Abstract
If a eukaryotic cell is to reproduce, it must duplicate its genetic information in the form of DNA, and faithfully segregate that information during a complex process of cell division. During this division process, the resulting cells inherit one, and only one, copy of each chromosome. Over thirty years ago, it was predicted that the segregation of sister chromosomes could occur non-randomly, such that a daughter cell would preferentially inherit one of the two sister chromosomes according to some characteristic of that chromosome’s template DNA strand. Although this prediction has been confirmed in studies of various cell-types, we know little of both the mechanism by which the asymmetric inheritance occurs and the significance it has to cells. In this essay, we propose a new model of non-random chromosome segregation – the mortal strand hypothesis – and discuss tests of the model that will provide insight into the molecular choreography of this intriguing phenomenon.
Keywords: sister chromatids, immortal strands, mortal strands, chromosomes, DNA damage, stem cells, asymmetric cell division
1. Introduction
In 1975, John Cairns proposed that “ stem cells would be protected against errors of duplication if it were so arranged that the immortal daughter cell always receives the DNA molecules which have the older of the two parental strands and the mortal daughter always collects the molecules with the younger parental strand.”1 Since then, Cairns’ “ immortal strand” hypothesis has been studied in various organisms and celltypes. Despite evidence in support of the hypothesis,2 it has met with well-reasoned skepticism arising from observations that fail to support Cairns and from a near-absence of mechanistic insight into the phenomenon.3 Here, using evidence gained from nearly forty years of study, we put forth a new theory of non-random chromosome segregation and we propose a model as to how it might occur. The main goals are to help guide the evaluation of existing studies of non-random chromosome segregation and to provide testable models for future investigation.
1.1 The “mortal strand” hypothesis
We hypothesize that the segregation of sister chromatids according to relative template strand age is a consequence of replication stress, which is defined as “ inefficient DNA replication that causes DNA replication forks to progress slowly or stall.”4 In our model (Figure 1), replication stress generates frank DNA damage, asymmetrically, in chromosomes bearing newer template DNA (the “ mortal” strands). This creates a situation in which it is advantageous to preferentially segregate chromosomes bearing newer template strands, in which there is DNA damage, to a single daughter cell. We further hypothesize that the preferential segregation is possible because DNA damage repair machinery recruited to sites of replication-derived damage signals to the mitotic spindle apparatus to direct the attachment of spindle microtubules. Recent advances in cell biology give us a framework for understanding how this might occur (see below). The preferential search-and-capture of sister chromatids may happen in concert with asymmetric centrosome behavior at the spindle pole, possibly related to the relative age of the mother centrioles. This centrosome asymmetry could thus direct the co-segregation of damaged chromosomes with fate determinants, such as the protein Numb, that direct the differentiation or cell-cycle exit of the daughter cell inheriting damaged chromosomes. Alternatively, the DNA damage inherited by one of the two daughter cells may by itself instruct the cell to adopt a particular fate. We predict that cells inheriting chromosomes bearing a large burden of DNA damage are prevented from replicating, at least until they are fully repaired. In our model, these daughters may differentiate, senesce, or die.
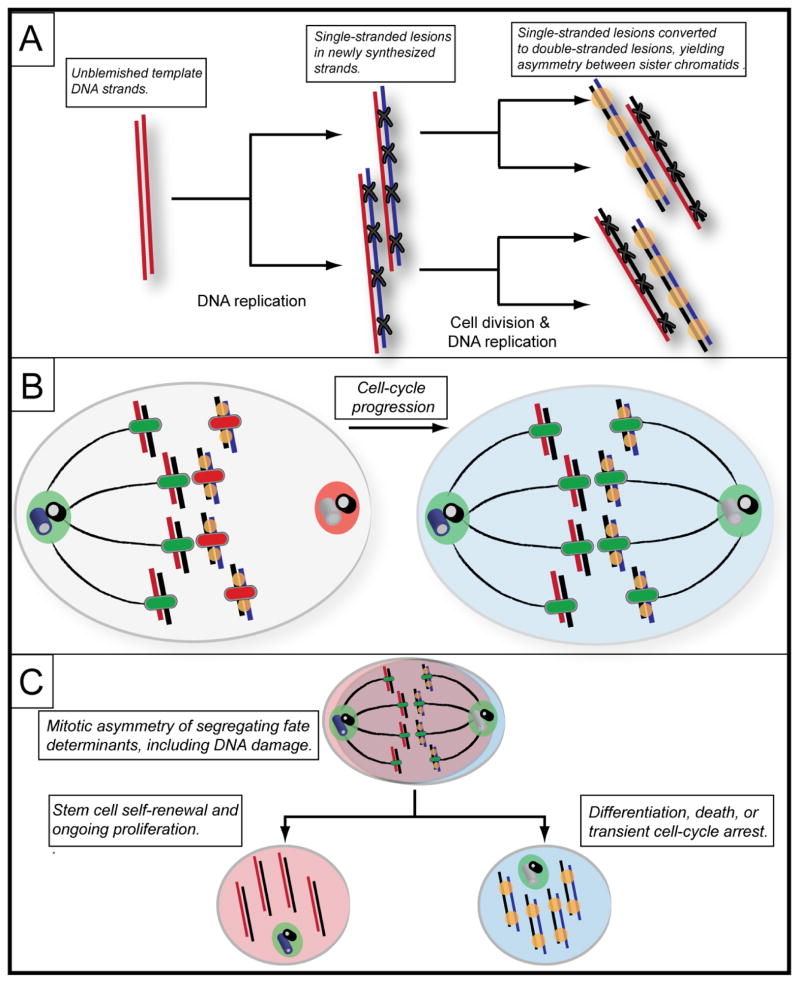
Figure 1. The mortal strand hypothesis.
