the smooth-GSB procedure (with λ = 0.6) and MAX-PC were 0.465, 0.448, and 0.420 respectively. The power of MAX3-PC was 4.5% (in absolute terms) lower than that of the GA trend test.
Table III.
Empirical power based on 10,000 replicates (the nominal FWER was 0.05).
| Modela | R1 | R2 | E-HWDTTb | GA trendc |
GSBd | S-GSBe |
MAX3- PCf |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| λ=.55 | λ=.6 | λ=.65 | λ=.7 | λ=.75 | λ=.8 | λ=.85 | λ=.9 | |||||||
| ADD | 1.2 | 1.4 | .000 | .101 | .065 | .090 | .091 | .093 | .094 | .096 | .098 | .098 | .099 | .079 |
| 1.3 | 1.6 | .000 | .465 | .379 | .445 | .448 | .451 | .454 | .456 | .458 | .460 | .462 | .420 | |
| 1.4 | 1.8 | .000 | .766 | .705 | .752 | .755 | .757 | .758 | .761 | .762 | .764 | .764 | .733 | |
| 1.5 | 2 | .000 | .888 | .855 | .882 | .883 | .883 | .884 | .885 | .886 | .887 | .888 | .873 | |
| DOM | 1.3 | 1.3 | .002 | .069 | .100 | .095 | .093 | .092 | .091 | .089 | .087 | .085 | .083 | .103 |
| 1.5 | 1.5 | .030 | .385 | .499 | .488 | .485 | .483 | .480 | .475 | .470 | .463 | .455 | .505 | |
| 1.7 | 1.7 | .153 | .556 | .672 | .657 | .655 | .652 | .650 | .647 | .644 | .638 | .631 | .687 | |
| 1.9 | 1.9 | .352 | .627 | .743 | .730 | .728 | .726 | .723 | .720 | .717 | .713 | .705 | .764 | |
| MUL | 1.2 | 1.44 | .000 | .158 | .104 | .146 | .148 | .150 | .151 | .153 | .154 | .155 | .156 | .126 |
| 1.3 | 1.69 | .000 | .640 | .547 | .621 | .623 | .626 | .629 | .631 | .634 | .636 | .637 | .597 | |
| 1.4 | 1.96 | .000 | .903 | .861 | .895 | .897 | .898 | .898 | .899 | .900 | .900 | .902 | .886 | |
| 1.5 | 2.25 | .000 | .971 | .958 | .967 | .968 | .968 | .969 | .969 | .969 | .970 | .971 | .966 | |
| REC | 1 | 1.3 | .002 | .070 | .093 | .092 | .091 | .090 | .089 | .088 | .087 | .085 | .083 | .104 |
| 1 | 1.5 | .027 | .438 | .542 | .534 | .531 | .528 | .525 | .520 | .515 | .512 | .503 | .555 | |
| 1 | 1.7 | .121 | .664 | .775 | .764 | .762 | .760 | .758 | .755 | .752 | .748 | .742 | .773 | |
| 1 | 1.9 | .276 | .750 | .857 | .847 | .845 | .843 | .841 | .838 | .835 | .831 | .825 | .853 | |
ADD: additive model; DOM: dominant model; MUL: multiplicative model; REC: recessive model
E-HWDTT = the extended HWD trend test
GA trend = the GA trend test
GSB = generalized sequential Bonferroni procedure
S-GSB = the smooth-GSB procedure with different λ from 0.55 to 0.9
MAX3-PC = the extended MAX3 with PCs as covariates.
Another advantage of the smooth-GSB procedure is that it is much faster than MAX3-PC. On a computer with CPU 3.47G Hz, with the principal components (PCs) already computed, to process 10,000+10 SNPs, our R implementation of the smooth-GSB procedure for GWAS took less than 15 seconds, while MAX3-PC implemented in the Robust SNP package (So and Sham 2011) took about 1,100 seconds if the absolute error of p-values was set to 10−6. It took about 2,500 seconds if the absolute error of p-values is set to 10−8. This means that for GWAS with about one million SNPs, the MAX3-PC will take about 70 hours with p-value accuracy level of 10−8, while the smooth-GSB procedure takes less than 25 minutes.
4. Application to WTCCC data
The WTCCC data contain seven different data sets of major diseases [13]. To evaluate the performance of the proposed method on real data, we chose four data sets on bipolar disorder (BD), coronary artery disease (CAD), Crohn’s disease (CD), and type 2 diabetes (T2D) from the WTCCC data. Each of the four data sets had the reported genomic control inflation factor > 1.06 [13], therefore there may be population stratification in each of these data sets. There were about 3,000 controls and about 2,000 cases in each data set.
