Abstract
Effects of cervid browsing on timber production, especially during winter, lead to economic losses in forest management. The aim of this study was to present an efficient DNA-based method which allows qualitative assessment of the winter diet from stools of moose (Alces alces), roe deer (Capreolus capreolus), and red deer (Cervus elaphus). The preliminary results of the diet composition of the three cervids from Poland were also presented with a special emphasis on moose. The electropherograms of the chloroplast intron trnL (UAA) P6 loop amplification products using g (fluorescence-labeled) and h primers revealed differences in the length of PCR products among various plant species eaten by these herbivores. In addition, the usage of species-specific primers allowed unambiguous identification of different gymnosperms and angiosperms. The preliminary moose diet analysis, based on winter fecal samples from the entire range of moose occurrence in Poland, revealed the presence of 15 to 24 tree, shrub, and herbaceous species. This fast, cost-efficient, and simple method proved also to be reliable for the diet analysis of red deer and roe deer. It may be a valuable tool in forest and conservation management, as well as a way of enhancing ecological studies focusing on the impact of herbivores on the ecosystems and their possible food niche overlap.
Electronic supplementary material
The online version of this article (doi:10.1007/s13364-013-0146-9) contains supplementary material, which is available to authorized users.
Keywords: Alces alces, Capreolus capreolus, Cervus elaphus, DNA metabarcoding, Ungulate diet analysis, Ungulate management
Introduction
The large herbivores, as keystone species, have a significant impact on ecosystems (Graham et al. 2010). Feeding on trees and shrubs affects the composition of plant communities, plant productivity, and nutrient flow (Danell et al. 1994). Cervids, especially moose (Alces alces), red deer (Cervus elaphus), and roe deer (Capreolus capreolus), cause considerable damage to tree stands by browsing, trampling, and bark stripping (Bobek et al. 1992; Graham et al. 2010). This leads to economic losses in forest management. Extensive knowledge about diet and feeding habits of particular species of cervids is required to prevent this damage. It also allows understanding of relationships between herbivores and plant communities, which in return will enable the design of effective management strategies and/or protection of these ungulate species (Putman 1996).
So far, studies of the herbivorous diet have been primarily based on methods of direct observation in the field, observation of feeding traces along transects, microscopic analysis of rumen contents, and identification of plant species from feces. However, all these methods may lead to erroneous results. The first method is feasible on a small group of animals only, and despite the immense work involved, it is not accurate in estimating the list and the amount of consumed species. During the observations of feeding traces along transects, it is often difficult to distinguish signs of use of the plants by different species of herbivores (Gębczyńska 1980; Wam and Hjeljord 2010). The analysis of the diet using microscopic examination is time consuming and requires appropriate training and an extensive reference database of food items (Holechek et al. 1982). In addition, a significant amount of the material is too fragmented and cannot be identified (Gębczyńska 1980). The development of molecular biology techniques allowed the use of DNA isolated from feces to determine the composition of the diet of the organism. In recent years, researchers focused on chloroplast trnL (UAA) intron, especially its P6 loop, to study the diet of herbivores as the P6 loop is a relatively short sequence that usually varies in length and nucleotide composition among different plant species (Taberlet et al. 2007). The use of high-throughput parallel pyrosequencing of DNA from feces can identify the plants in the diet of these animals (Valentini et al. 2009; Kowalczyk et al. 2011). Although next-generation sequencing (NGS) technology is currently the best solution and has become increasingly more accurate, it requires specialized equipment found in a limited number of laboratories and is still relatively expensive. Thus, there is a need, in research practice, especially in forest management for a fast, easy, and inexpensive method that can be used in laboratories with basic equipment for molecular biology for a general qualitative analysis of the diet composition in terms of the content of tree and shrub species.
The moose prefers moist or wet environments such as wetlands, alder (Alnus spp.) wood forests, and moors. It is usually present on marshes that are overgrown with willow (Salix spp.) (Dzięciołowski and Pielowski 1975). Moose is a concentrate selector, i.e., it needs easily digestible and nutritious food. Its morphological and anatomical structure is adapted for high selectivity (Hofmann 1989), which means that they actively search for the most preferred plant species (Dzięciołowski 1974). Previous studies show that during summer, moose mainly eats herbaceous plants, leaves of trees, and shrubs while winter is dominated by browsing (Dzięciołowski and Pielowski 1975). The species’ diet composition varies between seasons, and it is also variable in different parts of Europe. For example, in Norway, the winter diet was predominantly rowan (Sorbus aucuparia), birch (Betula spp.), and pine (Pinus sylvestris), whereas pine was not found in the summer diet, instead the willow was browsed (Wam and Hjeljord 2010). In Sweden, the winter diet was dominated by pine and birch, while in summer, birch, willow, and herbaceous plants were most abundant (Cederlund and Nystrom 1981). The study of Morow (1976) in the Augustów Forest in NE Poland showed that from December to March, at least 92 % of the consumed biomass was dominated by pine. In September, more species were recorded (at least seven), especially willow, hazel (Corylus avellana), and poplar (Populus spp.).
