Abstract
Background
Cortical inhibition plays a critical role in controlling and modulating cortical excitation and a more detailed understanding of the neuronal circuits contributing to each will provide more insight into their roles in complex cortical computations. Traditional neuronal tracers lack a means for easily distinguishing between circuits of inhibitory and excitatory neurons. To overcome this limitation, we developed a technique for retrogradely labeling inputs to local clusters of inhibitory or excitatory neurons, but not both, using neurotropic adeno-associated and lentiviral vectors, cell-type specific promoters and a modified rabies virus.
Results
Applied to primary visual cortex (V1) in mouse, the cell-type specific tracing technique labeled thousands of presynaptically connected neurons, and revealed that the dominant source of input to inhibitory and excitatory neurons is local in origin. Neurons in other visual areas are also labeled; the percentage of these inter-cortical inputs to excitatory neurons is somewhat higher (~20%) than to inhibitory neurons (<10%), suggesting that inter-cortical connections have less direct control over inhibition. The inputs to inhibitory neurons were also traced in cat V1, and when aligned with the orientation preference map, revealed for the first time that long-range inputs to inhibitory neurons are well tuned to orientation.
Conclusions
These novel findings for inhibitory and excitatory circuits in the visual cortex demonstrate the efficacy of our new technique and its ability to work across species, including larger-brained mammals such as the cat. This paves the way for better understanding the roles of specific cell-types in higher-order perceptual and cognitive processes.
Keywords: neural circuits, cell type specificity, adeno-associated virus, lentivirus, inhibitory neurons, rabies virus, glycoprotein, genetic targeting
The neocortex is comprised of a dense population of excitatory neurons that are balanced by a diverse and more sparsely distributed array of inhibitory neurons [1–4]. Interactions between local inhibitory and excitatory neurons provide gain control that works under myriad conditions throughout the cortex. In primary visual cortex (V1), for example, inhibition is essential for several basic functional characteristics, including a neuron’s preference for stimulus contrast, size, and orientation (i.e., [5–7]). Although inhibitory neurons serve to control excitation, it is not known whether local populations of inhibitory and excitatory cell-types are mediated through the same set of cortical circuits, and whether the functional selectivity of these circuits differs. This is largely due to technical limitations, since inhibitory and excitatory neurons are intermingled and the study of one population over the other is difficult to achieve. Circuits related to specific cell-types have been identified in part using various neurophysiological techniques in brain slice preparations [8–10] and in vivo [11, 12], yet these approaches are unable to provide a full-scale pattern of connectivity throughout the intact brain. On the other hand, traditional neuronal tracers can reveal widespread connection patterns, yet are unable to discriminate between inputs to inhibitory and excitatory neurons unless the ultrastructure of synapses are examined under electron microscopy [13–15] where only a fraction of the connections can be studied.
To overcome these limitations, we developed a technique for labeling presynaptic inputs to inhibitory or excitatory neurons that takes advantage of the neurotropism of recombinant viral vectors, cell-type specific promoters, and a modified rabies virus (RV). The modified rabies virus can be made to retrogradely trace the direct presynaptic inputs to a particular population of neurons through delivery of two key genes [16]. The most successful approaches thus far have used transgenic mouse lines to target and enhance the necessary gene expression [17–20]. However, key to our new technique is the design of two helper-viruses that enable cell-type specific gene delivery without relying on transgenic animals.
Tests in V1of wild-type mice confirmed the cell-type specificity of gene transduction of our helper-viruses and established the efficiency of modified rabies virus infection and retrograde spread. At the same time, these experiments allowed unprecedented comparison of wide-scale input patterns to inhibitory and excitatory V1 neurons. The technique was also used to target inhibitory neurons in cat V1, where, in combination with intrinsic signal optical imaging to reveal the orientation preference map we show for the first time that orientation preference of long-range lateral inputs to inhibitory neurons is well tuned. The technique’s ability to work in non-transgenic animals provides the first opportunity to explore cell-type specific connections in neocortex of higher-order mammals, paving the way for better understanding the roles of excitatory and inhibitory neurons in complex perceptual and cognitive processes.
RESULTS
Design of the Cell-Type Specific Tracing Technique
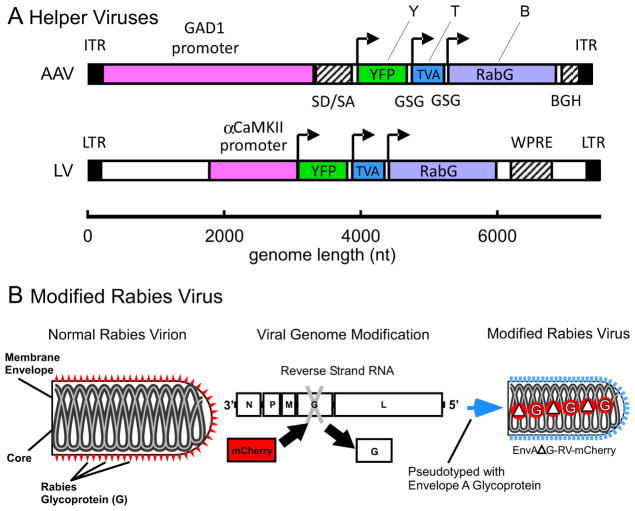
Our new method was developed to label the wide spread pre-synaptic inputs of local clusters of inhibitory or excitatory neurons in neocortex of any mammalian species. To do this, we designed two helper-viruses (Fig. 1A) to optimize for cell-type specific transduction of the three-gene construct, YTB [21], that allows for expression of two key genes, TVA (T) and RabG (B for the B-19 strain of rabies virus), necessary for initial infection and retrograde spread of the modified rabies virus (RV), as well as a third reporter gene, YFP, (Y). A recombinant adenos-associated virus (AAV) was constructed to express YTB specifically in inhibitory neurons under the control of a 3.1kb fragment of the GAD1 promoter (Fig. 1A, top); whereas a lentivirus (LV) (Fig. 1A, bottom) with the αCaMKII promoter [22, 23] was used to transduce excitatory cortical neurons with YTB.
Figure 1. Design of the Cell-Type Specific Vectors and Modified Rabies Virus.
(A) Schematic representation of the transfer vectors for recombinant adeno-associated virus, AAV/GAD1/YTB (top), and lentivirus, LV/αCaMKII/YTB (bottom). Each vector includes a cell-type specific promoter (magenta), GAD1 (glutamic acid decarboxylase 1) or αCaMKII (α-calcium/calmodulin-dependent protein kinase II), cis-regulatory sequences (hatched; SD/SA = splice donor and acceptor sequence from human β-globin, BGH = bovine growth hormone polyadenylation signal; WPRE = woodchuck hepatitis virus posttranscriptional regulatory element), and the YTB transgene. YTB codes for three gene products using 2A peptide mediated cleavage [52]: the yellow fluorescent protein reporter (YFP, or Y; yellow), the artificial TVA 950 receptor (TVA, or T; blue), and the glycoprotein from the B-19 strain of rabies (RabG, or B; purple). Arrows indicate the start codons for each gene. Inverted terminal repeats (ITR) and long terminal repeats (LTR) are indicated in black. (B) The rabies virus (RV) has been modified in two ways based on [16]: the glycoprotein gene (G) was deleted (ΔG) and replaced with the gene for mCherry, creating, ΔG-RV-mCherry; and it was subsequently pseudotyped with the envelope A glycoprotein, creating EnvA-ΔG-RV-mCherry.
