Abstract
Background
The plastome of embryophytes is known for its high degree of conservation in size, structure, gene content and linear order of genes. The duplication of entire tRNA genes or their arrangement in a tandem array composed by multiple pseudogene copies is extremely rare in the plastome. Pseudogene repeats of the trnF gene have rarely been described from the chloroplast genome of angiosperms.
Findings
We report the discovery of duplicated copies of the original phenylalanine (trnFGAA) gene in Solanaceae that are specific to a larger clade within the Solanoideae subfamily. The pseudogene copies are composed of several highly structured motifs that are partial residues or entire parts of the anticodon, T- and D-domains of the original trnF gene.
Conclusions
The Pseudosolanoid clade consists of 29 genera and includes many economically important plants such as potato, tomato, eggplant and pepper.
Electronic supplementary material
The online version of this article (doi:10.1186/2193-1801-2-459) contains supplementary material, which is available to authorized users.
Keywords: Chloroplast DNA (cpDNA), Gene duplications, Phylogeny, Plastome evolution, Tandem repeats, trnL-trnF, Solanaceae
Findings
The plastid trnT-trnF region has been widely applied to resolve phylogeny of embryophytes (Quandt and Stech 2004; Zhao et al. 2011) and to address various questions of population genetics since the development of universal primers by Taberlet et al. (1991). This marker is located in the large single copy region of the chloroplast genome and contains a co-transcribed region consisting of three highly conserved exons that code the transfer RNA (tRNA) genes for threonine (UGU), leucine (UAA) and phenylalanine (GAA). The region is interspersed by two intergenic spacers and by a group I intron intercalated within the first and second exon of the trnL(UAA) gene. Phylogenetic results obtained with the trnT-trnF region (or part of it) should be treated with caution. This is due to the fact that some recent studies (e.g. Koch et al. 2005; Pirie et al. 2007; Schmikl et al. 2009; Vivjerberg and Bachmann 1999) have shown that there are clearly several copies of certain parts of this region. If this is ignored, it will easily lead to situations where basic requirement of homology of the characters used for phylogenetic analyses is compromised. This might lead to false hypotheses of phylogeny, especially when they are based on the analyses of only this region.
Larger structural changes (>50 bp) rarely occur in the plastome. However, duplications of the rpl2 or rpl23 genes (Bowman et al. 1988) or even the duplication of tRNAs (pseudogenes) are occasionally reported. The later are extremely rare in angiosperms and so far they have only been described from Asteraceae (Vijverberg and Bachmann 1999; Witzell 1999), Annonaceae (Pirie et al. 2007), Brassicaceae (Ansell et al. 2007; Koch et al. 2007; Tedder et al. 2010) and Juncaceae (Drábkova et al. 2004). In our recent study we reported a tandem repeat comprising of two to four pseudogene copies upstream of the original trnF gene in four Solanum (Solanaceae) species (Poczai and Hyvönen 2011a). We have characterized these structural duplications and shown that they consist of several highly structured motifs, which are partial residues, or entire parts of the anticodon, T- and D-domains of the original gene, but all lack the acceptor stems at the 5′ or 3′. We were further interested to evaluate the possible occurrence of complete or partial trnF pseudogenes in Solanaceae. This family contains many economically important plant species, e.g., potato (Solanum tuberosum L.), tomato (Solanum lycopersicum L.) and paprika (Capsicum annuum L.) and is under intensive phylogenetic investigation and the trnT-F plastid marker is commonly used in these studies. These sequences together with the results of molecular breeding programs provide large amount of data that is available in GenBank. During data mining we concentrated on a structured dataset generated in previous phylogenetic studies (Fukuda et al. 2001; Garcia and Olmstead 2003; Santiago-Valentin and Olmstead 2003; Bohns 2004; Clarkson et al. 2004; Levin and Miller 2005; Levin et al. 2005; Weese and Bohns 2007; Olmstead et al. 2008) that contained 195 taxa and 390 sequences. This dataset provided the basis for the latest robust phylogenetic hypothesis of the Solanaceae including 89 from the 98 (Olmstead and Bohns 2007) recognized genera. Manual search using the anticodon domain of the original trnF gene and automated tRNA recognition by CENSOR (Kohany et al. 2006) indicated the presence of pseudogene repeats in numerous genera of Solanaceae.