The most abundant population of moose in Poland is located in the Biebrza Valley in NE Poland. As the animals found in this region exhibit typical seasonal migrations (Gębczyńska and Raczyński 1989) in winter, they are suspected to cause significant damages to managed forests, especially pine stands (Komenda 2001).
The roe deer is the most common cervid in Poland. It has similar nutritional requirements to moose, as it is also a concentrate selector (Hofmann 1989). Its spring and summer diet is dominated by herbs (about 80 %) (Gębczyńska 1980). In winter, more than 80 % of the diet consisted of shoots of trees and shrubs (Siuda et al. 1969). The proportion of spruce (Picea abies) was high, as well as pine, and hay taken from feeding stations during this season (Siuda et al. 1969; Cederlund and Nystrom 1981).
The red deer is a widespread species in Poland, but it has only been recorded in the Biebrza Valley since the late seventies of the twentieth century (Gębczyńska and Raczyński 1993). In comparison to moose and roe deer, it has much lower nutritional requirements, being of “intermediate type” (Hofmann 1989). In summer, its principal foods were grasses, herbaceous plants, young shoots, and leaves of trees and shrubs. In winter, the diet was dominated by shoots, the bark of shrubs, conifers, and deciduous trees (Bobek et al. 1992). As well as moose, both roe deer and red deer show variation in diet according to the season and location of the population (Siuda et al. 1969; Gębczyńska 1980; Bobek et al. 1992).
The aim of this study was to find a method for efficient DNA-based qualitative assessment of the winter diet from feces of three cervids: moose, red deer, and roe deer. We also present the preliminary results for winter diet composition of these three herbivorous species in Poland with a special emphasis on moose and also compare the obtained results with traditional methods.
Materials and methods
For this study, we used 29 samples of moose feces collected in winter (December to March) over the years 2006–2011 from nine different sites within the species’ range in Poland. We also collected 11 red deer and 11 roe deer stools in March and December 2012. To distinguish stools of these species, we considered their shape and size according to Jędrzejewski and Sidarowicz (2010). Stool droppings were frozen at −20 °C or stored in silica gel until processed. DNA was isolated by QIAamp DNA Stool Mini Kit (QIAGEN), and to assure accurate species identification, a fragment of mitochondrial DNA control region was amplified using primers from Hundertmark et al. (2002). mtDNA-cr amplification from stools of three herbivores and sequencing conditions were consistent with the descriptions provided by Świsłocka et al. (2008).
In order to obtain a basic database for tree and shrub species that could be eaten by the studied cervids, we analyzed samples of 36 plant species: shrub, trees, and herbaceous plants (Table 1) collected from March to May in 2012 in the Biebrza River Valley. The majority of these plant species were previously identified as moose food by microscopic examination of their gut contents or by direct observations of herbivores (Dzięciołowski 1974; Morow 1976; Belovsky 1981). Whole DNA was extracted from leaves and buds using Genomic Mini AX Plant (A&A Biotechnology) with elution in 50 μL of QIAGEN nuclease-free water. For each plant sample, we amplified the fragment of the chloroplast intron trnL (UAA) P6 loop using g and h primers (Taberlet et al. 2007) as they work extremely well with degraded samples such as stools (Valentini et al. 2009). The primer g was fluorescence-labeled using FAM as a dye. The PCR was carried out in a final volume of 5 μL with GeneAmp PCR System 9700 (Applied Biosystems). The PCR mixture contained 2 μL of extracted DNA as a template, 1.7 μL QIAGEN multiplex PCR master mix, 1 μL QIAGEN nuclease-free water, and 0.3 μL mix of primers (0.2 μM of each primer). The DNA polymerase was activated at 95 °C for 15 min, followed by 15 (plant DNA samples) or 35 (stool samples) cycles with denaturation at 94 °C for 30 s, annealing at 57 °C for 90 s, extension at 72 °C for 60 s, and final extension at 60 °C for 30 min. The PCR products were denatured with 10 μL Hi-Di formamide (Applied Biosystems) and 0.2 μL GeneScan 500-LIZ size standard (Applied Biosystems) at 95 °C for 5 min and immediately cooled in ice. Detection was carried out on ABI 3130 Genetic Analyzer (Applied Biosystems) using 36-cm capillaries and POP-7™ polymer. The fragment lengths were interpreted using the GeneMapper 4.0 software. PCR products for different plant species were run in separate wells/capillaries. In order to precisely estimate the possible peak overlap between several plant species, their PCR products were run separately and then mixed and run in a single capillary (Fig. 1a). After peak size detection (Fig. 1b, c), the number of fragments and their sizes for different stool samples were compared to our reference database that was constructed using the studied plant species.