(A) A chromosome consisting of two unblemished DNA strands (red), each of which serves as a template for synthesis of a new strand (blue) to form two sister chromatids. In our model, because of DNA replication stress, the newly synthesized strand contains lesions (X), such as singlestranded gaps or misincorporated nucleotides. Following sister chromatid separation and cell division, each of the two strands of the chromosome again serves as a template for synthesis of one of two sister chromatids. In this round of DNA synthesis, the replication fork encounters pre-existing, unrepaired lesions on the newer template DNA strand, causing the fork to collapse and form double-stranded breaks (yellow circle). The older, still unblemished template DNA strand again generates a chromatid with single-strand lesions (X) on the newly synthesized strand (black). The resulting sister chromatids are therefore distinguished by the DNA damage each bears. (B) The presence of doublestranded breaks on the sister chromatid containing the newer template strand results in epigenetic changes that are transmitted to the centromere of that chromatid (red centromere). These epigenetic changes transiently inhibit association of the mitotic microtubule spindle with chromatids bearing newer template strands. At the same time, an asymmetry in the spindle-forming capacity of the centrosomes, based on the relative age of the mother centrioles, generates a transient half-spindle. Because the undamaged chromatids bearing older template strands do not contain an inhibitory signal at the centromere (green centromere), the spindle microtubules of the dominant centrosome (green) preferentially attach to these chromatids. Later in the cell cycle, the inhibitory signal at the centromere of the chromatids containing newer template strands is lost, as the non-dominant centrosome forms the second half of the mitotic spindle. The microtubules emanating from the non-dominant “ younger” centrosome therefore form attachments with the chromatids with newer template strands. (C) The non-random segregation of template DNA strands is coordinated with asymmetric fate determination in the two daughter cells resulting from the division. DNA damage, having been segregated asymmetrically, can by itself initiate a signaling pathway that leads to the differentiation or death of the daughter cell inheriting the damage. Another possibility is that fate determinants, such as Numb, are distributed unequally between daughter cells (depicted by the different colors of the cells). The distribution of fate determinants may be coordinated with non-random chromosome segregation, or the two phenomena may occur independently.
2. Model of DNA damage inheritance during non-random chromosome Segregation
2.1 The origin of DNA damage asymmetry on sister chromatids
Our model identifies replication stress as a source of DNA damage that could give rise to frank DNA breaks that are asymmetrically localized between sister chromatids in a manner that is dependent on the relative age of the template strands. Replication stress encompasses defects in DNA synthesis associated with fluctuations in the availability of dNTPs or other factors required for DNA replication, increased or decreased firing of replication origins (hyper- or hypo-replication, respectively), and single-strand lesions on pre-existing template DNA.4 A key characteristic of DNA replication stress is that the stress is commonly brought about by changes in a cell’s environment. DNA hyper-replication, for example, can be induced by stimulating cells with growth factors.5 Single-strand lesions can result from a number of environmental agents, such as reactive oxygen species and UV radiation.4 If, as we propose, replication stress is a key initiator of asymmetries in sister chromatids, non-random chromosome segregation may be determined in part by the cellular environment. This prediction may have important implications for the interpretation of existing experimental analyses of non-random chromosome segregation, which we discuss later.
How might replication stress lead to asymmetries in the amount of DNA damage in sister chromosomes? Replication stress will initially generate single-stranded defects in the newly synthesized strands of both sister chromatids during S-phase. After the cell undergoes mitosis and cytokinesis, the resulting daughter cells will contain chromosomes in which the newly synthesized strand retains defects from the previous round of DNA replication, unlike the complementary, older strand. When S phase is initiated in either of the daughter cells, the synthesis of new strands differs in conjunction with the age of the template strand for each chromosome. For the older template strand, DNA synthesis proceeds as in the previous round, generating a new sister chromatid with single-stranded damage on the newly synthesized strand. By contrast, for the newer template strand, which contains defects from the previous round of DNA synthesis, the replication fork will collapse when it encounters pre-existing, unrepaired lesions, yielding double-stranded breaks.6 The end-result is a cell in G2 in which sister chromatids differ in terms of their burden of double-stranded DNA breaks.
Although our idea is akin to that originally proposed by Cairns – that newly synthesized DNA has errors from DNA replication and that it is advantageous to segregate damaged chromosomes away from the stem cell compartment – there are substantial differences. These differences may be important in guiding our thinking about past and future experimental tests of the immortal (or mortal) strand hypothesis. One major distinction between our model and that proposed by Cairns relates to the critical types of DNA errors attributed to the chromosome containing the newer template strand. While Cairns model focused on the spontaneous nucleotide misincorporation that always occurs during DNA synthesis, our model concerns chromosome breaks. This distinction has two important implications. First, double strand breaks are expected to induce growth-suppressing cellular phenotypes such as cell-cycle arrest, senescence, or death. The possibility that non-random chromosome segregation allows for a cell lineage to avoid such growth suppression suggests a functional significance unrelated to oncogenesis, as suggested by Cairns. Rather, it is analogous to asymmetric segregation of damaged macromolecules observed in many evolutionarily distant cell types (see below). Another important distinction is that double strand breaks may play a functional role in the process by which sister chromatids are inherited asymmetrically.