The modified RV, EnvA-ΔG-RV (Fig. 1B), has been pseudotyped with the envelope-A glycoprotein and can thus only infect cells containing the receptor, TVA [16]. Normally, TVA is only expressed in avian cells [24]. Yet, through selective delivery of the TVA gene to inhibitory or excitatory mammalian neurons we can target EnvA-ΔG-RV infection (Fig. 2). As indicated in Figure 2CD, EnvA infection occurs at the cell body where the TVA receptor becomes embedded. Furthermore, EnvA-ΔG-RV has been genetically modified so that the negative RNA strand coding for RabG has been removed (ΔG) and replaced with the sequence coding for mCherry (Fig. 1B). Without RabG, newly produced ΔG-RV virions in the starter cells cannot retrogradely spread and infect other neurons. The critical component for retrograde tracing using EnvA-ΔG-RV, therefore, is the delivery beforehand of RabG via our helper-viruses into starter cells (left panels in Fig. 2), enabling virions to spread from the host neuron to infect presynaptic terminals and be retrogradely transported to the soma (Fig. 2CE). The ΔG-RV cannot spread additional synapses because the RabG gene will not be available in presynaptically connected neurons, only starter cells [16]. Importantly, the helper-viruses express YFP and EnvA-ΔG-RV expresses mCherry, so that initially infected starter cells appear doubled-labeled with both YFP and mCherry, whereas presynaptically connected neurons can be distinguished as being labeled by mCherry-only (see right panels in Fig. 2).
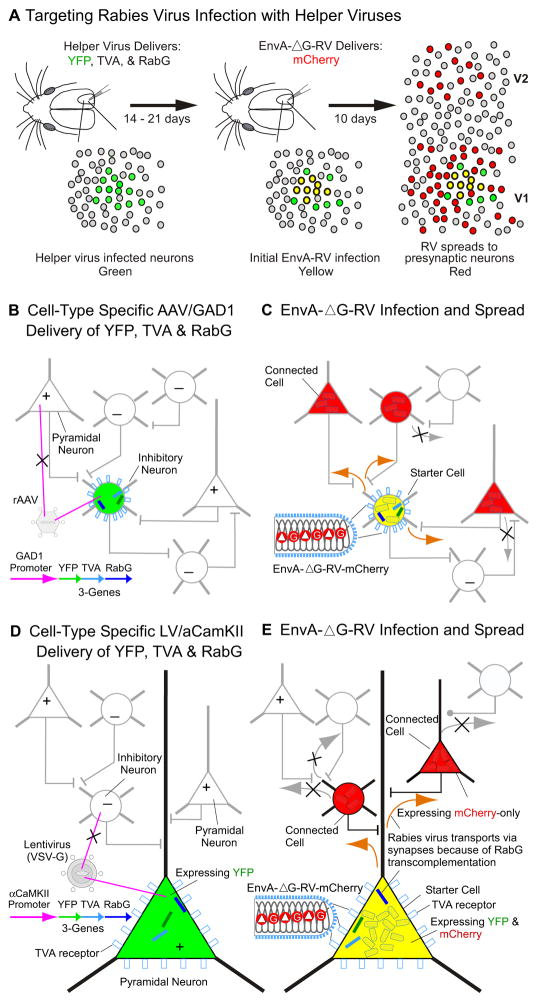
Figure 2. Schematic for Targeting Rabies Virus to Trace the Inputs to Inhibitory or Excitatory Neurons.
A simplified diagram of our tracing technique is shown in (A) and described in more detail in (B–E). In the left panel of (A), helper-virus is used to deliver three genes (YFP, TVA, and RabG; see Fig. 1A) to a local population of neurons in mouse V1 at the site of injection. As shown in (B) and (D) the helper-viruses infect directly through the cell soma (magenta lines). The cell-type specificity of gene expression depends on which helper-virus is used; AAV/GAD67/YTB (B) and LV/αCaMKII/YTB (D) vectors are designed to introduce the genes to inhibitory or excitatory neurons, respectively. The three gene products are produced by the single cell-type specific promoter (GAD1 or αCaMKII) using 2A peptide mediated cleavage [52]. Helper-virus infected neurons are depicted as green due to YFP expression. Following a 14–21 day survival period to allow for sufficient gene expression, the modified rabies virus, EnvA-ΔG-RV-mCherry (Fig. 1B), is injected in the same V1 location (middle panel in A). The TVA receptor now embedded in the membrane of the cell soma (blue rectangles on cell bodies in BE) enables direct EnvA infection at the soma as indicated in (C and E). These EnvA-ΔG-RV-mCherry superinfected neurons are depicted as yellow because they will now begin to express mCherry as well as YFP. Because RabG is also co-expressed via the helper-virus, over the next 10 days rabies virions produced in starter cells will incorporate RabG and spread retrogradely from the yellow ‘starter cells’ via the synaptic terminals of presynaptically connected neurons (orange arrows in C and E). Resulting ‘connected cells’ which can be found locally within V1 and at longer-range in other brain regions such as V2 (right panel in A) are depicted as red because they will only express mCherry delivered by the rabies virus. Black crosses (x) in (C and E) illustrate that the ΔG-RV virus cannot spread beyond these directly connected cells because they were not transduced with RabG.
Cell-type Specificity of the AAV/GAD1/YTB and LV/α CaMKII/YTB Helper-Viruses
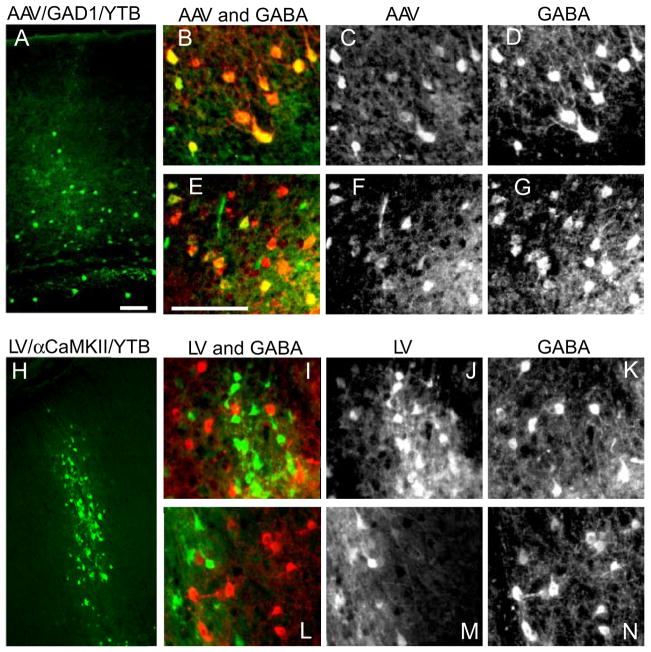
Following helper-virus production, we next determined whether each would lead to detectable levels of transgene expression and whether expression would be cell-type specific. Two weeks after injections of each helper-virus, histological examination revealed strong native YFP expression (Fig. 3). AAV/GAD1/YTB yielded a more sparsely distributed array of cells (Figs. 3ACF, S1A) characteristic of cortical inhibitory neurons [1, 3]. Importantly, every YFP positive cell (Fig. 3C and F) was also positive for the inhibitory neuron marker, GABA (Fig. 3D and G), verifying the selectivity of this helper virus (also see Fig. S1B-D).