Table 1.
The basic database of plant species with the absolute fragment length (in base pair) and size reads (in base pair) and the comparison of plant species in the moose’ diet obtained using different methods
| No. | Species | Absolute fragment length (bp) | Size read (bp) | Detected in studied moose | |
|---|---|---|---|---|---|
| Traditional methods | Molecular methodsa | ||||
| 1 | Abies alba | 86 | ~84 | − | − |
| 2 | Acer spp. | 95 | ~92 | 3 | − |
| 3 | Alnus glutinosa | 100 | ~97 | 1, 2 | + |
| 4 | Betula spp. | 100 | ~97 | 1, 2 | + |
| 5 | Calluna vulgaris | 86 | ~82 | 1, 2 | + |
| 6 | Carex remota | 121 | ~124 | 2 | − |
| 7 | Carpinus betulus | 100 | ~97 | 1, 2 | + |
| 8 | Corylus avellana | 100 | ~97 | 1 | + |
| 9 | Euonymus europaea | 90 | ~86 | 1, 2 | + |
| 10 | Fagus sylvatica | 103 | ~100 | 1 | + |
| 11 | Frangula alnus | 104 | ~101 | 1, 2 | + |
| 12 | Fraxinus excelsior | 78 | ~74 | 1 | − |
| 13 | Juniperus communis | 79 | ~77 | 1, 2 | + |
| 14 | Larix decidua | 85 | ~84 | − | − |
| 15 | Ledum palustre | 85 | ~81 | 1 | − |
| 16 | Ligustrum vulgare | 78 | ~74 | − | − |
| 17 | Padus spp. | 90 | ~87 | 2 | + |
| 18 | Phragmites australis | 91 | ~88 | − | + |
| 19 | Picea abies | 93 | ~91 | 1 | + |
| 20 | Pinus sylvestris | 84 | ~82 | 1, 2 | + |
| 21 | Poa pratensis | 93 | ~118 | 2 | − |
| 22 | Populus spp. | 107 | ~103 | 1, 2 | + |
| 23 | Prunus spp. | 90 | ~86 | 3 | + |
| 24 | Quercus spp. | 93 | ~89 | 1 | + |
| 25 | Ribes spp. | 101 | ~97 | 1, 2 | + |
| 26 | Rubus spp. | 91 | ~88 | 1, 2, 3 | + |
| 27 | Salix spp. | 95 | ~91 | 1, 2 | + |
| 28 | Sambucus racemosa | 89 | ~85 | − | + |
| 29 | Sorbus aucuparia | 90 | ~87 | 1, 2 | + |
| 30 | Syringa vulgaris | 78 | ~74 | − | − |
| 31 | Tilia spp. | 98 | ~95 | 1, 2 | + |
| 32 | Urtica dioica | 73 | ~70 | − | − |
| 33 | Vaccinium myrtillus | 86 | ~83 | 1 | + |
| 34 | Vaccinium uliginosum | 84 | ~81 | 1 | − |
| 35 | Vaccinium vitis-idaea | 89 | ~86 | 1 | + |
| 36 | Viburnum opulus | 79 | ~74 | − | − |
Fig. 1.