2.2 The biased segregation of damaged chromosomes
Double-stranded DNA breaks initiate local epigenetic responses that are transmitted throughout the cell by a DNA damage response network.7 This response network ultimately results in cellular phenotypes such as cell-cycle arrest, senescence, or programmed cell death.8 In order for non-random chromosome segregation to occur, sister chromatids must exhibit some molecular feature that reflects the relative age of the template DNA strand. Furthermore, this mark must have some potential to communicate – either directly or indirectly – with the mitotic spindle to promote or inhibit the attachment of spindle microtubules to that chromatid. This communication with the mitotic spindle is likely to take place at the centromere, the site of attachment of kinetochore microtubules.
Until now, only one specific model for the differential “ labeling” of sister chromosomes has been proposed.9 The hypothesis is based on the idea that the polarity of replication fork movement in a given region of DNA will determine whether that DNA is replicated by leading- or lagging-strand DNA synthesis. Leading- and lagging-strand synthesis differ in the recruitment of various replication factors, such as proliferating cell nuclear antigen (PCNA), that may remain associated with the chromatin after synthesis is complete. Although these replication factors may recruit others to the DNA, it is not clear from previous studies if these factors are able to communicate, directly or indirectly, with the mitotic spindle in a way that would allow for the selective association of sister chromatids with kinetochore microtubules. Also, we do not yet know whether replication fork movement across centromeres, which are large enough to require multiple origins of replication, is unidirectional for all chromosomes in cell-types in which non-random chromosome segregation occurs.
We hypothesize that the generation of asymmetric DNA damage on sister chromatids by replication stress could initiate a local epigenetic response that serves as a mark or identifier of sister chromatids bearing newer template DNA strands. In other words, the DNA damage, itself, emits a transient “ pick me” or “ don’t pick me” signal that is interpreted by the mitotic spindle. In this model, the double-stranded breaks generated by DNA replication on the newer template strand would recruit DNA damage response factors, including PI3 kinase family members ATM/R, which in turn would phosphorylate the histone H2A variant H2AX, yielding γ-H2AX. We speculate that γ-H2AX, enriched on the sister chromatid with the newer template strand, could function as a mark that distinguishes these chromatids from those having the older template strand. A recent elegant study in yeast demonstrated the feasibility of this crosstalk between DNA damage checkpoint and spindle assembly checkpoint pathways.10 James Haber and colleagues used an inducible double-stranded break system to demonstrate that just one double-stranded break can delay mitosis by a γ-H2AX-dependent mechanism. Even more remarkably, this mitotic checkpoint is dependent on the presence of the centromere of the chromosome in which the double-stranded break occurred.
These findings provide clear evidence that DNA breaks generate a signal that is conveyed to the centromere of the chromosome on which the break occurred and that is ultimately communicated to the mitotic spindle. In these studies, the presence of a single double-stranded break could be “ sensed” by the centromere even at a distance of 90 kb or more. How might a double-stranded break “ alert” the centromere to its presence? One possibility is that γ-H2AX spreads from the site of the double-stranded break into distant parts of the damaged chromosome. Indeed, using chromatin-immunoprecipitation, Haber and colleagues found that the level of γ-H2AX at the centromere increased as much as 10-fold following induction of a break 90 kb away.10 Similar γ-H2AX-spreading is known to occur in mammalian cells.11 The idea that the γ-H2AX signal spreads along chromosomes with double-stranded breaks is appealing because transmission of the “ don’t pick me” signal clearly occurs in cis to the damage. γ-H2AX may not be the sole DNA damage-associated modification communicating with distant sites on damaged chromosomes: chromatin-immunoprecipitation studies in human cells have shown that 53BP1 and MDC1 spread to an extent that mirrors spreading of γ-H2AX.12
How is the “ don’t pick me” signal propagated? Studies of γ-H2AX-spreading in yeast indicate that it depends on the serine/threonine-protein kinase Mec1p, the homolog of human ATR.13 Mec1p is also required for the centromere-dependent G2/M cell-cycle arrest induced by a double-stranded break.10 We propose that γ-H2AX is a substrate for and recruiter of ATM/R, generating a positive-feedback loop in which γ-H2AX recruits additional ATM/R that is capable of phosphorylating nearby H2AX and propagating the signal to neighboring chromatin. This positive-feedback loop may involve the DNA damage response protein MDC1 and the MRN complex, which may help to recruit ATM to H2AX. We hypothesize that γ-H2AX, once at the centromere, impairs kinetochore function either directly, by distorting chromatin structure, or indirectly, by recruiting other factors that inhibit kinetochore formation.