Figure 3. Cell-type Specificity of Helper-Virus Transduction in Mouse Neocortex.
Low magnification images show expression of YFP (green) in cortical neurons following AAV/GAD1/YTB (A; AAV) and LV/αCaMKII/YTB (H; LV) helper-virus injections in mouse cortex. Higher magnified black and white images of AAV infected neurons (C and F) co-localize with neurons labeled through GABA immunofluorescence (D and G), as can be seen when shown together in color (B and E; AAV is shown in green and GABA in red). In contrast, higher magnified black and white images of LV infected neurons (J and M) do not co-localize with neurons positive for GABA immunofluorescence (K and N), as can be seen when shown together in color (I and L; LV is shown in green and GABA in red). Scale bars = 100 μm. See also Fig. S1.
YFP expression from LV/αCaMKII/YTB (green in Fig. 3I and L), in contrast, was distinctly absent from GABA immuno-positive neurons (red in Fig. 3I and L), verifying excitatory neuron selectivity of this helper-virus. Moreover, LV transduced neurons formed a denser cluster along the injection track (Fig. 3H) consistent with excitatory neurons comprising 80% of the neuronal population in neocortex [1, 3].
Cell-Type Specific Infection and Spread of EnvA-Δ G-RV-mCherry
Tracing Pre-synaptic Inputs to Inhibitory Neurons
Having established the cell-type specificity of our helper-viruses, we next tested whether expression of TVA and RabG is sufficient to allow for EnvA-ΔG-RV-mCherry infection and monosynaptic retrograde spread (see schematics in Fig. 2). Data from three mice using AAV/GAD1/YTB and EnvA-ΔG-RV-mCherry are presented in Fig. 4. In one case (M11–16; Fig. 4A–G), a relatively large, 1.0 μl injection of AAV/GAD1/YTB was made into a single location in mouse V1 and followed 3 weeks later by a single 0.4 μl injection of EnvA-ΔG-RV-mCherry to within ~300 μm of the same location (Fig. 4A). In the two other cases, smaller AAV injections of 0.3 μl (M12–17) and 0.5 μl (M11–20) were made, also followed 3 weeks later by 0.4 μl injections of EnvA-ΔG-RV-mCherry (Fig. 4H; Fig. S2A–C). After post-EnvA injection survival times of 8–14 days, we found that AAV infection led to robust YFP expression (i.e., Fig. 4A) in a relatively sparse array consistent with the distribution of inhibitory neurons (see close up images in Figs. 4CFI and S2F). GABA immunofluorescence was also used to confirm inhibitory cell specificity in every fourth section, as already demonstrated (Fig. 3A–G). In addition, in one case a series of sections were processed for the calcium binding protein parvalbumin (PV), a marker of a large subpopulation of inhibitory cells, showing a high, though not exclusive overlap (Fig. S2E–G). AAV spread as observed through YFP expression was restricted to within ~500 μm for the two smaller injections (see examples in Fig 4I and Fig. S2B), but extended more than 2 mm in superficial layers 1–3 for the larger injection (Fig. 4A).
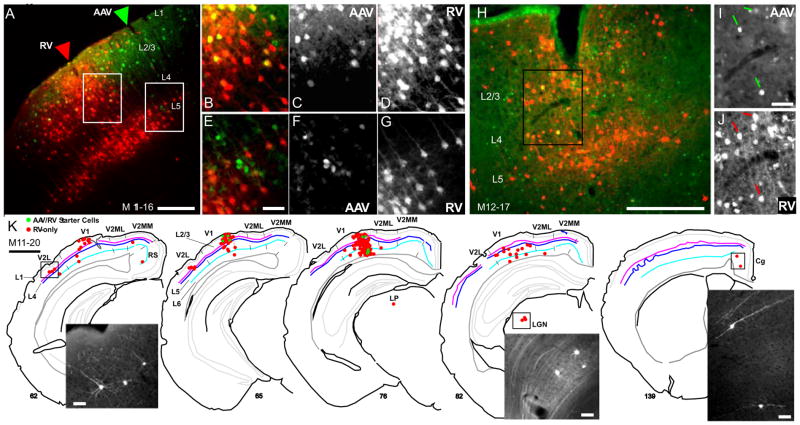
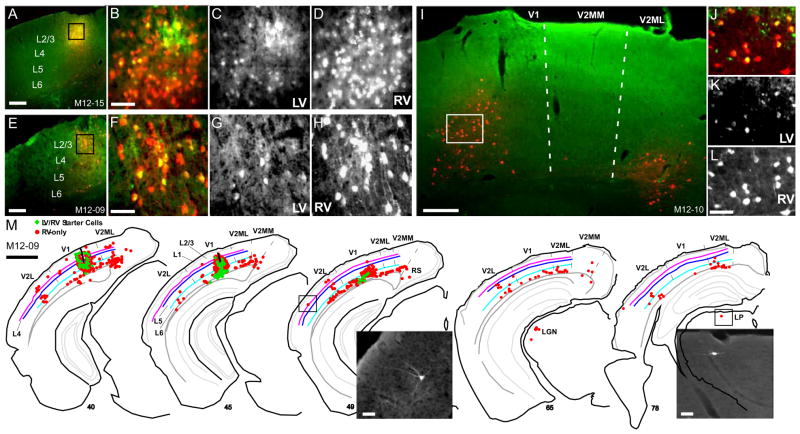
Figure 4. Targeting EnvA-Δ G Rabies Virus (RV) infection to and Monosynaptic Retrograde Spread from Inhibitory Neurons in Mouse V1 using the AAV/GAD1/YTB Vector.
The local pattern of RV infected inputs to AAV infected inhibitory neurons is shown in coronal sections through V1 in three mice, M11–16 (A–G), M12–17 (H–J) and M11–20 (K). In (A), the AAV and EnvA-ΔG-RV-mCherry (RV) injection sites are indicated by green and red arrow heads, respectively. In (H), injection sites overlap. Starter cells appear yellow due to transduction of the cell first with AAV leading to YFP expression (green) and subsequent infection by EnvA-ΔG-RV-mCherry due to the presence of the TVA receptor, leading to mCherry expression (red; see schematic in Fig. 2BC). Neurons presynaptically connected to starter cells that were retrogradely infected by RV following RabG complementation within starter cells are identified by expression of mCherry-only (red; see Fig. 2C). Higher magnified images of the regions outlined by white and black rectangles in (A and H) are shown in (B–G, and IJ). AAV infected neurons expressing YFP (Green; C, F and I) and RV infected neurons expressing mCherry (red; D, G and J) are shown together in (B and E) and separately in (C, F and I; AAV) and (D, G and J; RV). (K) A representative reconstruction of the brain wide pattern of RV infected neurons (mCherry-expressing; red) providing direct inputs to inhibitory starter cells (green) found in layers 1–6 of V1 is shown for mouse M11–20. The AAV and RV injection sites are located between sections 65 and 76 (see Fig. S2A–C). Sections are ordered from posterior to anterior. Inset images show mCherry expression in long range connected neurons in V2L, LGN, and cingulate cortex (cg). For all sections in (A–K), left is lateral, up is dorsal. Scale bars = 200 μm in (A and H), 50 μm in (E, I and K inset), and 1mm in (K). See also Fig. S2.