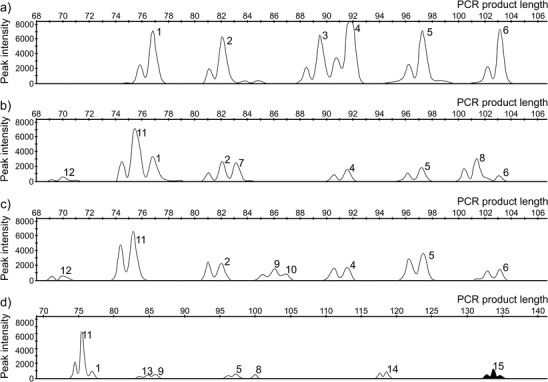
Examples of multipeak profiles obtained after capillary electrophoresis of the PCR products of P6 loop amplified using fluorescence-labeled primers. a Amplified DNA of several plant species mixed and run in a single capillary. b, c Examples of plant species present in moose winter diet based on DNA analysis from feces. d An example of multiplex PCR products using g (FAM) and h and SaF_trnL (VIC) and SaR_trnL primers. Filled peak represents willow (Salix spp.). 1 Juniperus communis; 2 Pinus sylvestris/Calluna vulgaris; 3 Quercus spp.; 4 Picea abies/Salix spp.; 5 Alnus glutinosa, Betula spp., Carpinus betulus, Corylus avellana, and Ribes spp.; 6 Populus spp.; 7 Vaccinium myrtillus; 8 Frangula alnus; 9 Euonymus europaea, Prunus spp., and Vaccinium vitis-idaea; 10 Sorbus aucuparia and Padus spp.; 11 Solanum tuberosum; 12 Urtica dioica; 13 Sambucus racemosa; 14 Poa pratense; 15 Salix spp.
The preliminary analysis showed that the willow and spruce had identical size reads (91 bp), despite obvious differences in sequence composition and absolute fragment lengths (95 and 93 bp, respectively, Table 1). A similar problem with peak overlap occurred for pine and heather (Calluna vulgaris) and several other species (Table 1). To overcome this, we designed species-specific primer pairs for the following gymnosperms: the spruce (PAF_ trnL and PAR_ trnL), the pine (PSF_trnL and PSR_trnL), and the juniper (Juniperus communis) (JCF_trnL and JCR_trnL, Table 2) with the FastPCR software (Kalendar et al. 2009). The same was done for angiosperms: the heather (CVF_trnL and CVR_trnL) and the birch (BeF_trnL and BeR_trnL). The PCR procedure using species-specific primers from stool samples was the same as for g and h primers, but with 42 cycles and annealing at 57 °C for spruce and juniper and 59 °C for pine, heather, and birch (Table 2). The PCR products were visualized on 2 % agarose gel after the 30-min electrophoresis, while DNA sequences were obtained according to Świsłocka et al. (2008).
Table 2.
Sequences of the species-specific primer pairs for gymnosperms and angiosperms species
| Name | Sequence 5′–3′ | Amplified species | Amplified sequence length (bp) |
|---|---|---|---|
| PAF_trnL | ATACTGTATTTATAGAACGCTC | Picea abies | 113 |
| PAR_trnL | TCTTACGAATAGATGCTTGG | Picea abies | 113 |
| PSF_trnL | TGTTTCTTCTCCTAAGATAGGAAGGG | Pinus sylvestris | 172 |
| PSR_trnL | AACGAGTTCTGATCGCCAG | Pinus sylvestris | 172 |
| JCF_trnL | ACGAATCAACATAATTTGGATTGG | Juniperus communis | 260 |
| JCR_trnL | GAAGAGAAGCAGACATTGGTC | Juniperus communis | 260 |
| CVF_trnL | TCGCAAACAAACAAAGATTCCGA | Calluna vulgaris | 99 |
| CVR_trnL | CTTCTACCAACGTAGCCAAATCG | Calluna vulgaris | 99 |
| BeF_trnL | GGATAGGTGCAGAGACTCAATGG | Betula spp. | 194 |
| BeR_trnL | GTCGGTATTGACATGTAGAATGGGAC | Betula spp. | 194 |
| SaF_trnLa | TAATGACAACCGACCCAAATCTGT | Salix spp. | 136-137 |
| SaR_trnL | ATCCTTTCGTCAACTTGGAATCGA | Salix spp. | 136-137 |
aFluorescence-labeled primer using VIC
We also designed a primer pair (SaF_trnL and SaR_trnL, Table 2) to selectively amplify willow. The primer SaF_trnL was fluorescence-labeled using VIC as a dye to test possible multiplexing using universal and species-specific primers in a single PCR. First, we performed PCR using the SaF_trnL and SaR_trnL primer pair under the same conditions as with g and h primers. Then, we simultaneously amplified DNA in a single reaction with the same conditions as before but with 0.2 μM of g and h as well as 0.6 μM of SaF_trnL and SaR_trnL primers. The presence of willow was visualized as a green peak (VIC) and interpreted using the GeneMapper 4.0 software (Fig. 1d).