Once all chromatids with the newest template strands have been “ identified” as bearing DNA damage, they must be recognized by and attached to one of the two centrosomes. Owing to their obvious age-related asymmetry and biased segregation in stem cells,14,15 centrosomes have received attention for their possible role in non-random chromosome segregation.16 One of the more parsimonious explanations for the cosegregation of template DNA strands with centrosomes of a particular age relates to observations of differential microtubule-nucleating activity between “ mother” and “ daughter” centrosomes, which are composed respectively of the older and younger mother centrioles. In Drosophila larval brain neuroblasts, a model system for studying asymmetric stem cell division, the two centrioles separate during interphase, but only one remains capable of organizing a microtubule aster.17,18 This “ dominant” centrosome remains stationary at the cell’s apical cortex and goes on to form one pole of the mitotic spindle. In contrast, the other centriole loses microtubule-nucleating activity and moves extensively throughout the cell during interphase. Just prior to mitosis, this centriole moves to the basal cell cortex, regains microtubule-organizing activity, and forms the second pole of the mitotic spindle. Remarkably, the “ dominant” centriole, which never loses microtubule-nucleating activity, is stereotypically inherited by the self-renewing stem cell. The more motile centriole is segregated to the differentiating daughter cell. In our model, different microtubule-organizing capabilities of mother and daughter centrosomes could account for the coordinated inheritance of sister chromatids with DNA damage. Because one centrosome forms its half of the mitotic spindle before the other, a situation arises in which sister centromeres are “ competing” for attachment to astral microtubules. If the set of newer template strands contain factors, possibly recruited by γ-H2AX or other DNA damage response proteins, that discourage astral microtubule attachment, the spindles of the older, dominant centrosome will prefer to attach to the undamaged sister chromatids. Later, after the older centrosomeundamaged chromatid relationship is established, the newer centrosome will become active, and will at that point have the opportunity to form kinetochores at the centromeres only of chromatids with newer template DNA. These attachments may be aided by a cell-cycle-dependent signal, such as dephosphorylation of γ-H2AX, that promotes attachment to the once-inhibited centromeres.
2.3 The fate of cells inheriting DNA damage
In many cases in which non-random chromosome segregation has been observed, there has been evidence in support of Cairns’ hypothesis that the biased template segregation would occur in conjunction with asymmetric cell fate determination.19–23 As in the immortal strand hypothesis, we propose that the cells inheriting chromosomes with newer template DNA strands adopt a fate that is distinct from its sister cell. This fate may range from differentiation to death.
In our model, the inheritance of DNA damage, itself, can account for the coordination of asymmetric cell fate with non-random chromosome segregation. It is well-known that DNA damage can activate signaling pathways that lead to cell-cycle arrest or death. In stem cell populations, a number of studies have also linked DNA damage-response pathways to the molecular regulation of stem cell self-renewal or differentiation. In hematopoietic stem cells, DNA damage due to ionizing radiation induces a “ differentiation checkpoint” that turns on a differentiation program and thus limits self-renewal.24 A similar process occurs in melanocyte stem cells in response to DNA damage, ultimately leading to the loss of melanocytes and hair-graying.25 It is thus possible that the DNA damage inherited by one of the daughter stem cells induces the differentiation of that cell. It will be interesting in future studies to analyze the effect of DNA damage signaling pathways on the adoption of cell fates during non-random chromosome segregation. This and other analyses of non-random chromosome segregation during asymmetric cell division would clearly benefit from novel techniques to monitor template DNA strand segregation using videomicroscopy of living cells.
Although previous studies have combined videomicroscopy with fixed-cell imaging to retrospectively study the fates of cells exhibiting non-random chromosome segregation,20 a technique to study non-random chromosome segregation in real-time would enable observation of cell fates, like cell death, that could not be studied using retrospective approaches.
A more complex model that explains the coordination of non-random chromosome segregation with asymmetric cell fate determination involves the directed segregation of cell-fate determinants. One possibility, given the model of asymmetric centrosome behavior and inheritance discussed earlier, is that a stereotyped centrosome asymmetry occurs in conjunction with the inheritance of cell-fate determinants. In other words, the centrosome-dependent asymmetry of the mitotic spindle may work in parallel with the spatial segregation of cell-fate determinants, such as Numb. We already know that the asymmetric distribution of Numb correlates with non-random chromosome segregation and with asymmetries in centrosome characteristics.20 An interesting question for future study is whether non-random chromosome segregation occurs on the background of asymmetric cell fate determination, or whether the two processes are co-regulated such that they occur in unison. Aurora-A, a mitotic kinase that regulates the asymmetric segregation of Numb26 and is regulated by DNA damage response pathways,27 is a possible nexus of coordinated regulation of the two asymmetries. There is already evidence that asymmetric segregation of Numb is related to the activity of DNA damage response pathways.28
3. Evidence of a role for DNA damage in non-random chromosome segregation
3.1 Non-random chromosome segregation as a quality control mechanism
In our model of non-random chromosome segregation, conditions that lead to inefficient DNA replication generate DNA damage in chromosomes with newer template strands. In a more traditional view of cell biology, this damage would be expected to initiate a signaling cascade that inhibits cell-cycle progression or even lead to the death of the cell. An alternative outcome, which would enable the preservation of the cell lineage, is the preferential segregation of the entire complement of sister chromatids with newer template strands to one of the two daughter cells.
The asymmetry we describe is analogous to lineage-preserving segregation phenomena that have been observed in cell-types ranging from E. coli to human embryonic stem cells. The spatial segregation of damaged or misfolded proteins in budding yeast is perhaps the most thoroughly studied of these phenomena. In this example, aggregates of oxidatively damaged proteins, arising from an acute oxidative stress or as a consequence of advanced replicative age, are positioned within cells such that all of the aggregates are trafficked to the mother cell rather than the daughter cell during cell division.29 This amazing “ quality control” mechanism is enabled by tethering aggregates of damaged proteins to an array of actin with a polarized flow. Because damaged proteins (like damaged DNA) can accumulate to such a level that they impair cell function, the segregation of protein aggregates can be viewed as a mechanism for rejuvenating, or simply preserving, a cell lineage.30 An analogous mechanism for discarding damaged DNA would seem to serve a similar purpose in maintaining cell lineages.