We used the core trnL-F dataset to map the occurrence of pseudogenic repeats on the phylogenetic tree of Solanaceae. As presented in Figure 1 the distribution of pseudogenic duplications is in congruence with the previously published phylogeny of the Solanaceae (Olmstead et al. 2008), and it is obvious that the first pseudogenic copy evolved only once at the base of a highly supported clade within the subfamily Solanoideae. Among the members of this lineage, referred here as the Pseudosolanoid clade, the anticodon domain of the trnF gene exhibits extensive gene duplications with one to seven tandemly repeated copies in close 5′-proximity of the original functional gene (Table 1). The size of each pseudogenic copy ranged between 32 and 73 bp and the anticodon domain was identified as the most conserved element. A common ATT(G)n motif is of particular interest and its modifications were found to border the 5′ of the duplicated regions in the same way as found in Brassicaceae (Ansell et al. 2007; Koch et al. 2005 and 2007; Schmikl et al. 2009; Tedder et al. 2010). Other motifs were partial residues or entire parts of the T- and D-domains. The residues of the 3′ and 5′ acceptor stems were rarely found among the copies (see Table 1). The D-domain was more conserved than the T-domain among the copies and other internal repeats (AT, AAT, ATT, AATCC) were intercalated within this region for example in genus Lycianthes (Dunal.) Hassl. In addition to these newly discovered pseudogenes we were also able to characterize putative promoter motifs showing high similarity to a sigma70-type bacterial promoter. These two elements (−35 TTGACA/-10 GAGGAT) are consistently found in the trnL-F spacer region of embryophytes, and they are believed to represent the ancient and original trnF gene promoter (Quandt et al. 2004). Interestingly, pseudogenic repeats were found to be exclusively inserted after such motifs in Solanaceae, contrary to Brassicaceae, where similar pseudogenic repeats were found only between promoter motifs in the trnL-F intergenic spacer region (Koch et al. 2005). The later finding lead Koch et al. (2005) to support the conclusion by Kanno and Hirtai (1993) that these elements should be non-functional due to the intercalated position of pseudogenes between promoters. However, this may be challenged by the position of Solanaceae pseudogenes following the −10 and −35 promoters, which are also variable in number and composition.
Figure 1.
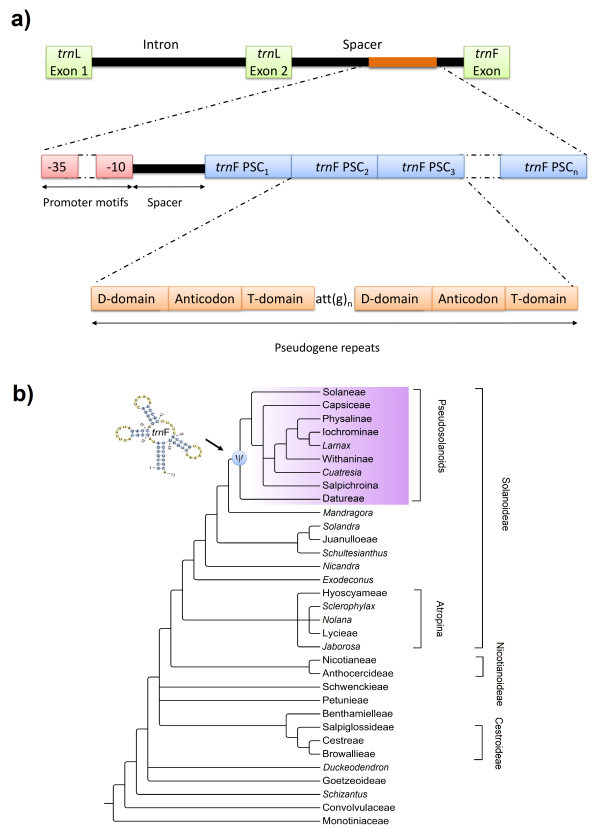
Phylogeny of Solanaceae and the distribution and schematic structure oftrnF pseudogene copies. a) Suprageneric groups recognized are indicted to the right on the tree, while major clades are collapsed at the base node and their names follow Olmstead et al. (2008). The new Pseudosolanoid clade united by the presence of pseudogenic trnF gene duplication is marked with ‘ψ’ in the Solanoideae subfamily. b) The schematic representation of the plastidic trnL-F spacer region in Solanaceae and the intercalated pseudogene copies (PSC) in the intergenic spacer region close to 5′ of the trnF gene. Pseudogene repeats are variable in number and structure and are found after the putative promoter motifs that are also variable among species. The spacer region between the first PSC and promoter motifs consists of intergenic repeats of variable length. Each PSC is separated by a common bordering motif (ATTG) at the 5′end.