The repeatability of the methods using universal and species-specific primer pairs was verified by two independent DNA isolations for different feces using two different droppings for a given stool and at least two independent PCRs. We also compared consistency of PCR results for two samples of the same stool: one frozen and one dried in silica gel. Samples stored for 2 weeks and up to 2 years in silica gel were analyzed. These extractions were carried out with QIAamp DNA Stool Mini Kit (QIAGEN). In this way, we compared different storage method of feces and their sensitivity to time lag between sampling and DNA isolation.
Results
Reference database
We obtained expected products for all 36 studied plant species (Table 1). The amplified sequences generally differed in absolute length. Their range varied from 73 bp for nettle (Urtica dioica) to 121 bp for Carex remota. The peak size reads obtained after capillary electrophoresis corresponding to the fluorescent PCR products of the studied plants ranged from 70 to 124 bp.
When the absolute length of the sequence for several species was the same, size reads of these fragments overlapped (for example, spindle [Euonymus europaea], cherry [Prunus spp.], and lingonberry [Vaccinium vitis-idaea]) and such species were not distinguished (Table 1). On the other hand, the plant pairs willow and maple (Acer spp.) as well as spruce and oak (Quercus spp.), despite the fact their sequences had the same absolute length, still could be distinguished from each other as they showed different mobility during electrophoresis resulting from different nucleotide composition (Fig. 1a). Several plants had the same size reads of the P6 loop, despite their sequences differing in the absolute length and base pair composition (for example, spruce and willow, pine and heather; Table 1). In such a case, the species-specific primer pairs were successfully used to detect plant species in winter diet analysis of herbivores from their stools. For example, we obtained PCR products of 172 bp long for DNA isolates from pine needles using the pine-specific primer pair while there were no such products for other plant species, including heather, which overlapped with pine using g and h primers. Thus, for the stool samples, PCR products using species-specific primer pair were only obtained and visualized on agarose gels if the DNA of a given plant species was present in the studied stool sample. Species-specific primers that were designed in this study (Table 2) allowed the unambiguous detection of juniper (260 bp), pine (172 bp), spruce (113 bp), birch (194 bp), heather (99 bp), and willow (136–137 bp) in the studied herbivores’ diet that was not possible using g and h primers. Using willow-specific primers, white poplar (Populus alba) and aspen (Populus tremula) were occasionally visible; however, PCR products for these species differed from willow in size reads (125 and 124 bp, respectively).
The repeatability of the method
Two independent DNA isolations from two different droppings from one moose stool gave identical results. Two independent PCRs conducted, using DNA from each of those two isolations, allowed us to obtain the same plant composition in the diet. However, a time and a method of samples storage proved to be an important factor in the qualitative diet analysis. The analysis of DNA from feces stored for a relatively long period of time (over 2 years) in silica gel that was not changed showed that the resulting number of plant species seen as peaks on electropherogram was reduced by a half when compared to samples isolated shortly after sampling from the field (samples frozen and stored no longer than 1 month). For example, if we obtained six peaks from a given moose stool sample that was frozen, as few as three peaks were usually obtained in the corresponding sample that stayed in silica gel for 2 years prior to DNA extraction. If samples were stored for a short period of time, freezing or drying did not affect the plant species composition as revealed by PCRs.
Analysis of the winter diet
PCR products of plant DNA using g and h primers that were obtained after 35 cycles for each stool sample (moose, red deer, and roe deer) gave in total 26 different peaks corresponding to 26–40 plant species (Tables 1 and 2 of the Supplementary Material). By making the reference database of size reads for the studied plants, we were able to identify at least 72 % of peaks that were amplified from the cervid samples. Seven peaks (55, 59, 72, 78, 79, 80, and 90 bp) were not assigned to any plant species present in our database (Tables 1 and 2 of the Supplementary Material). However, for as many as 93 % of moose samples, all peaks were identified. The peak of 76 bp was detected on electropherograms for all the analyzed droppings of moose, red deer, and roe deer. This peak most likely corresponds to the potato Solanum tuberosum that came from a pill included in QIAamp DNA Stool Mini Kit (QIAGEN) as suggested by Valentini et al. (2009). In addition, the potato gave a faint additional peak of 70 bp that fully overlapped with the nettle peak. The potato and nettle peaks were ignored in diet assessment of cervids. Since potato and juniper peaks were very close (76 and 77 bp, respectively), to avoid the risk of possible errors, it was necessary to perform another PCR using juniper-specific primers.