Extension of the work in budding yeast has led to the discovery of examples of asymmetric segregation of damaged macromolecules in evolutionarily distant cell types. In E. coli, which were once thought to divide into phenotypically identical progeny, daughter cells inheriting the older of the two cell poles exhibit a decline in fitness, decreased replicative potential and increased cell death.31 Just as in yeast, this observation has been linked to the segregation of protein aggregates to the cell inheriting the older pole.32 The situation is remarkably similar in the dividing cells of higher eukaryotes. In cultured human embryonic stem cells, polyubiquinated proteins, soon to be degraded, are segregated in a biased way to one daughter cell.33 Notably, this cellular refuse is physically associated with the centrosome, where the proteasomal machinery resides.34 It has been proposed that this asymmetry may relate to the age, and associated functional properties, of the centrosomes.33 If the segregation of damaged proteins is coordinated with non-random chromosome segregation, it may provide some insight into the significance, if not the mechanism, of co-segregation of centrosomes and template DNA strands.
3.2 Influence of cell context on non-random chromosome segregation
Our model relates to Cairns’ in that it associates DNA replication with errors in template DNA strands. However, our model identifies replication stress as a source of DNA damage, which occurs as a result of abnormalities in DNA synthesis often associated with cell growth conditions. Therefore, our model, unlike the immortal strand hypothesis, predicts that non-random chromosome segregation is precipitated by cellular context. This simple fact may help us to understand why non-random chromosome segregation occurs in some settings and not in others. It also might help to address certain shortcomings of the immortal strand hypothesis, including the observation that non-random chromosome segregation does not occur with great frequency during the homeostatic turnover of epidermal35 or hematopoietic stem cells36, but occurs frequently in the setting of acute tissue injury as in skeletal muscle.19,20 We predict that characteristics of the injured skeletal muscle niche, like oxidative or nutrient stress, promote replication stress, leading to the observed non-random chromosome segregation. Similar environmental stressors may affect cell behavior in a transplantation model used to study non-random chromosome segregation in mammary gland epithelium.37 It would be interesting to determine to what extent this and other transplantation models resemble acute injury or homeostatic turnover in terms of the frequency of non-random chromosome segregation and the level of replication stress induced in the cell population of interest. In general, our model could be tested by correlating the level of replication stress with the frequency of non-random chromosome segregation in these various contexts. Eventually, identification of the sources of replication stress would allow one to test whether modulating the level of replicationassociated DNA damage influences the level of non-random chromosome segregation.
An interesting question for future investigation is whether perturbations of the cell environment, either in culture or in vivo, have any impact on non-random chromosome segregation. An example of such a study, which examined non-random chromosome segregation in cultured lung epithelial cancer cells, found that the frequency of template strand asymmetries is influenced by the conditions in which the cells are cultured.38 This study identified cell density, serum concentration, and oxygen tension as environmental parameters that influence the frequency with which cells segregate DNA non-randomly.
In addition to determining whether these principles of environmental control of asymmetric cell division extend to other cell types, it will be interesting in the future to study the mechanisms by which changes in growth conditions impact non-random chromosome segregation. One possibility is that the environmental changes lead to differentiation of cells into cell types that do not divide asymmetrically with respect to template DNA strand age. An alternative explanation, related to our model, is that these changes in the environment induce cellular stresses leading to replication-dependent DNA damage or to changes in the DNA damage response. Changes in serum concentration and oxygen availability are indeed environmental factors that may be expected to induce the sort of replication stress that underlies our model. If manipulations of the cell environment cause changes in the frequency of non-random chromosome segregation that correlate with changes in the level of replicationassociated DNA damage, further manipulations of DNA damage signaling pathways may provide mechanistic insight.
3.3 Regulation of non-random chromosome segregation and cell growth kinetics by p53
One of the most intriguing connections between DNA damage and non-random chromosome segregation involves the regulation of biased DNA segregation by the quintessential DNA damage-response protein, p53 (Figure 2). Pioneered by James Sherley, this work has examined the regulation of non-random chromosome segregation, and other characteristics of asymmetric cell division, in cultured cell populations in which p53 is conditionally activated in the background of p53 nullizygosity.39 Specifically, Sherley and coworkers have engineered p53-deficient mouse cell lines harboring Zn-dependent p53-expression alleles. In these cells, pharmacological activation of p53 expression by administration of Zn induced cosegregation of template DNA according to relative age, as monitored by BrdU pulsechase analyses.
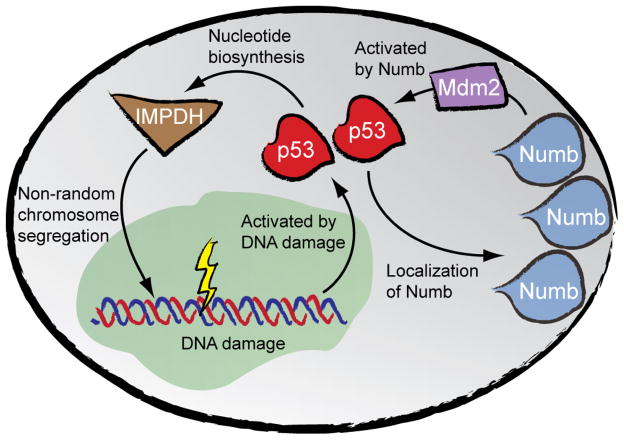
Figure 2. p53 in non-random chromosome segregation and asymmetric cell division.
p53 induces asymmetric segregation of the fate determinant Numb in dividing mouse mammary progenitors.28 Separately, Numb physically interacts with and stabilizes p53 by preventing its Mdm2-dependent degradation. The activation of p53 alters nucleotide biosynthesis secondary to p53’s transcriptional regulation of the gene encoding inosine 5′-monophosphate dehydrogenase (IMPDH), an enzyme required for guanine nucleotide biosynthesis. The effect of p53 on IMPDH induces non-random chromosome segregation by a mechanism that is currently unknown. We propose that the p53-dependent regulation of IMPDH results in replication-associated DNA damage that leads to non-random chromosome segregation. In its canonical role, p53 is activated in response to DNA damage and thus induces cell cycle arrest or cell death.