We did the quality control according to the description in the WTCCC paper [13] to filter out some individuals and SNPs. The total number of SNPs after quality control was about 400,000. Following the suggestion of the WTCCC paper [13], we used only two top PCs as covariates to correct for population stratification in the GA trend test and MAX3-PC. These PCs were calculated based on about 190,000 SNPs which was obtained by pruning the whole genome SNPs using the software PLINK [24]. In each of the three single marker-based tests: the extended HWD trend test, the GA trend test and MAX3-PC, we can calculate a p-value at each of the about 400,000 SNPs. But in the smooth-GSB procedure, we can only calculate an adjusted p-value (see Section 2.3) for each SNP. In order to compare the adjusted p-value with the p-values from the three single marker-based tests, we define a p-value for each SNP in the smooth-GSB procedure as the adjusted p-value divided by the number of tests (m); however, if the adjusted p-value at a SNP is 1, then we will not define a p-value for that SNP because the null hypothesis will not be rejected at the SNP. When assuming the independence of markers, for the smooth-GSB procedure, comparing the adjusted p-value with the nominal FWER α is equivalent to comparing the new defined p-value with α/m.
Following the suggestion of the WTCCC paper [13], we used the threshold of 5×10−7 for individual SNPs. Table IV lists the significant SNPs identified by at least one of the three methods: the GA trend test, the smooth-GSB procedure, and MAX3-PC. For each significant region, only the SNP with the smallest p-value is listed. We can see that all the SNPs detected by the GA trend test, were also detected by the smooth-GSB procedure with different values of λ: 0.6, 0.7, and 0.8; the only exception is that SNP rs10761659 was not identified by the smooth-GSB procedure with λ = 0.6 (p-value=5.01×10−7,which is very close to the threshold 5×10−7). For the bipolar disorder, SNP rs420259 was detected by both the smooth-GSB procedure and MAX3-PC, but not by the GA trend test. This was reasonable because the best-fitting genetic model for SNP rs420259 was recessive [13]. As we described earlier, the GA trend test has low power under the recessive model. In addition, two SNPs, rs11747270 (for CD) and rs10806665 (for T2D), and their corresponding regions, were detected by both the GA trend test and the smooth-GSB procedure, but were not detected by MAX3-PC. The reason for this might be that the underlying disease models at these two SNPs are additive and MAX3-PC method has relatively lower power than the other two methods under the additive model. We note that the SNP rs10806665 (for T2D) was not identified significant by WTCCC [13]. We also note that for the coronary artery disease, two SNPs, rs4854090 and rs5007171, were detected by MAX3-PC but not by the GA trend test and smooth-GSB procedure. The underlying disease models at these two SNPs might be recessive or dominant, because under these two models, MAX3-PC has higher power than the other two methods. These two SNPs and their corresponding regions were not reported as significant by WTCCC [13].
Table IV.
P-values for the most significant SNPs in the regions identified by the GA trend test, the smooth-GSB procedure or MAX3-PC in four WTCCC data sets with population stratification.