The moose diet analysis was based on 29 winter fecal samples from the entire range of occurrence of moose in Poland. Fifteen peaks were found that correspond to at least 15 plant species (24 maximum). Among plant species detected from stools, there were gymnosperms and angiosperms that have been previously reported in moose diet (Dzięciołowski 1974; Morow 1976; Belovsky 1981) by traditional methods (Table 1). One peak (59 bp) was not identified. The number of peaks differed between individual stool samples, from 2 to 8. The most common were droppings with two and seven peaks, and plant species composition differed between samples (Table 1 of the Supplementary Material). The most frequent species was pine and five species that had the same size reads: alder (Alnus glutinosa), birch, currant (Ribes spp.), hazel, and hornbeam (Carpinus betulus, present in 69 % of the stools), followed by spruce (65.5 %). In about half of winter fecal samples (55.2 %), willow and alder buckthorn (Frangula alnus, 51.7 %) were present. Other commonly occurring species were also oak (44.8 %), birch (37.9 % estimated using species-specific primers), and blueberry (Vaccinium myrtillus, 34.5 %). Species of moderate occurrence were Padus spp. with rowan, juniper, and poplar (27.6 %). The least frequently consumed species were raspberry (Rubus spp.) and/or reed (Phragmites australis), and red-berried elder (Sambucus racemosa)—occurring in single fecal sample each (Table 1 of the Supplementary Material).
In roe deer diet, we distinguished 19 peaks that correspond to at least 19 plant species (32 maximum). The most frequent was pine (90.9 %) and blueberry (81.8 %). Heather, spindle, cherry, and lingonberry were present in half of the stools. Four peaks (78, 79, 80, and 90 bp) were not identified. In the case of red deer, we also found 19 peaks that correspond to 19 plant species (29 maximum). Again, pine had the highest frequency of occurrences, and it was found in 81.8 % of the stools. The same frequency was found for heather, blueberry, spindle, cherry, and lingonberry. In 63.6 % of the stool samples, there occurred red-berried elder, oak, and species with 97 bp read lengths: birch, hazel, hornbeam, alder, and currant. Birch, distinguished by species-specific primers, was present in 45.5 % of the red deer samples (Table 2 of the Supplementary Material). Six peaks (55, 59, 72, 78, 79, and 90 bp) were not identified.
Discussion
Our study showed that the multipeak profiles obtained after electrophoresis of the fluorescent PCR products using the g and h primers (the P6 loop trnL (UAA) intron) (Taberlet et al. 2007) for fecal samples can be successfully used to identify the winter diet composition of the three cervids. This simple method proved to be reliable and consistent among different PCRs using DNA isolated from the stools of moose, red deer, and roe deer. The method is qualitative, and it allows a quick identification of the majority of tree and shrub species eaten by those herbivores. Due to low costs, it may be an appropriate method for any pilot study that will allow better decision making concerning the necessity (or not) of much more advanced (and expensive) analyses such as NGS (Pompanon et al. 2012). This cost-efficient method could be also a primary step in herbivore diet analysis when no data are available for a given species in a new study area and a large number of samples have to be studied. It could be especially important that if at least two cervid species occur together, it might be interesting to survey if they browse on the same number of tree and shrub species to test the possibility for food niche overlap.
In this method, multiple plant species were successfully amplified within the same PCR from fecal samples because primers used to amplify the P6 loop (g and h) are very well conserved from gymnosperms to angiosperms (Taberlet et al. 2007; Valentini et al. 2009). Moreover, the fragments amplified using these primers for the identified species are very short (70–103 bp, Table 1). Indeed, Taberlet et al. (2007) suggested that the P6 loop will become the marker of choice for degraded DNA and diet studies based on feces (see also Kowalczyk et al. 2011; Pompanon et al. 2012). As many as 15 different peaks were identified from moose stools that correspond to 15–24 plant species (Table 1 of the Supplementary Material), including gymnosperms and angiosperms that have been previously reported in its diet (Dzięciołowski 1974; Morow 1976; Belovsky 1981). Different plant species were also successfully identified in the roe deer and red deer stool samples (19–32 and 19–29, respectively). After electrophoresis of PCR products, none of the samples gave a single peak, indicating diverse diets and providing evidence for the sensitivity and utility of the method.
Unfortunately, the method has several drawbacks. Taberlet et al. (2007) pointed out that the resolution of the trnL (UAA) intron is relatively low. Indeed, we were not able to distinguish between, for example, hazel, hornbeam, currant, and alder due to peak overlap of identical sequences. Moreover, despite some sequences differing in both size and nucleotide composition, we observed peak overlaps for spruce/willow, pine/heather, and reed/raspberry species pairs. Although reed and raspberry rarely occurred in the studied cervids’ diets, the pine/heather and spruce/willow overlap challenges moose diet analysis considerably.