Strikingly, the induction of p53-dependent non-random chromosome segregation coincides with a change in cell proliferation kinetics from symmetric, exponential growth to asymmetric, linear growth.40 The asymmetric growth kinetics are explained by the adoption of a senescence-like growth arrest of one of the daughters resulting from divisions subsequent to the induction of p53.40 This outcome would be accounted for by our model of DNA damage inheritance, which predicts the birth of a “ mortal” – here senescent – daughter during each cell division. As illustrated by these studies, it may be mortality, not immortality, that defines the cellular outcome of non-random chromosome segregation.
What is the mechanism by which p53 activation brings about non-random chromosome segregation and asymmetric cell-cycle kinetics? A possibility was suggested by studies in which it was discovered that treatment of p53-expressing cells with purine, but not pyrimidine, nucleotide precursors (hypoxanthine or xanthosine) could reverse p53-dependent asymmetric cell kinetics.41 The effect of p53 on asymmetric cell division could therefore be linked to its role in purine biosynthesis. Additional insight was gained from analysis of the relative rate of production of guanine and adenine bases before and after induction of p53 expression. The observation that the ratio of the rate of production of guanine bases to the rate of production of adenine bases decreased upon p53 induction further pinpointed the biosynthetic defect of p53-over-expressing cells to the guanine synthesis branch of the pathway.41 The guanine synthesis branch of the purine nucleotide synthesis pathway consists of two steps: inosine 5′-monophosphate (IMP) is first converted by inosine 5′-monophosphate dehydrogenase to xanthosine 5′-monophosphate (XMP), which is then converted to guanosine 5′-monophosphate (GMP) by guanine 5′-monophosphate synthetase. In order to understand which step of the pathway is regulated by p53, the flow of radioactively labeled inosine from IMP into XMP and then into GMP was studied. Since these studies showed that labeled inosine accumulated in IMP, and not XMP or GMP, it was concluded that p53 regulated cellgrowth kinetics by suppression of IMP-dehydrogenase, the rate-limiting enzyme in guanine nucleotide biosynthesis. Subsequent studies provided evidence that activation of p53 suppressed transcription of the gene encoding IMP-dehydrogenase, and that p53-dependent changes in cell-cycle dynamics could be negated by over-expression of IMP-dehydrogenase. More recently, studies of the effects of modulating purine biosynthetic pathways on various somatic progenitor cell populations have shown that these manipulations can be used to control the kinetics of stem cell division and, in some cases, non-random chromosome segregation in diverse cell-types.21,42 An interesting aim for future study will be to understand how manipulations of purine biosynthesis in cultured stem and progenitor cells relates to the functional biology of these cells in vivo.
The question of how nucleotide biosynthesis could relate to non-random chromosome segregation brings us back to the model proposed here. As indicated earlier, a critical element of the model is that DNA replication stress produces replication-dependent DNA damage that yields an asymmetry in the amount of DNA damage on sister chromosomes. A well-studied cause of DNA replication stress is the lack of availability of the ingredients of DNA synthesis, including nucleotides.4
Connecting the experimental data to our model, then, we propose that the regulation of non-random chromosome segregation by p53 and IMP-dehydrogenase is related to the availability of nucleotides for DNA synthesis, which in turn determines the efficiency of DNA replication and the amount of replication stress. Consistent with this idea, the p53- stabilizing drug Nutlin-3 can induce DNA breaks in cultured cells and this damage seems to be generated during S-phase 43, albeit by an unknown mechanism. Future studies could be designed to test the hypothesis that p53-dependent non-random chromosome segregation is related to the induction of genotoxic replication stress. Inducible p53- expressing cells would thus be valuable tools with which to study non-random chromosome segregation as it relates to DNA damage.
It is important to note that in skeletal muscle stem cells, the absence of p53 does not alter the frequency with which cells segregate DNA non-randomly.20 These data indicate that p53 is not absolutely required for non-random chromosome segregation and suggest that the replication stress leading to non-random chromosome segregation in muscle stem cells is not generated by the activity of p53. Still, it would be interesting to test whether the activation of p53 alters the frequency with which muscle stem cells segregate template DNA in a biased manner. We hypothesize that p53 is not required for non-random chromosome segregation, but can induce mitotic asymmetries secondary to its induction of DNA replication stress or other DNA damage responses.