| Disease | Chr | SNP* | MAF | E-HWDTTa | GA trendb | S-GSBc |
MAX3-PCd | ||
|---|---|---|---|---|---|---|---|---|---|
| λ=0.6 | λ=0.7 | λ=0.8 | |||||||
| BD | 16 | rs420259 | .282 | 6.93×10−06 | 7.64×10−04 | 2.34×10−07 | 3.12×10−07 | 4.68×10−07 | 4.69×10−09 |
| CAD | 2 | rs4854090 | .251 | 4.31×10−06 | 6.97×10−02 | NAe | NA | NA | 3.26×10−08 |
| 3 | rs5007171 | .210 | 5.15×10−07 | 1.21×10−01 | NA | NA | NA | 1.31×10−08 | |
| 7 | rs17146094 | .016 | 1.01×10−03 | 6.46×10−06 | 3.01×10−07 | 3.95×10−07 | 5.74×10−07 | 7.10×10−05 | |
| 9 | rs1333049 | .474 | 3.05×10−01 | 6.66×10−15 | 9.96×10−15 | 8.86×10−15 | 7.98×10−15 | 1.21×10−14 | |
| CD | 1 | rs11805303 | .317 | 8.03×10−01 | 8.24×10−13 | 1.29×10−12 | 1.13×10−12 | 1.00×10−12 | 4.71×10−12 |
| 2 | rs10210302 | .481 | 2.79×10−02 | 5.12×10−14 | 2.94×10−14 | 3.29×10−14 | 3.73×10−14 | 1.58×10−13 | |
| 5 | rs17234657 | .125 | 7.38×1001 | 2.19×10−14 | 3.40×10−14 | 2.99×10−14 | 2.66×10−14 | 5.17×10−13 | |
| 5 | rs11747270 | .068 | 7.13×10−01 | 2.57×10−07 | 3.99×10−07 | 3.51×10−07 | 3.13×10−07 | 1.51×10−06 | |
| 10 | rs10761659 | .461 | 8.95×10−01 | 3.18×10−07 | 5.01×10−07 | 4.38×10−07 | 3.89×10−07 | 6.14×10−07 | |
| 10 | rs10883365 | .477 | 2.74×10−01 | 3.39×10−08 | 4.73×10−08 | 4.30×10−08 | 3.95×10−08 | 7.82×10−08 | |
| 16 | rs2076756 | .242 | 4.36×10−01 | 3.77×10−15 | 2.83×10−15 | 3.02×10−15 | 3.24×10−15 | 5.73×10−14 | |
| 18 | rs2542151 | .163 | 3.54×10−01 | 1.09×10−08 | 1.59×10−08 | 1.43×10−08 | 1.29×10−08 | 6.75×10−08 | |
| T2D | 6 | rs10806665 | .077 | 8.03×10−01 | 2.83×10−07 | 4.48×10−07 | 3.92×10−07 | 3.47×10−07 | 1.29×10−06 |
| 10 | rs4506565 | .324 | 9.94×10−01 | 8.54×10−13 | 1.36×10−12 | 1.19×10−12 | 1.05×10−12 | 3.52×10−12 | |
| 16 | rs7193144 | .396 | 1.23×10−01 | 3.91×10−07 | 4.84×10−08 | 4.57×10−08 | 4.33×10−08 | 5.53×10−08 | |
Only the SNP with the smallest p-value in each significant region is listed.
E-HWDTT: the extended HWD trend test
GA trend: the GA trend test
The smooth-GSB procedure with different λ values; P-values for S-GSB was calculated as the adjusted p-values divided by the number of test
MAX3-PC: the extended MAX3 adjusting for top PCs.
the adjusted p-value was equal to 1 and therefore the adjusted p-value for individual SNPs was not calculated. The p-values in bold mean they do not reach the genome-wide significant level 5×10−7. The SNP IDs with underscore and in italic style denote that the corresponding regions are not listed in Table 3 of the WTCCC paper, which lists the regions of the strongest association signals.
5. Discussion
In this study, we proposed a fast and robust association test procedure (the smooth-GSB procedure) which can be applied to GWAS data with case-control designs in the presence of subpopulations. As showed in the simulation studies, the smooth-GSB procedure can have significantly higher power than the GA trend test implemented in EIGENSTRAT when the underlying genetic model is dominant or recessive. It can also have power comparable to the GA trend test when the underlying genetic model is additive or multiplicative. Our simulation studies indicate that the existing robust test, MAX3-PC, can be computationally intensive for GWAS with a large number of SNPs and can have relatively lower power than the GA trend test when the underlying disease model is additive or multiplicative. Our proposed method can be viewed as an extension to account for population stratification of our previously proposed generalized sequential Bonferroni (GSB) procedure for GWAS in homogeneous populations [25].
The proposed smooth-GSB procedure is not appropriate for analysis of rare variants (for example with minor allele frequency (MAF) less than 0.05). When MAF < 0.05, the extended HWD trend test may not produce accurate p-values.