Critically, in all 3 cases, injections of EnvA-ΔG-RV-mCherry led to a hundred or more AAV infected neurons (YFP-positive) co-expressing mCherry (see starter cells in Table 1). This confirms sufficient AAV expression of the TVA receptor since EnvA infection cannot occur in its absence. The observed EnvA superinfection rate of TVA-expressing neurons was high, over 75%, particularly in regions where the EnvA and AAV injection sites were highly overlapping (see examples in Fig. 4B–D, I and J and Fig. S2A–C). In this regard, despite the variability in spread of AAV/GAD1/YTB infected neurons, double-labeled starter cells were confined to within a 250 μm radius of the EnvA-ΔG-RV-mCherry injection track (red arrow in Figs. 4A and S2A), likely due to the more limited extra-cellular spread of EnvA-ΔG-RV-mCherry.
Table 1.
Numbers of EnvA-ΔG-RV Infected Starter Cells in V1 and Presynaptically Connected Cells Located in V1 (Intrinsic V1), Higher Visual Cortex (Feedback), Cortex in the Opposite Hemisphere (Callosal), LGN, LP, Other Thalamic (OT) and Hypothalamic (HT) nuclei, and Cingulate Cortex (Cg). See also Fig. S4.
| Vector | Species | Case# | Starters | Connected | V1 | Feedback | Callosal | RS | Cg | LGN | LP | OT | HT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAV | Mouse | M11–16 | 537 | 7851 | 7620 | 193 (2.5%) | 6 | 8 | 0 | 16 | 6 | 2 | 0 |
| M11–20 | 70 | 1752 | 1510 | 148 (8.4%) | 10 | 14 | 8 | 22 | 9 | 8 | 23 | ||
| M12–17 | 390 | 9359 | 8433 | 667 (7.9%) | 4 | 244 | 0 | 8 | 0 | 2 | 1 | ||
| LV | Mouse | M12–09 | 404 | 4647 | 3225 | 1057 (23%) | 57 | 8 | 1 | 7 | 3 | 7 | 0 |
| M12–10 | 300 | 3727 | 2140 | 1314 (35%) | 13 | 268 | 4 | 0 | 0 | 0 | 0 | ||
| M12–15 | 1032 | 21358 | 15749 | 5029 (24%) | 344 | 221 | 3 | 4 | 1 | 6 | 1 | ||
| ΔG-RV | Mouse | M12–13mCh | NA | 3456 | 3315 | 756 (22%) | 40 | 6 | 12 | 3 | 2 | 1 | 2 |
| M12–13GFP | NA | 4188 | 3102 | 832 (20%) | 24 | 220 | 2 | 3 | 1 | 1 | 3 | ||
| M12–21GFP | NA | 6919 | 5367 | 1274(18%) | 21 | 95 | 26 | 52 | 19 | 13 | 51 | ||
| M12–22GFP | NA | 1975 | 1755 | 181 (9.2%) | 2 | 4 | 4 | 16 | 2 | 3 | 8 | ||
| AAV | Cat | C12–02R | 336 | 9174 | 8916 | 258 (2.8%) | NA | NA | NA | NA | NA | NA | NA |
| C12–02L | 12 | 4149 | 3945 | 192 (2.2%) | NA | NA | 0 | 0 | 12 | 0 | 0 |
Finally, several thousand pre-synaptically connected neurons expressing mCherry-only were identified locally within V1 (Fig. 4AH) and throughout the brain (Fig. 4K), confirming RabG expression was sufficient enough to allow for retrograde spread of ΔG-RV from starter cells. Counts show that the vast majority of connected neurons are within V1, ranging from 86–97% (Table 1). Feedback-like inputs from neurons in higher visual areas (V2L, V2ML, and V2M) were far fewer in number, but represented the next largest source to V1 inhibitory neurons, ranging from 2.5–8.4% (Table 1; left image in Fig. 4K, and Fig. S2D). Inputs from the dorsal lateral geniculate nucleus (LGN; Fig. 4K, center inset), lateral posterior nucleus (LP), retrosplenial cortex (RS), cingulate cortex (Cg; Fig. 4K, right inset), hypothalamus (HT), and the opposite cortical hemisphere made up the remainder of the mCherry-labeled inputs (Table 1). These results demonstrate that our AAV EnvA vector mediated technique is effective at labeling the inputs to GABAergic neurons. Moreover, the sensitivity of this technique reveals that the dominant source of inputs to V1 inhibitory cells is local in origin, with only a small percentage coming from higher visual areas and subcortical thalamic nuclei.
Tracing Pre-synaptic Inputs to Excitatory Neurons
We next evaluated the efficacy of EnvA-ΔG-RV-mCherry complementation in neurons transduced by the LV/αCaMKII/YTB vector. In three mice, LV/αCaMKII/YTB injection volumes of 1.0 μl (M12–15, Fig. 5A–D) and 0.5 μl (M12–09, Fig. 5E-H; M12–10, Fig. 5I-L) were used, and followed three weeks later by a 0.4 μl injection of EnvA-ΔG-RV-mCherry at or near the same location. After an additional 11–14 day survival time, TVA expression was confirmed as all three cases exhibited high rate of superinfected starter cells (>75%; see examples in Fig. 5A-L). In addition, thousands of pre-synaptically connected neurons expressing mCherry-only were also found indicating adequate expression of RabG in starter cells (see red-only neurons in Fig. 5). As found for inhibitory starter cells, the majority of neurons projecting to excitatory starter cells were found locally within V1 (Fig. 5M), with only a small percentage in LGN, LP (Fig. 5M, right inset), RS, cg, and the opposite cortical hemisphere. However, in comparison to V1 inhibitory starter cells where only 3–8% of the inputs come from higher visual areas, the percentage to V1 excitatory neurons is higher averaging over 20% (Table 1; see examples in Fig. 5IM). While factors such as differences in the concentration of starter cells by cortical layer could affect these percentages and are not accounted for in our current analysis, the higher percentage of ‘feedback-like’ projections to excitatory neurons is nevertheless consistent with ultrastructural analysis in rat [14, 24, 25] and monkey [13, 15] showing that a greater percentage of V2 feedback-synapses are onto excitatory neurons in V1.
Figure 5. Targeting EnvA-Δ G-RV-mCherry Infection to and Retrograde Spread from Excitatory Neurons in Mouse V1 using the LV/α CaMKII/YTB Vector.