Table 1.
Distribution oftrnF pseudogenes among Solanaceae and number of multiplicatedtrnF anticodon domains
| Taxa | GenBank | Tribe | Copy number |
|---|---|---|---|
| Acnistus arborescens | EU580954 | Physaleae | 2a,b |
| Aureliana fasciculata | EU580961 | Physaleae | 2 |
| Brachistus stramonifolius | EU580963 | Physaleae | 3 |
| Brugmansia aurea | EU580965 | Datureae | 1c |
| Brugmansia sanguinea | EU580966 | Datureae | 1c |
| Capsicum baccatum | EU580969 | Capsiceae | 4a,b |
| Capsicum chinense | EU603443 | Capsiceae | 4d |
| Capsicum minutiflorum | EU580970 | Capsiceae | 6d |
| Capsicum pubescens | AY348982 | Capsiceae | 6b,d |
| Capsicum rhomboideum | EU580971 | Capsiceae | 1e |
| Chamaesaracha coronopus | EU580978 | Physaleae | 4 |
| Chamaesaracha sordida | EU580979 | Physaleae | 4 |
| Cuatresia exiguiflora | EU580981 | Physaleae | 2 |
| Cuatresia riparia | EU580982 | Physaleae | 2 |
| Datura leichhardtii | EU580983 | Datureae | 1f |
| Datura stramonium | EU580984 | Datureae | 1f |
| Deprea sylvarum | EU580985 | Physaleae | 3 |
| Discopodium penninervum | EU580986 | Physaleae | 4 |
| Dunalia solanacea | EU580988 | Physaleae | 4 |
| Eriolarynx lorenzii | EU580990 | Physaleae | 4 |
| Iochroma australe | EU580999 | Physaleae | 4 |
| Iochroma cardenasianum | EU581000 | Datureae | 1f |
| Iochroma fuchsioides | EU581001 | Physaleae | 2 |
| Iochroma umbellatum | EU581002 | Physaleae | 2 |
| Jaltomata auriculata | EU581006 | Solaneae | 2f |
| Jaltomata grandiflora | EU581007 | Solaneae | 2f |
| Jalotmata procumbens | AY098695 | Solaneae | 1a,b,f |
| Jaltomata sinuosa | DQ180418 | Solaneae | 2f |
| Larnax subtriflora | EU581009 | Physaleae | 3 |
| Leucophysalis grandiflora | EU581013 | Physaleae | 2 |
| Leucophysalis nana | EU581014 | Physaleae | 2 |
| Lycianthes biflora | EU581015 | Capsiceae | 2g |
| Lycianthes ciliolata | EU581016 | Capsiceae | 4h,i |
| Lycianthes glandulosa | EU581017 | Capsiceae | 3g,i |
| Lycianthes heteroclita | DQ180414 | Capsiceae | 2g,i |
| Lycianthes inaequilatera | EU581018 | Capsiceae | 6h,i |
| Lycianthes multiflora | EU581019 | Capsiceae | 3g,i |
| Lycianthes peduncularis | EU581020 | Capsiceae | 4h,i |
| Lycianthes shanesii | EU581021 | Capsiceae | 1g,h,i |
| Margaranthus solanaceus | EU581025 | Physaleae | 5i |
| Nectouxia formosa | EU581031 | Salpichroina* | 1a,b |
| Nothocestrum latifolium | EU581037 | Physaleae | 2 |
| Nothocestrum longifolium | EU581038 | Physaleae | 3 |
| Oryctes nevadensis | EU581039 | Physaleae | 3 |
| Physalis alkekengi | DQ180420 | Physaleae | 2 |
| Physalis carpenteri | EU581042 | Physaleae | 2 |
| Physalis heterophylla | EU581043 | Physaleae | 2 |
| Physalis peruviana | EU581044 | Physaleae | 4a,b |
| Physalis philadelphica | EU581045 | Physaleae | 5a,b |
| Quincula lobata | EU581051 | Physaleae | 1a,b |
| Salpichroa origanifolia | EU581052 | Salpichroina* | 2a,b |
| Saracha punctata | EU581053 | Physaleae | 4 |
| Solanum abutiloides | AY266236 | Solaneae | 1f |