In our proposed smooth-GSB procedure, the parameter λ (0 ≤ λ ≤ 1) should be set empirically. If we choose λ = 1, then the smooth-GSB procedure becomes the Holm’s sequential rejective procedure [20] using the GA trend test and is approximately equivalent to the GA trend test with Bonferroni correction (if the number of markers is large enough such as in GWAS analysis). When λ = 1 the smooth-GSB procedure has the highest power if the underlying disease models are additive or multiplicative, but has relatively low power if the underlying disease models are recessive or dominant. On the other hand, in the smooth-GSB procedure, if we choose λ = 0, then it becomes the GSB procedure (see Section 2.4), which has the highest power when the disease models are recessive or dominant, but has lower power than the GA trend test if the underlying disease models are additive or multiplicative. In real data analysis, usually we don’t know the true disease models. Since most significant SNPs reported in the literature so far have demonstrated additive effects, based on our simulation studies, we would suggest choosing the value of λ such that 0.6 ≤ λ ≤ 0.85. For example, we can set λ = 0.7. As indicated by the results of our simulation studies, when the values of λ change from 0.6 to 0.85, the power of our smooth-GSB procedure is: a) very robust to different values of λ under different disease models, b) very close to that of the GA trend test if the underlying disease model is additive or multiplicative, and c) much higher than that of the GA trend test if the underlying disease model is recessive or dominant.
We note that in the extended HWD trend test, any proper clustering algorithm to partition the individuals into subpopulations can be applied, such as the more computationally intensive method proposed by Prichard et al. [26]. The k-means clustering method used in this paper has the advantage of low computation cost.
In the extended HWD trend test, we applied the k-means clustering algorithm to the top PCs of the genotype data to partition the individuals in the case-control data into several clusters corresponding to homogeneous subpopulations. This k-means clustering may not be appropriate for data from admixed populations, for which different partitioning methods may be needed. For example, we can partition individuals by the number of alleles at a marker inherited from an ancestral population. This partitioning method was used by Shriner et al. [27] in association studies in admixed populations.
In this article, we assume independence of markers in GWAS by ignoring the dependence among markers. The proposed methods under this assumption can be relatively conservative when applied to GWAS data analysis due to ignoring the LD among markers. Accounting for linkage disequilibrium among dense markers may increase power of association study methods in detecting causal variants. In our future study, we plan to incorporate SNP set (or gene)-based association tests, e.g., SKAT [28], which can account for dependence among markers, into our smooth-GSB procedure.
Acknowledgements
This research was supported by the National Institutes of Health grants: R01GM073766 from the National Institute of General Medical Sciences, U01HL101064 from the National Heart, Lung, and Blood Institute, and R01LM011177 from the National Library of Medicine. The research was also partly supported by the National Center for Advancing Translational Sciences (grant UL1TR000058). This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475.
Appendix: Evaluation of FWER of the GSB procedures of Holm (1979) for multiple testing with weights weakly correlated with the tests
Here we show by simulation studies that under some situations, in the GSB procedure of Holm (1979) [20] for m independent hypotheses H1, H2, …, Hm, even though the weights are weakly correlated with the m tests, the GSB procedure can still control FWER well (see also section 2.3). Assuming all the null hypotheses H1, H2, …, Hm are true, the p-value pj from the j-th test corresponding to hypothesis Hj follows the uniform distribution U[0,1] (j =1,…,m). Suppose we calculate a weight wj for this p-value pj by wj=1/qj, where qj is a random variable whose marginal distribution is also the uniform distribution U[0,1], and qj is weakly correlated with pj, with qj and pj jointly following a bivariate uniform distribution. The correlation between the two variables qj and pj can be simulated by the bivariate copulas [29]. Suppose there were 1,000 tests (m=1,000). For the j-th test, we simulated a pair of q-value and p-value (qj and pj) from a bivariate uniform distribution with dependence between qj and pj by the Gaussian copula. We used Spearman’s rank correlation coefficient ρs to measure the dependency between the pair of q-value and p-value (qj and pj).
Once the pair of q-value and p-value was simulated for each test, we had 1,000 pairs of q-value and p-value corresponding to 1,000 tests. We calculated weights by using the q-values and then we applied the GSB procedure to the 1,000 tests. We replicated this simulation procedure 107 times and therefore generated 107 replicated “data sets”, each consisting of 1,000 pairs of q-value and p-values. We estimated the FWER of the GSB procedure over these 107 replicated “data sets” with the nominal FWER as 0.01, 0.05 and 0.1. The FWER was estimated as the proportion of replicates with at least one false discovery. The results are showed in Table V. From the results we can see that the FWER of the GSB procedure was well controlled when the Spearman’s rank correlation coefficient ρs is equal to or less than 0.01 (i.e., q-value and p-value were weakly correlated).
Table V.
Empirical FWERs of the GSB procedures of Holm (1979) for 1,000 tests with weights weakly correlated with the tests#.
| Nominal FWER (α) |
|||
|---|---|---|---|
| ρs* | 0.01 | 0.05 | 0.1 |
| 0.005 | 0.0103 | 0.0505 | 0.0987 |
| 0.01 | 0.0101 | 0.0495 | 0.0965 |
based on 107 replicated datasets.