Examples of the local and long range pattern of RV infected neurons (red) providing inputs to excitatory starter neurons (yellow) are shown in coronal sections through V1 in three mice, M12–15 (A–D), M12–09 (E–H), and M12–10 (I–L). In (A, E, and I), overlapping injection sites of LV/αCaMKII/YTB (green) and EnvA-ΔG-RV-mCherry (red) are shown at low magnification; Higher magnified images of the regions outlined by black or white rectangles are shown in (B, F, and I). Double labeled starter cells (yellow in B, F, and J) co-infected by LV (C, G and K) and RV (D, H and L), express YFP from LV infection and mCherry from RV infection. (M) (K) A representative reconstruction of the brain wide pattern of RV infected neurons (mCherry-expressing; red) providing direct inputs to excitatory starter cells (green) found in layers 2–6 of V1 is shown for mouse M12–09. The LV and RV injection sites are marked by thick black lines in sections 40 and 45. Inset images show RV infected neurons in V2L (sect. 49) and the lateral posterior nucleus (sect. 78). Other conventions are as in Fig. 4. Scale bars = 200 μm in (A, E and I), 50 μm in (B, F, L and M inset), and 1 mm in (M). See also Fig. S3.
Comparison of Vector Complementation Techniques with Unpseudotyped ΔG-RV
To help evaluate the efficacy of our transfer vector mediated EnvA-ΔG-RV-mCherry tracing technique, we made small V1 injections of the unpseudotyped ΔG-RV in separate mice. The results are summarized in Table 1 and discussed in detail in the Supplemental Information (Fig. S3; see Additional Histology and Data Analysis).
Tracing Presynaptic Inputs to Inhibitory Neurons in Cat V1
We next applied our AAV helper-virus to evaluate the functional input to inhibitory neurons in cat V1 by correlating the pattern of long-range horizontal connections to the orientation preference map. Orientation preference maps are only present in highly visual mammals such as carnivores [26], tree shrews [27] and primates [28], and long-range horizontal connections across these maps have been shown to link regions, or domains, of V1 neurons preferring the same orientation. Furthermore, in cat and monkey these tuned long-range intrinsic inputs have been indirectly implicated in the orientation tuning of the suppressive surround [29–31]. The tuning of which largely depends on the ‘near’ component of the surround [29], a relatively narrow region nearest to the classical receptive field that can be accounted for by monosynaptic intrinsic connections ranging from 0.5 to 2.0 mm within carnivore and monkey V1 [32–34]. However, even though much of the evidence from intracellular recording indicates that inhibitory and excitatory inputs to cat V1 neurons are similarly tuned to orientation (i.e., [6, 7, 35]), it has not been shown directly that inhibitory neurons, which can provide the local source of suppression, receive tuned long-range horizontal inputs. To add to this uncertainty, other studies report that a subgroup of cat V1 inhibitory neurons lack orientation tuning [36, 37]. Moreover, there is mounting evidence from mouse V1, where transgenic lines allow for clearer identification of inhibitory neurons, that orientation tuning of many inhibitory neurons is much broader than for excitatory neurons (i.e., [38–40]). Thus, an examination of the orientation preference of inputs to inhibitory neurons in cat is needed.
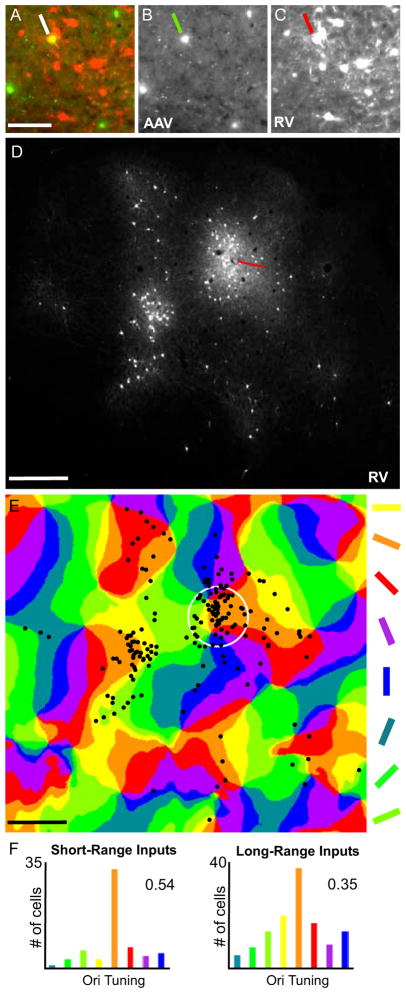
The inhibitory cell-type specificity of AAV/GAD1/YTB in cat visual cortex was first confirmed (Fig. S2) followed by demonstration that it allowed for initial infection and subsequent spread of EnvA-ΔG-RV-mCherry (Fig. S4; Table 1). Moreover, in three cats, the orientation preference map of area 17 (V1) was first obtained through intrinsic signal optical imaging and used to guide the AAV injection. For example, in the left hemisphere of C12–02 the large ‘orange’ domain near the center of the imaged V1 region was identified (white circle in Fig. 6E) and a small, ~0.4 μl, injection of AAV/GAD1/YTB was followed three weeks later by a ~0.4 μl injection of EnvA- ΔG-RV-mCherry. Resulting YFP/mCherry starter cells were confined to the domain and several hundred mCherry-only connected neurons within V1 were found locally (<250 μm radius) and at long-range (>250 μm; see Figs. 6 and S5). Notably, the pattern of connected cells in superficial cortex (<600 μm from the pial surface) was quite patchy in appearance (Fig. 6D) and when aligned with the orientation preference map (Fig. 6E), showed tuning to the preferred orientation (112.5°; orange; Fig. 6F). This observed orientation tuning preference of inputs to inhibitory neurons was found in two additional cases (Fig. S5A-H).
Figure 6. Correlating Orientation Preference of Inputs to V1 Inhibitory Neurons in Cat.
(A) As shown in the high magnification image of a superficial section of flattened V1 cortex, AAV/GAD1/YTB transduction (green) allowed for initial infection with the EnvA-ΔG-RV-mCherry creating a double-labeled starter cell (yellow; indicated by the white line) and leading to RV spread to presynaptically connected neurons (red). AAV/GAD1/YTB and EnvA-ΔG-RV-mCherry infected neurons are shown separately in (B and C). In (D), the pattern of EnvA-ΔG-RV- mCherry infected neurons is shown in the low magnification image of a superficial section of flattened V1 cortex adjacent to that shown in (A–C). A reconstruction of this pattern is overlaid on the orientation preference map in (E). The numbers of cells present in each colored domain in (E) were counted for short- (within the white circle) and long-range intrinsic distances and displayed in the histograms in (F). Corresponding orientation selectivity index (OSI) values of 0.54 and 0.35 calculated using the formula provided in Supplemental Information are also given. The red line in (D) points to the location of the EnvA-ΔG-RV-mCherry injection site. The white circle in (E) represents a 250 μm radius around the approximate center of the location of the starter cell identified in (A). Scale bars = 100 μm in (A), and 500 μm in (D and E). See also Fig. S5.
Quantitatively, the normalized and averaged orientation preference of long-range intrinsic connections for the three cases indicates that while inputs come mainly from the preferred orientation, there are substantial inputs from non-preferred orientations which leads to a moderate orientation selectivity index (OSI) of 0.25±0.12 (FigS5I, top right; for more detailed analysis see Additional Histology and Data Analysis in Suppl. Info.). Thus, if involved in the suppressive surround of V1 neurons as suggested above [29–32], such inputs should provide suppression that is tuned but not exclusive to the preferred orientation. This is what has been reported. In fact, in cat the average OSI for the suppressive-surround of V1 domain neurons is 0.26±0.19 [29], quiet similar to the OSI for our long-range inputs.