| Solanum aviculare | HM006836 | Solaneae | 2a,b,f |
| Solanum betaceum | DQ180426 | Solaneae | 1f |
| Solanum dulcamara | HM006840 | Solaneae | 1f |
| Solanum herculeum | DQ180466 | Solaneae | 2f |
| Solanum lycopersicum | NC007898 | Solaneae | 1f |
| Solanum melongena | EU176149 | Solaneae | 2h,i |
| Solanum pseudocapsicum | DQ180436 | Solaneae | 1 |
| Solanum torvum | AY266246 | Solaneae | 4i |
| Solanum trisectum | JN130370 | Solaneae | 2 |
| Solanum wendlandii | DQ180440 | Solaneae | 1 |
| Tubocapsicum anomalum | EU581066 | Physaleae | 7 |
| Vassobia dichotoma | EU581067 | Physaleae | 4 |
| Witheringia cuneata | EU581070 | Physaleae | 2 |
| Witheringia macrantha | EU581071 | Physaleae | 5 |
| Witheringia meiantha | EU581072 | Physaleae | 4 |
| Witheringia mexicana | EU581073 | Physaleae | 5 |
| Witheringia solanacea | EU581074 | Physaleae | 3 |
Taxonomic classification and a GenBank accession number is provided for each species. *Unranked informal clade name.
aPartial pseudogenic copy at the 3′ end. bMissing original trnF gene. cIntact 5′acceptor stem present. dThree copies of −35 promoter (TTGACA) motif. eFour copies of −35 promoter (TTGACA) motif. fOne copy of −10 promoter (GAGGAT) motif present. gPseudogene repeats are separated by long internal repeats after the promoter motifs. hOnly one copy of −35 promoter (TTGACA) motif. i3′ acceptor stem present.
The occurrence of pseudogenes provides strong evidence of relationships among some groups that had low support values in the previous analyses (e.g. Olsmtead et al. 2008). This event robustly separates the (1) Atropina (Hyoscyameae, Lycieae, Jabrosa, Latua, Nolana and Scleraphylax) and (2) Juanulloeae clades from the Pseudosolanoid clade composed by (3) Solaneae, Capsiceae, Physaleae and Datureae and (4) Salpichroina (Salpichroa Miers and Nectouxia Kunth). In clades (1) and (2) pseudogenes are absent while they appear at the basal node of clade (3) and (4). This lineage where pseudogene copies have been found includes 29 genera; here belongs also the clade of Solanum L. and Capsicum L. with many economically important plant species. However, sequence information was lacking for the genera Mellissia Hook. f. and Athenaea Adans. to confirm the presence of trnF pseudogenes. This is not surprising as available plant material of these taxa is very restricted. For example Mellissia is a genus with a single species, Mellissia begoniifolia (Roxb.) Hook. f. which is critically endangered and endemic to the island of Saint Helena. The larger clade of Solanoideae also includes several branches with low support values composed of small genera (Exodeconus Raf., Mandragora L., Nicandra (L.) Gaerten., Schultesianthus Hunz., Solandra Sw.) in the phylogeny proposed by Olmstead et al. (2008). These lineages are from the early diversification of the Solanoideae with no close relatives and all lack pseudogene repeats that could be informative to trace their ancestry.