The moose affects timber production by browsing on pine stands, and reliable data on diet analysis are of primary importance. Due to large uncertainty associated with peaks overlapping when using universal (g and h) primers, we strongly recommend performing agarose gel electrophoresis of PCR products and/or classical sequencing approaches based on plant-specific primers designed for pine, heather, spruce, willow, and birch as well as for juniper due to the proximity of the potato peak on electropherograms. In order to further simplify this method, one should consider a multiplex PCR approach using a set of g and h and species-specific primers (fluorescence-labeled with different dyes) in a similar way as it was successfully applied in this study for willow (Fig. 1d). This may allow proper estimation of the percentage of occurrences in stools for the above-mentioned problematic plant species. Such traditional approaches are still used to complement the NGS methodology (Pompanon et al. 2012).
Another difficulty comes from the fact that kits for DNA isolation from stools may often contain plant DNA. Indeed, in all the studied samples of moose, red deer, and roe deer isolated using QIAamp DNA Stool Mini Kit (QIAGEN), we found a peak of 76 bp that most probably corresponds to the potato. Since the size of a peak of juniper is 77 bp, the presence of potato makes the peak count more difficult, especially when the juniper peak is not very high. The potato peaks (70 and 76 bp) were absent in PCR products on electropherograms when plant DNA was isolated from stools using DNeasy Blood & Tissue Kit (QIAGEN). Thus, caution should be taken when QIAamp DNA Stool Mini Kit (QIAGEN) is used as it may cause a potato peak (Valentini et al. 2009) overlap with other plant species eaten by a given herbivore. Caution should also be taken as DNA isolated from stools stored in silica gel, which were stored over 2 years and did not change, always yielded considerable losses of peak numbers (up to 50 %) when compared to stools that were isolated immediately after collection or frozen. Due to utilization of degraded DNA from stools and peak overlap among some plant species, we strongly recommend to perform at least two independent multiplex PCR reactions with g and h and species-specific (fluorescence-labeled) primers to obtain reliable results.
This study is the first DNA-based approach of moose, red deer, and roe deer winter diet in Poland. As the number of species eaten from spring to autumn may be larger and the method was not tested for other stools than those collected in winter, future work is needed to test its applicability for diet analysis in other seasons. The method seems however to be a very valuable tool in forest and conservation management as the vast majority of forest damage is done by herbivores in winter (Dzięciołowski and Pielowski 1975; Bobek et al. 1992). In addition, in winter, the studied species occupy the same habitats; thus, one should expect more niche overlap among them. Indeed, our study demonstrated that pine was one of the most frequent plant species eaten by all three herbivores: moose, roe deer, and red deer. This study based on a limited number of stool samples may confirm moose preferences to pine in winter (Morow 1976). However, the number of deciduous plant species in winter moose diet seems to be surprisingly high. Future research should concentrate on moose diet analysis in different areas of the species’ occurrence, including those with extensive forest damage and those without any apparent damage made by the species. Since the method offers fast and cost-efficient investigation of several issues concerning the impact of herbivores on ecosystems, it may be used for the rapid collection of sound data sets from different ecosystems and on large spatial scales. Adjusting forest management to the occurrence of moose will be of primary importance in lowering the conflict between abundant moose and timber production.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 173 kb)
Acknowledgments
We are grateful to the Head of the Polish State National Forest Holding for kindly allowing us to collect moose stool samples from Poland and Ada Wróblewska for the help in plant species identification. The moose winter diet study was financed by the Ministry of Science and Higher Education (N N304 280940 to M. Ratkiewicz) and University of Białystok for the other cervid species (BMN 164 to M. Czernik).