Qualitatively, the observed patchy, orientation-tuned connectivity is similar to that in cat, monkey and tree shrew that has been repeatedly revealed through traditional neuronal tracers (i.e., [7, 26–28, 41]). However, traditional tracers could not distinguish between inputs to excitatory and inhibitory neurons. Thus, our results provide the first anatomical evidence that long-range intrinsic inputs to V1 inhibitory neurons can be tuned to the preferred orientation and may explain recent results that indirectly link long-range intrinsic connections to the orientation tuning of surround suppression [29, 30].
DISCUSSION
Here we demonstrate a novel method for independent anatomical tracing of inputs to inhibitory or excitatory neurons in mammalian neocortex. The technique takes advantage of neurotropic adeno-associated and lentiviral vectors, cell-type specific promoters, and a genetically modified and pseudotyped rabies virus. Our viral vector/promoter design drove cell-type specific expression of a three-gene construct (YTB) that enabled targeted initial infection and subsequent spread of a custom-made EnvA pseudotyped and glycoprotein deleted rabies virus (EnvA-ΔG-RV) [16] that labeled thousands of presynaptically connected neurons with a fluorescent reporter (mCherry). Our AAV/GAD1/YTB vector led to cell-type specific RV tracing of inputs to inhibitory cells, whereas the LV/αCaMKII/YTB vector led to RV tracing of inputs to excitatory neurons. Since traditional neuronal tracers cannot distinguish between inputs to these two types of neocortical neurons, this new technique allows for unprecedented exploration of cell-type specific circuitry. For example, we demonstrated here how the method can be used to compare the local and long range connections of inhibitory and excitatory neurons in primary visual cortex (V1), showing that while both cell-types receive the dominant share of their inputs locally from within V1, the percentage of long-range inputs from higher visual cortex is larger for excitatory neurons, suggesting that excitation is controlled more directly through inter-cortical connections than is inhibition. Moreover, in combination with intrinsic signal optical imaging of the V1 orientation map in the cat we provide the first anatomical evidence that the orientation preference of intrinsic inputs to inhibitory neurons is well tuned.
While our demonstrations were in visual cortex, this method can be used to investigate the connections to inhibitory or excitatory neurons of any neocortical system due to the flexibility of the viral vector based delivery of YTB. Moreover, as demonstrated the cell-type specific tracing method can be applied to multiple mammalian species because our viral vector design is not cre-dependent, opening the door to animal models with more complex cortical organization that may be better suited for studying higher order perceptual and cognitive processes.
Efficacy of the Cell-Type Specific Vectors
For our technique to work it was imperative that our vector/promoter combinations selectively transduce either inhibitory or excitatory neurons, but not both; And that transgene expression driven by the promoters be high enough to visualize the transduced neurons and interact effectively with the EnvA-ΔG-RV, enabling initial infection and subsequent spread.
Cell-Type Specificity
To optimize cell-type specificity we used AAV with an inhibitory cell promoter to target inhibitory neurons and LV with an excitatory cell promoter to target excitatory neurons. In mice, inhibitory cell specificity was confirmed by showing complete overlap with GABA immunostaining, likewise, excitatory cell specificity was confirmed through an absence of GABA labeling (Fig. 3). In cat, inhibitory cell specificity was further confirmed through an absence of double labeling with the antibody for αCaMKII (Fig. S1E-G). The observed selectivity of our LV/αCaMKII/YTB vector is consistent with published reports showing that the same LV/αCaMKII vector/promoter combination leads to excitatory cell specific expression of enhanced GFP in somatosensory cortex of mouse [22] and of channelrhodopsin-2 fused to GFP in frontal cortex of monkey [23]. Furthermore, our inhibitory cell-type specific results observed for AAV/GAD1/YTB are consistent with recent work by Nathanson and colleagues [42] showing that AAV used in conjunction with a 2.6 kb fragment of the somatostatin inhibitory cell promoter leads to selective expression of GFP in inhibitory neurons of somatosensory and motor cortex in mouse, rat, and monkey.
Importantly, we chose not to use AAV with an excitatory cell promoter or LV with an inhibitory cell promoter because the same Nathanson study [42] showed such combinations led to non-cell-type specific transduction. Moreover, in a separate second study [43] they found that LV pseudotyped with VSV-G has an endogenous tropism for excitatory cortical neurons and AAV a tropism for inhibitory cortical neurons; the latter, particularly being the case when AAV titers were low. In this regard, it is worth noting that because our GAD1 promoter and YTB insert totaled ~7.4 kb (Fig. 1A, top) there is likely an overall reduction in effective titer of our AAV virus. This is because the packaging capacity of AAV is only ~5 kb [44]. Recent evidence indicates that genomes as large as 8.7kb are packaged in AAV as smaller fragments that can be reassembled following transduction into the full transgene, however, this process invariably reduces the effective titer of the virus [45].
Tracing Efficacy of the Vector Mediated Approach
Regarding efficiency of expression of the three-gene YTB construct, for both vectors YFP expression was high enough to be visualized without antibody amplification. In addition, expression of TVA, the EnvA receptor, was adequate enough to enable EnvA-ΔG-RV-mCherry infection in ~75% of YTB transduced cells. Furthermore, expression of RabG, which is necessary for ΔG-RV spread to pre-synaptically connected neurons, was also deemed adequate as several thousand connected neurons were identified all over the brain in regions known to project to V1.
With our new helper-virus based approach the great majority of RV connected neurons are found locally within V1 (>80%; Table 1). The implication here that local V1 circuits provide the dominant input to both inhibitory and excitatory neurons is consistent with several other lines of evidence (see [2, 46]). Moreover, results for inhibitory neurons showing that only 3–8% of inputs arise from extrinsic cortical connections are similar to one example shown for mouse somatosensory cortex using a PV-cre line to target TVA and RabG expression to the inhibitory cell subtype and subsequent infection and spread with EnvA-ΔG-RV [19]. There, Wall et al. found that only ~3% of RV connected neurons were not local (20 out of 620 in counts of every 6th section). Given that our AAV/GAD1/YTB infected inhibitory neurons included the PV-positive subpopulation (Fig. S2E-H), these results corroborate our findings.
Conclusion
Understanding the connectivity of neocortical networks is essential for explaining complex cortical computations. Decades of research using ‘traditional’ neuronal tracers have been spent piecing together the connectivity, but without a means for easily distinguishing between circuits of inhibitory and excitatory cell-types. Cortical inhibition plays a critical role in controlling and modulating cortical excitation and a more detailed understanding of the neuronal circuits contributing to each will provide more insight into the interplay between these two major classes of neurons. Our viral vector mediated technique provides a new way for addressing inhibitory and excitatory circuits through retrograde labeling of presynaptic inputs, and when paired with global measures of functional selectivity, such as intrinsic signal optical imaging, can be used to infer functional properties of these circuits. Beyond this, genes that allow for cell monitoring through calcium imaging [47] or cell manipulation through light gated ion channels [48, 49] can also be delivered with the ΔG-RV virus [50]. Thus, not only can inputs to excitatory and inhibitory neurons now be distinguished anatomically by our new approach, future applications using this method should provide the opportunity to observe and manipulate the function of each circuit in vivo.