The latest large scale phylogenetic analysis of the Solanaceae (Olmstead et al. 2008) established major clades of the family but sampling in some of the lineages can still be improved. Goldberg et al. (2010) analyzed a larger data set but they did not focus on taxonomic relationships but rather on the evolution of self-compatibility. Some studies have attempted to calibrate a molecular clock for various groups within Solanaceae, but all of these used the same (Paape et al. 2008; Poczai and Hyvönen 2011b), or only few fossil records (Dillon et al. 2009; Tu et al. 2010). Fossil record of the Solanaceae has not been reviewed recently. This urges for the re-assessment of the specimens and could potentially provide more robust calibration points for the family (Särkinen, personal communication). Latest current estimates show the age of the Pseudosolanoids to be approximately 20 My (Särkinen, personal communication), and thus the origin of the pseudogene duplications of Solanaceae to be approximately of the same Miocene age as in Brassicaceae (16–21 My; Koch et al. 2005).
Conclusions
Despite of the extensive studies based on sequence level characters the taxonomy of the Solanaceae is not yet completely understood. However, there is ongoing work on different levels by multiple groups to resolve phylogenetic relationships (Fukuda et al. 2001; Garcia and Olmstead 2003; Santiago-Valentin and Olmstead 2003; Bohs 2004; Clarkson et al. 2004; Levin and Miller 2005; Levin et al. 2005; Weese and Bohs 2007; Olmstead et al. 2008). There are a number of questions that should be answered regarding the discovery of trnF pseudogenes, for example: How did the duplications originate? Are the pseudogene copy numbers a useful character for phylogenetic inference? To what extent does the number of pseudogene copies vary within a single species? The evolution and structure of pseudogenic copies should be compared with others reported from different plant families especially from Brassicaceae. The potential of trnF pseudogenes as phylogenetic markers need to be investigated further in the future for better understanding of the evolution of Solanaceae. These investigations could answer what are the wider implications of the pseudogene repeats for Solanaceae studies that utilize the trnL-F spacer region.
Methods
Solanaceae sequence dataset
For the Solanaceae and several outgroups we used the trnL-F spacer data assembled by Olmstead et al. (2008). This dataset contained 195 taxa and 390 sequences generated in previous phylogenetic studies (Fukuda et al. 2001; Garcia and Olmstead 2003; Santiago-Valentin and Olmstead 2003; Bohs 2004; Clarkson et al. 2004; Levin and Miller 2005; Levin et al. 2005; Weese and Bohs 2007; Olmstead et al. 2008) and this was used to align and mask pseudogenic copies. The goal was to map the taxonomic distribution of pseudogenes at family level sampling as many genera as possible. This dataset and representative trees used in our study were previously deposited in TreeBASE (ID S2191). This alignment was also used to demonstrate copy number distribution corresponding to the published phylogenetic hypothesis that was not only based on the trnL-F spacer information but relied on sequence data from the ndhF region.