ρs is the Spearman’s rank correlation coefficient between the test statistics, respectively.
The nominal FWER used to reject null hypothesis.
References
- 1.Reich DE, Goldstein DB. Detecting association in a case-control study while correcting for population stratification. Genetic Epidemiology. 2001;20:4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 2.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 4.Campbell CD, Ogburn EL, Lunetta KL, Lyon HN, Freedman ML, Groop LC, Altshuler D, Ardlie KG, Hirschhorn JN. Demonstrating stratification in a European American population. Nat. Genet. 2005;37:868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- 5.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 6.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am. J. Hum. Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freidlin B, Zheng G, Li Z, Gastwirth JL. Trend tests for case-control studies of genetic markers: power, sample size and robustness. Hum. Hered. 2002;53:146–152. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- 8.González JR, Carrasco JL, Dudbridge F, Armengol L, Estivill X, Moreno V. Maximizing association statistics over genetic models. Genet. Epidemiol. 2008;32:246–254. doi: 10.1002/gepi.20299. [DOI] [PubMed] [Google Scholar]
- 9.Wang K, Sheffield VC. A constrained-likelihood approach to marker-trait association studies. Am. J. Hum. Genet. 2005;77:768–780. doi: 10.1086/497434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joo J, Kwak M, Zheng G. Improving power for testing genetic association in case-control studies by reducing the alternative space. Biometrics. 2010;66:266–276. doi: 10.1111/j.1541-0420.2009.01241.x. [DOI] [PubMed] [Google Scholar]
- 11.Armitage P. Tests for linear trends in proportions and frequencies. Biometrics. 1955;11:375–386. [Google Scholar]
- 12.So HC, Sham PC. Robust association tests under different genetic models, allowing for binary or quantitative traits and covariates. Behavior Genetics. 2011;41:768–775. doi: 10.1007/s10519-011-9450-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song K, Elston RC. A powerful method of combining measures of association and Hardy-Weinberg disequilibrium for fine-mapping in case-control studies. Stat. Med. 2006;25:105–126. doi: 10.1002/sim.2350. [DOI] [PubMed] [Google Scholar]
- 15.MacQueen J. Some methods for classification and analysis of multivariate observations. In: Cam LM Le, Neyman J., editors. Proc. of the fifth Berkeley Symposium on Mathematical Statistics and Probability. Berkeley, CA: 1967. pp. 281–297. [Google Scholar]
- 16.Hennig C. fpc: Flexible procedures forclustering. 2010 R package version 2.0-3. http://CRAN.R-project.org/package=fpc.
- 17.Duda RO, Hart PE. Pattern classification and scene analysis. 1st ed. New York: John Wiley & Sons; 1973. [Google Scholar]
- 18.Calinski RB, Harabasz J. A dendrite method for cluster analysis. Communications in Statistics. 1974;3:1–27. [Google Scholar]
- 19.Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
- 21.Westfall PH, Yong SS. Resampling-Based Multiple Testing: Examples and Methods for p-Value Adjustment. New York: John Wiley and Sons; 1993. [Google Scholar]
- 22.Roeder K, Devlin B, Wasserman L. Improving power in genome-wide association studies: weights tip the scale. Genetic Epidemiology. 2007;31:741–747. doi: 10.1002/gepi.20237. [DOI] [PubMed] [Google Scholar]
- 23.Joo J, Kwak M, Ahn K, Zheng G. A robust genome-wide scan statistic of the Wellcome Trust Case-Control Consortium. Biometrics. 2009;65:1115–1122. doi: 10.1111/j.1541-0420.2009.01185.x. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao G, Kang G, Wang J, Chen W, Qin H, Jiang B, Li Q, Sun C, Liu N, Archer KJ, Allison DB. A generalized sequential bonferroni procedure using smoothed weights for genome-wide association studies incorporating information on hardy-weinberg disequilibrium among cases. Hum. Hered. 2011;73:1–13. doi: 10.1159/000332916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shriner D, Adeyemo A, Rotimi CN. Joint ancestry and association testing in admixed individuals. PLoS Comput Biol. 2011;7:e10002325. doi: 10.1371/journal.pcbi.1002325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare variant association testing for sequencing data with the sequence kernel association test (SKAT) Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelsen RB. An introduction to copulas. 2nd ed. New York: Springer; 2006. [Google Scholar]