EXPERIMENTAL PROCEDURES
Generation of AAV/GAD1/YTB
A four step cloning procedure (see Supplemental Information) was used to generate our final plasmid, AAV/GAD1/YTB, (11.0 kb). The viral genome to be packed into infectious particles encompasses 7382 bp (including 5′- and 3′-inverted terminal repeat; Fig. 1, top). Serotype 9 AAV particles were prepared and purified with iodixanol by the viral vector core facility at the Salk Institute (La Jolla, CA), yielding a titer of 9 × 109 genome copies/ml.
Generation of LV/α CaMKII/YTB
For details on LV subcloning see Supplemental Information. The resultsing LV/αCaMKII/YTB, (12.3 kb) plasmid has a viral genome to be packaged of 7.5 kb (including 5′- and 3′-long terminal repeat; Fig. 1, bottom). Purified VSV-G pseudotytped lentiviral particles were prepared by the viral vector core facility at the Salk Institute yielding a titer of 2 × 1010 transducing units/ml
Rabies Viruses
The modified RV, EnvA-ΔG-RV expressing mCherry, as well as the ΔG-RV expressing either mCherry or GFP were produced and concentrated following protocols described in [16, 51].
Animal Procedures
All animal procedures were approved by the UC Irvine Institutional Animal Care and Use Committee and the Institutional Biosafety Committee, and followed the National Institutes of Health guidelines. Pressure injections of viruses were made directly into V1 of anesthetized mouse and cat using sterile procedures. Intrinsic signals in cat were imaged by a 12-bit video camera with a 50 mm/50 mm tandem-lens combination feeding into the Optical Imager 3001 system [29, 30]. See Supplemental Information for more details.
Histology
After the full survival period, animals were deeply anesthetized with Euthasol and perfused transcardially, first with saline, then followed by 4% paraformaldehyde in phosphate buffer (PB; pH 7.4). For most animals, 1.5% glutaraldehyde was also included. Brains were removed and cryoprotected in 30% sucrose for ~48 hours prior to sectioning. Sectioned brains were processed for GABA, αCaMKII, or DAPI. See Supplemental Information for more details.
Supplementary Material
Highlights.
Cell-type specific gene delivery achieved through viral vector/promoter design
Gene expression targets rabies to trace inputs to inhibitory or excitatory neurons
Inhibitory neurons in V1 receive fewer feedback inputs than excitatory neurons
Intrinsic inputs to inhibitory V1 neurons are tuned to orientation preference
Acknowledgments
We thank Ed Callaway for the YTB construct, cell lines for RV production, and access to facilities for cloning the GAD1 promoter; Daniel Gibbs for helpful discussions regarding AAV vectors; Maziar Hashemi-Nezhad for initial assistance with cat surgery and intrinsic signal optical imaging; the Whitehall Foundation (2009-12-44: D.C.L.) and National Institutes of Health (NS072948: D.C.L) for support.
Footnotes
Supplemental Information includes additional Experimental Procedures and five figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beaulieu C. Numerical data on neocortical neurons in adult rat, with special reference to the GABA population. Brain Res. 1993;609:284–292. doi: 10.1016/0006-8993(93)90884-p. [DOI] [PubMed] [Google Scholar]
- 2.Burkhalter A. Many specialists for suppressing cortical excitation. Front Neurosci. 2008;2:155–167. doi: 10.3389/neuro.01.026.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeFelipe J. Neocortical neuronal diversity: chemical heterogeneity revealed by colocalization studies of classic neurotransmitters, neuropeptides, calcium-binding proteins, and cell surface molecules. Cereb Cortex. 1993;3:273–289. doi: 10.1093/cercor/3.4.273. [DOI] [PubMed] [Google Scholar]
- 4.Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 5.Adesnik H, Bruns W, Taniguchi H, Huang ZJ, Scanziani M. A neural circuit for spatial summation in visual cortex. Nature. 2012;490:226–231. doi: 10.1038/nature11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson JS, Carandini M, Ferster D. Orientation tuning of input conductance, excitation, and inhibition in cat primary visual cortex. J Neurophysiol. 2000;84:909–926. doi: 10.1152/jn.2000.84.2.909. [DOI] [PubMed] [Google Scholar]
- 7.Mariño J, Schummers J, Lyon DC, Schwabe L, Beck O, Wiesing P, Obermayer K, Sur M. Invariant computations in local cortical networks with balanced excitation and inhibition. Nat Neurosci. 2005;8:194–201. doi: 10.1038/nn1391. [DOI] [PubMed] [Google Scholar]
- 8.Fino E, Yuste R. Dense Inhibitory Connectivity in Neocortex. Neuron. 2011;69:1188–1203. doi: 10.1016/j.neuron.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X, Callaway EM. Laminar Specificity of Functional Input to Distinct Types of Inhibitory Cortical Neurons. Journal of Neuroscience. 2009;29:70–85. doi: 10.1523/JNEUROSCI.4104-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshimura Y, Dantzker JL, Callaway EM. Excitatory cortical neurons form fine-scale functional networks. Nature. 2005;433:868–873. doi: 10.1038/nature03252. [DOI] [PubMed] [Google Scholar]
- 11.Bock DD, Lee WC, Kerlin AM, Andermann ML, Hood G, Wetzel AW, Yurgenson S, Soucy ER, Kim HS, Reid RC. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko H, Hofer SB, Pichler B, Buchanan KA, Sjostrom PJ, Mrsic-Flogel TD. Functional specificity of local synaptic connections in neocortical networks. Nature. 2011;473:87–91. doi: 10.1038/nature09880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson JC, Martin AC. The Synaptic Connections between Cortical Areas V1 and V2 in Macaque Monkey. Journal of Neuroscience. 2009;29:11283–11293. doi: 10.1523/JNEUROSCI.5757-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson P, Burkhalter A. Microcircuitry of forward and feedback connections within rat visual cortex. Journal of Comparative Neurology. 1996;368:383–398. doi: 10.1002/(SICI)1096-9861(19960506)368:3<383::AID-CNE5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 15.Rockland KS. Elements of cortical architecture: Hierarchy revisited. In: Rockland KS, Kaas JH, Peters A, editors. Cerebral Cortex. Vol. 12. New York: Plenum Publishing Co; 1997. pp. 243–293. [Google Scholar]