Recognition and copy number assessment of the trnF(GAA) pseudogenes
The complete chloroplast genome of Solanum bulbocastanum Dunal (DQ347958) was used to select the corresponding loci of the trnL-trnF spacer region (bp positions 48,854 to 49,382), to annotate ambiguous sequences regions, and to ensure that our interpretations are based on homologous positions. Putative pseudogene repeats were identified with screening using Repbase (Jurka 2000) with the “mask pseudogenes” and “report simple repeats” options of the online tool CENSOR (Kobany et al. 2006). This was done to identify repetitive elements by comparing our sequences to known eukaryotic repeats and prototypic sequences stored in Repbase utilizing WU-BLAST. A second search was conducted with FastPCR (Kalendar et al. 2009) using the repeat search option of the program. Under “type of repeats” we checked for simple, direct, inverted, direct antisense, and direct reverse repeats, respectively. Default values were used under a kMers repeat screening. After each search, repetitive motifs and sequences were recorded and compared with the results obtained from the Repbase search. After repeats were identified in the trnL-F IGS sequences, further structural trnF(GAA) gene elements or residues were annotated manually using the anticodon domain as reference. The annotated sequence alignment is shown in Additional file 1.
Sequence annotation and alignment
Masked pseudogenic copies were further edited using Geneious v.4.8.5 (Biomatters Ltd.). We used the Nicotiana tabacum L. complete chloroplast genome (NC001879; bp positions 49,840 to 50,318) for comparisons and to determine the subunits of pseudogenic repeats as this species lacks these gene duplications. Sequence break points were examined manually to determine the cut off points of pseudogenic copies and to identify bordering motifs. Identified copies were aligned with MUSCLE (Edgar 2004) as implemented in Geneious v.4.8.5 using default settings. The sequence alignment in FASTA format is available as Additional file 2.
Electronic supplementary material
Additional file 1: Annotated sequence alignment of pseudogene repeats found in Solanaceae. Major parts of the trnF gene are marked as D- and T-domains and anticodon in the middle together with bordering 5′ and 3′ acceptor stems. The trnF gene of Nicotiana tabacum is used as a reference sequence to align different pseudogenes. (PDF 4 MB)
Additional file 2: Sequence alignment of pseudogene copies. (FASTA 41 KB)
Acknowledgments
PP gratefully acknowledges support from the Marie Curie Fellowship Grant (PIEF-GA-2011-300186) under the seventh framework program of the European Union. We thank Neil Bell for discussions on the manuscript.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PP conceived the study and drafted the early version of the manuscript and performed database search. JH commented on the manuscript, revised the text and structure, and outlined it several times together with PP. Both authors approved the final manuscript.
Contributor Information
Péter Poczai, Email: peter.poczai@gmail.com.
Jaakko Hyvönen, Email: jaakko.hyvonen@helsinki.fi.
References
- Ansell SW, Schneider H, Pedersen N, Grunmann M, Russell SJ, Vogel JC. Recombination diversifies chloroplast trnF pseudogenes Arabidopsis lyrata. J Evol Biol. 2007;20:2400–2411. doi: 10.1111/j.1420-9101.2007.01397.x. [DOI] [PubMed] [Google Scholar]
- Bohs L. A chloroplast DNA phylogeny of Solanum section Lasiocarpa. Syst Bot. 2004;29:177–187. doi: 10.1600/036364404772974310. [DOI] [Google Scholar]
- Bowman CM, Barker RF, Dyer TA. The location and possible evolutionary significance of small dispersed repeats in wheat ctDNA. Curr Genet. 1988;10:931–941. doi: 10.1007/BF00398291. [DOI] [PubMed] [Google Scholar]