3.4 Regulation of asymmetric cell-fate determination by p53
p53’s connection to non-random chromosome segregation becomes more interesting, and complex, when one considers more recent studies linking p53 to asymmetric cell fate determination. In these studies, ErbB2-expressing mouse mammary cancer stem cells were observed to divide symmetrically with greater frequency than their wild-type, non-cancerous counterparts.28 Activation of p53 in cancer stem cells (using the Mdm2 inhibitor Nutlin-3) was sufficient to enhance the frequency of asymmetric cell division and thus limit the growth of the cancer. Furthermore, the symmetry of mammary cancer stem cell divisions was monitored by analyzing the distribution of the fate determinant Numb, the asymmetric distribution of which has been previously correlated with non-random chromosome segregation and asymmetric cell divisions in muscle stem cells.20,44 Remarkably, p53 influenced the frequency with which cancer stem cells divided asymmetrically, as evidenced by an increase in the frequency with which these cell populations segregated Numb in a polarized fashion.28
Numb is particularly interesting in the context of non-random chromosome segregation not only because its asymmetric distribution correlates with that of template DNA, but also because previous studies have characterized Numb as a tumor suppressor that functions by stabilizing p53.45 Classically, Numb is associated with the inhibition of Notch signaling due to its ability to ubiquitinate the intracellular domain of the activated Notch receptor46 or to direct the endocytosis of the Notch receptor.47 In its tumor suppressor role, Numb stabilizes p53 by preventing the Mdm2-dependent ubiquitination and subsequent degradation of p53.45
What, then, might be the role of Numb in non-random chromosome segregation? One possibility is that the inheritance of Numb is coordinated with the inheritance of chromosomes containing newer template DNA. This would enable both the asymmetric stabilization of p53 and the localized inhibition of Notch signaling within the daughter cell inheriting damaged DNA. In this way, Numb would account for the cell-cycle arrest, DNA damage repair, and directed differentiation of the damage-inheriting daughter predicted by our model. To test this hypothesis, it will be important in future studies to clarify whether Numb co-segregates with newer or older template DNA, which is difficult to ascertain from nucleotide analog pulse-chase studies. Furthermore, with the advancement of techniques to analyze cell fate in conjunction with non-random chromosome segregation, it will be interesting to study how Numb loss-of-function alters the fate of asymmetrically dividing cells.
4. Conclusion
Non-random chromosome segregation is a phenomenon that has fascinated cell biologists since it was first observed. Much of this fascination stems from the mysteries that surround it, namely, how it occurs and what role it plays. Solving these mysteries has proven difficult, at least in part, because we lack even a framework for understanding how a cell might recognize and partition chromosomes according to the relative age of template DNA strands. Although testable models like the one presented here may help to guide future work, advances in our understanding of non-random chromosome segregation will likely come only with the advent of new techniques for isolating, analyzing, and manipulating stem and progenitor cell populations.
Because non-random chromosome segregation may be a unique characteristic of stem cells, or even a subset of stem cells, the development of methods to identify and purify these cells will be key to future work. At the very least, the ability to prospectively isolate populations of cells that exhibit non-random chromosome segregation will improve the efficiency of experiments. One can also envision unbiased assays of gene and protein expression in prospectively isolated cell populations, which may help to identify factors important for the mechanism of non-random chromosome segregation. In the past, non-random chromosome segregation has been associated with subpopulations of label-retaining cells.20,37 Recently, the development of mice carrying inducible alleles encoding fluorescently tagged histone proteins has enabled the prospective isolation of label-retaining cells from various tissues.48 Transient expression of the tagged histone followed by a period in which the histone label is chased enables identification of label-retaining cells by flow cytometry or microscopy. To the extent that non-random chromosome segregation occurs in label-retaining cells, these transgenic mice may be used to purify and study the rare cells that exhibit the phenomenon. A variation of this technique, in which a histone protein is fused not only to a fluorescent protein, but also to a portion of Cre recombinase, might even allow for genetic manipulation of the subpopulations of interest in vivo.49
A second technique that will be important for advancing our understanding of non-random chromosome segregation is live-cell videomicroscopy. As mentioned above, one important application of this technology would be in tracing the fate of cells exhibiting non-random chromosome segregation in real-time. Such studies would provide critical insight into the identity of cells that segregate DNA non-randomly. These studies might also provide data relating to the significance of the asymmetric segregation by allowing observation of the behavior of cells before and after asymmetric cell divisions. The ability to observe non-random chromosome segregation in living cells would also provide a new means of studying its mechanism. By analogy, videomicroscopy of living cells has been a critical tool in recent efforts to discern the mechanisms of asymmetric segregation of protein aggregates in budding yeast.50
Regarding the mechanism proposed here, live-cell imaging could be used to test the hypothesis that asymmetries in mitotic spindle assembly underlie the biased association of one of the spindle poles with one of the sets of sister chromatids. The development of techniques for live-imaging non-random chromosome segregation would benefit from the identification of traceable chromatin-associated proteins that also segregate in a biased manner and from the development of new nucleotide analogs that can be delivered to and imaged in living cells. With these and other technical breakthroughs on the horizon, and many mysteries to solve, studies of non-random chromosome segregation promise to yield exciting and important insights in the years to come.
Highlights.