- 16.Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haubensak W, Kunwar PS, Cai H, Ciocchi S, Wall NR, Ponnusamy R, Biag J, Dong HW, Deisseroth K, Callaway EM, et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–276. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamichi K, Amat F, Moussavi F, Wang C, Wickersham I, Wall NR, Taniguchi H, Tasic B, Huang ZJ, He Z, et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc Natl Acad Sci U S A. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weible AP, Schwarcz L, Wickersham IR, Deblander L, Wu H, Callaway EM, Seung HS, Kentros CG. Transgenic targeting of recombinant rabies virus reveals monosynaptic connectivity of specific neurons. J Neurosci. 2010;30:16509–16513. doi: 10.1523/JNEUROSCI.2442-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting Single Neuronal Networks for Gene Expression and Cell Labeling In Vivo. Neuron. 2010;67:562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dittgen T, Nimmerjahn A, Komai S, Licznerski P, Waters J, Margrie TW, Helmchen F, Denk W, Brecht M, Osten P. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc Natl Acad Sci U S A. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han X, Qian X, Bernstein JG, Zhou HH, Franzesi GT, Stern P, Bronson RT, Graybiel AM, Desimone R, Boyden ES. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62:191–198. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonchar Y, Burkhalter A. Differential subcellular localization of forward and feedback interareal inputs to parvalbumin expressing GABAergic neurons in rat visual cortex. J Comp Neurol. 1999;406:346–360. doi: 10.1002/(sici)1096-9861(19990412)406:3<346::aid-cne4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 25.Gonchar Y, Burkhalter A. Distinct GABAergic targets of feedforward and feedback connections between lower and higher areas of rat visual cortex. J Neurosci. 2003;23:10904–10912. doi: 10.1523/JNEUROSCI.23-34-10904.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert CD, Wiesel TN. Columnar specificity of intrinsic horizontal and corticocortical connections in cat visual cortex. J Neurosci. 1989;9:2432–2442. doi: 10.1523/JNEUROSCI.09-07-02432.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosking WH, Zhang Y, Schofield B, Fitzpatrick D. Orientation selectivity and the arrangement of horizontal connections in tree shrew striate cortex. J Neurosci. 1997;17:2112–2127. doi: 10.1523/JNEUROSCI.17-06-02112.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malach R, Amir Y, Harel M, Grinvald A. Relationship between intrinsic connections and functional architecture revealed by optical imaging and in vivo targeted biocytin injections in primate striate cortex. Proc Natl Acad Sci U S A. 1993;90:10469–10473. doi: 10.1073/pnas.90.22.10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashemi-Nezhad M, Lyon DC. Orientation tuning of the suppressive extraclassical surround depends on intrinsic organization of V1. Cereb Cortex. 2012;22:308–326. doi: 10.1093/cercor/bhr105. [DOI] [PubMed] [Google Scholar]
- 30.Liu YJ, Hashemi-Nezhad M, Lyon DC. Sharper orientation tuning of the extraclassical suppressive-surround due to a neurons location in the V1 orientation map emerges late in time. Neuroscience. 2013;229:100–117. doi: 10.1016/j.neuroscience.2012.10.071. [DOI] [PubMed] [Google Scholar]
- 31.Shushruth S, Mangapathy P, Ichida JM, Bressloff PC, Schwabe L, Angelucci A. Strong recurrent networks compute the orientation tuning of surround modulation in the primate primary visual cortex. J Neurosci. 2012;32:308–321. doi: 10.1523/JNEUROSCI.3789-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angelucci A, Levitt JB, Walton EJ, Hupe JM, Bullier J, Lund JS. Circuits for local and global signal integration in primary visual cortex. J Neurosci. 2002;22:8633–8646. doi: 10.1523/JNEUROSCI.22-19-08633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cantone G, Xiao J, McFarlane N, Levitt JB. Feedback connections to ferret striate cortex: direct evidence for visuotopic convergence of feedback inputs. J Comp Neurol. 2005;487:312–331. doi: 10.1002/cne.20570. [DOI] [PubMed] [Google Scholar]
- 34.Salin PA, Girard P, Kennedy H, Bullier J. Visuotopic organization of corticocortical connections in the visual system of the cat. J Comp Neurol. 1992;320:415–434. doi: 10.1002/cne.903200402. [DOI] [PubMed] [Google Scholar]
- 35.Cardin JA, Palmer LA, Contreras D. Stimulus feature selectivity in excitatory and inhibitory neurons in primary visual cortex. J Neurosci. 2007;27:10333–10344. doi: 10.1523/JNEUROSCI.1692-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirsch JA, Martinez LM, Pillai C, Alonso JM, Wang Q, Sommer FT. Functionally distinct inhibitory neurons at the first stage of visual cortical processing. Nat Neurosci. 2003;6:1300–1308. doi: 10.1038/nn1152. [DOI] [PubMed] [Google Scholar]
- 37.Nowak LG, Sanchez-Vives MV, McCormick DA. Lack of orientation and direction selectivity in a subgroup of fast-spiking inhibitory interneurons: cellular and synaptic mechanisms and comparison with other electrophysiological cell types. Cereb Cortex. 2008;18:1058–1078. doi: 10.1093/cercor/bhm137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerlin AM, Andermann ML, Berezovskii VK, Reid RC. Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron. 2010;67:858–871. doi: 10.1016/j.neuron.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li YT, Ma WP, Li LY, Ibrahim LA, Wang SZ, Tao HW. Broadening of inhibitory tuning underlies contrast-dependent sharpening of orientation selectivity in mouse visual cortex. J Neurosci. 2012;32:16466–16477. doi: 10.1523/JNEUROSCI.3221-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sohya K, Kameyama K, Yanagawa Y, Obata K, Tsumoto T. GABAergic neurons are less selective to stimulus orientation than excitatory neurons in layer II/III of visual cortex, as revealed by in vivo functional Ca2+ imaging in transgenic mice. J Neurosci. 2007;27:2145–2149. doi: 10.1523/JNEUROSCI.4641-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buzas P, Kovacs K, Ferecsko AS, Budd JM, Eysel UT, Kisvarday ZF. Model-based analysis of excitatory lateral connections in the visual cortex. J Comp Neurol. 2006;499:861–881. doi: 10.1002/cne.21134. [DOI] [PubMed] [Google Scholar]
- 42.Nathanson JL, Jappelli R, Scheeff ED, Manning G, Obata K, Brenner S, Callaway EM. Short Promoters in Viral Vectors Drive Selective Expression in Mammalian Inhibitory Neurons, but do not Restrict Activity to Specific Inhibitory Cell-Types. Front Neural Circuits. 2009;3:19. doi: 10.3389/neuro.04.019.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nathanson JL, Yanagawa Y, Obata K, Callaway EM. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience. 2009;161:441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Büeler H. Adeno-associated viral vectors for gene transfer and gene therapy. Biol Chem. 1999;380:613–622. doi: 10.1515/BC.1999.078. [DOI] [PubMed] [Google Scholar]
- 45.Hirsch ML, Agbandje-McKenna M, Samulski RJ. Little vector, big gene transduction: fragmented genome reassembly of adeno-associated virus. Mol Ther. 2010;18:6–8. doi: 10.1038/mt.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Douglas RJ, Martin KA. Recurrent neuronal circuits in the neocortex. Curr Biol. 2007;17:R496–500. doi: 10.1016/j.cub.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 47.Akerboom J, Chen TW, Wardill TJ, Tian L, Marvin JS, Mutlu S, Calderon NC, Esposti F, Borghuis BG, Sun XR, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 49.Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 50.Osakada F, Mori T, Cetin AH, Marshel JH, Virgen B, Callaway EM. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron. 2011;71:617–631. doi: 10.1016/j.neuron.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickersham IR, Sullivan HA, Seung HS. Production of glycoprotein-deleted rabies viruses for monosynaptic tracing and high-level gene expression in neurons. Nat Protoc. 2010;5:595–606. doi: 10.1038/nprot.2009.248. [DOI] [PubMed] [Google Scholar]
- 52.Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.