- Clarkson JJ, Knapp S, Garcia VF, Olmstead RG, Leitch AR, Chase MW. Phylogenetic relationships in Nicotiana (Solanaceae) inferred from multiple plastid DNA regions. Mol Phylogenet Evol. 2004;33:75–90. doi: 10.1016/j.ympev.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Dillon MO, Tu T, Xie L, Quipuscoa Silvestre V, Wen J. Biogeographic diversification in Nolana (Solanaceae), a ubiquitous member of the Atacama and Peruvian Deserts along the western coast of South America. J Syst Evol. 2009;47:457–476. doi: 10.1111/j.1759-6831.2009.00040.x. [DOI] [Google Scholar]
- Drábkova L, Kirschner J, Vlček Č, Paček V. TrnL-trnF intergenic spacer and trnL intron define major clades within Luzula and Juncus (Juncaceae): importance of structural mutations. J Mol Evol. 2004;59:1–10. doi: 10.1007/s00239-004-2598-7. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Yokoyama J, Ohashi H. Phylogeny and biogeography of the genus Lycium (Solanaceae): inferences from chloroplast DNA sequences. Mol Phylogenet Evol. 2001;19:246–258. doi: 10.1006/mpev.2001.0921. [DOI] [PubMed] [Google Scholar]
- Garcia VF, Olmstead RG. Phylogenetics of tribe Anthocercideae (Solanaceae) based on ndhF and trnL/F sequence data. Syst Bot. 2003;28:609–615. [Google Scholar]
- Goldberg EE, Kohn JR, Lande R, Robertson KA, Smith SA, Igic B. Species selection maintains self-incompatibility. Science. 2010;330:459–460. doi: 10.1126/science.1194513. [DOI] [PubMed] [Google Scholar]
- Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;9:418–420. doi: 10.1016/S0168-9525(00)02093-X. [DOI] [PubMed] [Google Scholar]
- Kalendar R, Lee D, Schulman AH. FastPCR software for PCR primer and probe design and repeat search. Genes Genomes Genomics. 2009;3:1–14. [Google Scholar]
- Kanno A, Hirtai A. A transcription map of the chloroplast genome from rice (Oryza sativa) Curr Genet. 1993;23:166–174. doi: 10.1007/BF00352017. [DOI] [PubMed] [Google Scholar]
- Koch MA, Dobeš C, Matschinger M, Bleeker W, Vogel J, Kiefer M, Mitchell-Olds T. Evolution of the trnF(GAA) gene in Arabidopsis relatives and the Brassicaceae family: monophyletic origin and subsequent diversification of a plastidic pseudogene. Mol Biol Evol. 2005;22:1032–1043. doi: 10.1093/molbev/msi092. [DOI] [PubMed] [Google Scholar]
- Koch MA, Dobeš C, Kiefer C, Schmickl R, Klimeš L, Lysak MA. Supernetwork identifies multiple events of plastid trnF(GAA) pseudogene evolution in the Brassicaceae. Mol Biol Evol. 2007;24:63–73. doi: 10.1093/molbev/msl130. [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka I. Annotation, submission and screening of repetitive elements in repbase: RepbaseSubmitter and censor. BMC Bioinf. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin RA, Miller JS. Relationships within tribe Lycieae (Solanaceae): paraphyly of Lycium and multiple origins of gender dimorphism. Am J Bot. 2005;92:2044–2053. doi: 10.3732/ajb.92.12.2044. [DOI] [PubMed] [Google Scholar]
- Levin RA, Watson K, Bohs L. A four-gene study of evolutionary relationships in Solanum section Acanthophora. Am J Bot. 2005;92:603–612. doi: 10.3732/ajb.92.4.603. [DOI] [PubMed] [Google Scholar]
- Olmstead RG, Bohs L. A summary of molecular systematic research in Solanaceae: 1982–2006. In: Spooner DM, Bohs L, Giovannoni J, Olmstead RG, Shibata D, editors. Solanaceae VI: genomics meets biodiversity. Proceedings of the sixth international Solanaceae conference. Leuven: Acta Horticulturae 745. International Society for Horticultural Science; 2007. pp. 255–268. [Google Scholar]