Little is known about the mechanism of non-random chromosome segregation
We propose a new model of non-random chromosome segregation
In our model, DNA damage due to replication stress is segregated asymmetrically
Unequal levels of DNA damage on sister chromatids leads to their biased segregation
Tests of our
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 2.Rando TA. The immortal strand hypothesis: segregation and reconstruction. Cell. 2007;129:1239–1243. doi: 10.1016/j.cell.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 3.Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 4.Burhans WC, Weinberger M. DNA replication stress, genome instability and aging. Nucl Acids Res. 2007;35:7545–7556. doi: 10.1093/nar/gkm1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 6.Kuzminov A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. PNAS. 2001;98:8241–8246. doi: 10.1073/pnas.131009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price BD, D’Andrea AD. Chromatin Remodeling at DNA Double-Strand Breaks. Cell. 2013;152:1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 9.Lew DJ, Burke DJ, Dutta A. The immortal strand hypothesis: how could it work? Cell. 2008;133:21–23. doi: 10.1016/j.cell.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 10.Dotiwala F, Harrison JC, Jain S, Sugawara N, Haber JE. Mad2 prolongs DNA damage checkpoint arrest caused by a double-strand break via a centromere-dependent mechanism. Curr Biol. 2010;20:328–332. doi: 10.1016/j.cub.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iacovoni JS, et al. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. The EMBO Journal. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meier A, et al. Spreading of mammalian DNA-damage response factors studied by ChIP-chip at damaged telomeres. EMBO J. 2007;26:2707–2718. doi: 10.1038/sj.emboj.7601719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tajbakhsh S, Gonzalez C. Biased segregation of DNA and centrosomes: moving together or drifting apart? Nat Rev Mol Cell Biol. 2009;10:804–810. doi: 10.1038/nrm2784. [DOI] [PubMed] [Google Scholar]
- 17.Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rebollo E, et al. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467– 474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 19.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shinin V, Gayraud-Morel B, Gomès D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 21.Huh YH, King J, Cohen J, Sherley JL. SACK-expanded hair follicle stem cells display asymmetric nuclear Lgr5 expression with non-random sister chromatid segregation. Sci Rep. 2011;1:176. doi: 10.1038/srep00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karpowicz P, et al. Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro. J Cell Biol. 2005;170:721–732. doi: 10.1083/jcb.200502073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karpowicz P, et al. The germline stem cells of Drosophila melanogaster partition DNA non-randomly. Eur J Cell Biol. 2009;88:397–408. doi: 10.1016/j.ejcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, et al. A differentiation checkpoint limits hematopoietic stem cell selfrenewal in response to DNA damage. Cell. 2012;148:1001–1014. doi: 10.1016/j.cell.2012.01.040. [DOI] [PubMed] [Google Scholar]
- 25.Inomata K, et al. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 2009;137:1088–1099. doi: 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
- 26.Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krystyniak A, Garcia-Echeverria C, Prigent C, Ferrari S. Inhibition of Aurora A in response to DNA damage. Oncogene. 2006;25:338–348. doi: 10.1038/sj.onc.1209056. [DOI] [PubMed] [Google Scholar]
- 28.Cicalese A, et al. The Tumor Suppressor p53 Regulates Polarity of Self- Renewing Divisions in Mammary Stem Cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 29.Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 30.Charville GW, Rando TA. Stem cell ageing and non-random chromosome segregation. Philos Trans R Soc Lond, B, Biol Sci. 2011;366:85– 93. doi: 10.1098/rstb.2010.0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart EJ, Madden R, Paul G, Taddei F. Aging and Death in an Organism That Reproduces by Morphologically Symmetric Division. PLoS Biol. 2005;3:e45. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. doi: 10.1073/pnas.0708931105. at < http://www.pnas.org>. [DOI] [PMC free article] [PubMed]
- 33.Fuentealba LC, Eivers E, Geissert D, Taelman V, Robertis EMD. Asymmetric mitosis: Unequal segregation of proteins destined for degradation. PNAS. 2008;105:7732–7737. doi: 10.1073/pnas.0803027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wigley WC, et al. Dynamic Association of Proteasomal Machinery with the Centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sotiropoulou PA, Candi A, Blanpain C. The majority of multipotent epidermal stem cells do not protect their genome by asymmetrical chromosome segregation. Stem Cells. 2008;26:2964–2973. doi: 10.1634/stemcells.2008-0634. [DOI] [PubMed] [Google Scholar]
- 36.Kiel MJ, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith GH. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development. 2005;132:681–687. doi: 10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- 38.Pine SR, Ryan BM, Varticovski L, Robles AI, Harris CC. Microenvironmental modulation of asymmetric cell division in human lung cancer cells. Proc Natl Acad Sci USA. 2010;107:2195–2200. doi: 10.1073/pnas.0909390107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rambhatla L, et al. Cellular Senescence: Ex Vivo p53-Dependent Asymmetric Cell Kinetics. J Biomed Biotechnol. 2001;1:28–37. doi: 10.1155/S1110724301000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherley JL. Asymmetric cell kinetics genes: the key to expansion of adult stem cells in culture. Stem Cells. 2002;20:561–572. doi: 10.1634/stemcells.20-6-561. [DOI] [PubMed] [Google Scholar]
- 41.Sherley JL. Guanine nucleotide biosynthesis is regulated by the cellular p53 concentration. J Biol Chem. 1991;266:24815–24828. [PubMed] [Google Scholar]
- 42.Paré JF, Sherley JL. Culture environment-induced pluripotency of SACK-expanded tissue stem cells. J Biomed Biotechnol. 2011;2011:312457. doi: 10.1155/2011/312457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verma R, Rigatti MJ, Belinsky GS, Godman CA, Giardina C. DNA damage response to the Mdm2 inhibitor nutlin-3. Biochem Pharmacol. 2010;79:565–574. doi: 10.1016/j.bcp.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 45.Colaluca IN, et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 46.McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- 47.Berdnik D, Török T, González-Gaitán M, Knoblich JA. The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev Cell. 2002;3:221–231. doi: 10.1016/s1534-5807(02)00215-0. [DOI] [PubMed] [Google Scholar]
- 48.Fuchs E, Horsley V. Ferreting out stem cells from their niches. Nat Cell Biol. 2011;13:513–518. doi: 10.1038/ncb0511-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buczacki SJA, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 50.Liu B, et al. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140:257–267. doi: 10.1016/j.cell.2009.12.031. [DOI] [PubMed] [Google Scholar]