- Olmstead RG, Bohs L, Migid HA, Santiago-Valentin E, Garcia VF, Collier SM. A molecular phylogeny of the Solanaceae. Taxon. 2008;57:1159–1181. [Google Scholar]
- Paape T, Igic B, Smith SD, Olmstead R, Bohs L, Kohn JR. A 15-myr-old genetic bottleneck. Mol Biol Evol. 2008;25:655–663. doi: 10.1093/molbev/msn016. [DOI] [PubMed] [Google Scholar]
- Pirie MD, Vargas MPB, Botermans M, Bakker FT, Chatrou LW. Ancient paralogy in the cpDNA trnL-F region in Annonaceae: implications for plant molecular systematics. Am J Bot. 2007;94:1003–1016. doi: 10.3732/ajb.94.6.1003. [DOI] [PubMed] [Google Scholar]
- Poczai P, Hyvönen J. Identification and characterization of plastid trnF(GAA) pseudogenes in four species of Solanum (Solanaceae) Biotechnol Lett. 2011;33:2317–2323. doi: 10.1007/s10529-011-0701-x. [DOI] [PubMed] [Google Scholar]
- Poczai P, Hyvönen J. Phylogeny of kangaroo apples (Solanum subg. Archaesolanum, Solanaceae) Mol Biol Rep. 2011;38:5243–5259. doi: 10.1007/s11033-011-0675-8. [DOI] [PubMed] [Google Scholar]
- Quandt D, Stech M. Molecular evolution of the trnTUGU-trnFGAA region in bryophytes. Plant Biol. 2004;6:545–554. doi: 10.1055/s-2004-821144. [DOI] [PubMed] [Google Scholar]
- Quandt D, Müller K, Stech M, Frahm J-P, Frey W, Hilu KW, Borsch T. Molecular evolution of the chloroplast trnL-F region in land plants. Monogr Syst Bot Missouri Bot Gard. 2004;98:13–37. [Google Scholar]
- Santiago-Valentin E, Olmstead RG. Phylogenetics of the Antillean Goetzeoideae (Solanaceae) and their relationships within the Solanaceae based on chloroplast and ITS DNA sequence data. Syst Bot. 2003;28:452–460. [Google Scholar]
- Schmickl R, Keifer C, Dobeš C, Koch MA. Evolution of trnF(GAA) pseudogenes in cruciferous plants. Plant Syst Evol. 2009;282:229–240. doi: 10.1007/s00606-008-0030-2. [DOI] [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tedder A, Hoebe PN, Ansell SW, Mable BK. Using chloroplast trnF pseudogenes for phylogeography in Arabidopsis lyrata. Diversity. 2010;2:653–678. doi: 10.3390/d2040653. [DOI] [Google Scholar]
- Tu T, Volis S, Dillon MO, Sun H, Wen J. Dispersal of Hyoscyameae and Mandragoreae (Solanaceae) from the New World to Eurasia in the early Miocene and their biogeographic diversification within Eurasia. Mol Phyl Evol. 2010;57:1226–1237. doi: 10.1016/j.ympev.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Vijverberg K, Bachmann K. Molecular evolution of tandemly repeated trnF(GAA) gene in the chloroplast genomes of Microseris (Asteraceae) and the use of structural mutations in phylogenetic analysis. Mol Biol Evol. 1999;16:1329–1340. doi: 10.1093/oxfordjournals.molbev.a026043. [DOI] [PubMed] [Google Scholar]
- Weese TL, Bohs L. A three-gene phylogeny of the genus Solanum (Solanaceae) Syst Bot. 2007;32:445–463. doi: 10.1600/036364407781179671. [DOI] [Google Scholar]
- Wittzell H. Chloroplast DNA variation and reticulate evolution in sexual and apomictic sections of dandelions. Mol Ecol. 1999;8:2023–2035. doi: 10.1046/j.1365-294x.1999.00807.x. [DOI] [PubMed] [Google Scholar]
- Zhao T, Wang Z-T, Branford-White CJ, Xu H, Wang C-H. Classification and differentiation of the genus Peganum indigenous to China based on chloroplast trnL-F and psbA-trnH sequences and seed coat morphology. Plant Biol. 2011;13:940–947. doi: 10.1111/j.1438-8677.2011.00455.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Annotated sequence alignment of pseudogene repeats found in Solanaceae. Major parts of the trnF gene are marked as D- and T-domains and anticodon in the middle together with bordering 5′ and 3′ acceptor stems. The trnF gene of Nicotiana tabacum is used as a reference sequence to align different pseudogenes. (PDF 4 MB)
Additional file 2: Sequence alignment of pseudogene copies. (FASTA 41 KB)