References
- Belovsky GE. Food plant selection by a generalist herbivore: the moose. Ecology. 1981;62:1020–1030. doi: 10.2307/1937001. [DOI] [Google Scholar]
- Bobek B, Morow K, Perzanowski K, Kosobucka M (1992) Jeleń. Monografia przyrodniczo – łowiecka. Wydawnictwo Świat, Warsaw
- Cederlund G, Nyström A. Seasonal differences between moose and roe deer in ability to digest browse. Holarct Ecol. 1981;4:59–65. [Google Scholar]
- Danell K, Bergström R, Edenius L. Effects of large mammalian browsers on architecture, biomass and nutrients of woody plants. J Mammal. 1994;75:833–844. doi: 10.2307/1382465. [DOI] [Google Scholar]
- Dzięciołowski R. Selection of browse twigs by moose. Acta Theriol. 1974;19:273–281. [Google Scholar]
- Dzięciołowski R, Pielowski Z (1975) Łoś. Państwowe Wydawnictwo Rolnicze i Leśne, Warsaw
- Gębczyńska Z. Food of the roe deer and red deer in the Białowieża Primeval Forest. Acta Theriol. 1980;25:487–500. [Google Scholar]
- Gębczyńska Z, Raczyński J. Distribution, population structure, and social organization of moose in the Biebrza Valley, Poland. Acta Theriol. 1989;34:195–217. [Google Scholar]
- Gębczyńska Z, Raczyński J. Problems of protection and management of the moose and other game in the Biebrza National Park. Park Nar Rezerw Przyr. 1993;12:5–36. [Google Scholar]
- Graham RT, Jain TB, Kingery JL. Ameliorating conflicts among deer, elk, cattle and/or other ungulates and other forest uses: a synthesis. Forestry. 2010;83:245–255. doi: 10.1093/forestry/cpq003. [DOI] [Google Scholar]
- Hofmann RR. Evolutionary steps of ecophysiological adaptation and diversification of ruminants: a comparative view of their digestive system. Oecologia. 1989;78:443–457. doi: 10.1007/BF00378733. [DOI] [PubMed] [Google Scholar]
- Holechek JL, Varva M, Pieper RD. Botanical composition determination of range diets: a review. J Range Manag. 1982;35:309–315. doi: 10.2307/3898308. [DOI] [Google Scholar]
- Hundertmark KJ, Shields GF, Udina IG, Bowyer RT, Danilkin AA, Schwartz CC. Mitochondrial phylogeography of moose (Alces alces): late Pleistocene divergence and population expansion. Mol Phylogenet Evol. 2002;22:375–387. doi: 10.1006/mpev.2001.1058. [DOI] [PubMed] [Google Scholar]
- Jędrzejewski W, Sidarowicz V. The art of tracking animals. Białowieża: Mammal Research Institute Polish Academy of Sciences; 2010. [Google Scholar]
- Kalendar R, Lee D, Schulman AH. FastPCR software for PCR primer and probe design and repeat search. Genes, Genomes and Genomics. 2009;3:1–14. [Google Scholar]
- Komenda E (2001) Wpływ wzbogacania bazy żerowej na populację łosi i drzewostany Doliny Biebrzy. Dissertation, Warsaw University of Life Sciences - SGGW
- Kowalczyk R, Taberlet P, Coissac E, Valentini A, Miquel C, Kamiński T, Wójcik JM. Influence of management practices on large herbivore diet—case of European bison in Białowieża Primeval Forest (Poland) For Ecol Manag. 2011;261:821–828. doi: 10.1016/j.foreco.2010.11.026. [DOI] [Google Scholar]
- Morow K. Food habits of moose from Augustów Forest. Acta Theriol. 1976;21:101–116. [Google Scholar]
- Pompanon F, Deagle BE, Symondson WOC, Brown DS, Jarman SN, Taberlet P. Who is eating what: diet assessment using next generation sequencing. Mol Ecol. 2012;21:1931–1950. doi: 10.1111/j.1365-294X.2011.05403.x. [DOI] [PubMed] [Google Scholar]
- Putman RJ. Ungulates in temperate forest ecosystems: perspectives and recommendations for future research. For Ecol Manag. 1996;88:205–214. doi: 10.1016/S0378-1127(96)03878-9. [DOI] [Google Scholar]
- Siuda A, Żurowski W, Siuda H. The food of the roe-deer. Acta Theriol. 1969;14:247–262. [Google Scholar]
- Świsłocka M, Ratkiewicz M, Borkowska A, Komenda E, Raczyński J. Mitochondrial DNA diversity in the moose, Alces alces, from northeastern Poland: evidence for admixture in a bottlenecked relict population in the Biebrza valley. Ann Zool Fenn. 2008;45:360–365. doi: 10.5735/086.045.0419. [DOI] [Google Scholar]
- Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucl Acids Res. 2007;35:1–8. doi: 10.1093/nar/gkl938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini A, Miquel C, Nawaz MA, Bellemain E, Coissac E, Pompanon F, Gielly L, Cruaud C, Nascetti G, Wincker P, Swenson JE, Taberlet P. New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: the trnL approach. Mol Ecol Resour. 2009;9:51–60. doi: 10.1111/j.1755-0998.2008.02352.x. [DOI] [PubMed] [Google Scholar]
- Wam HK, Hjeljord O. Moose summer and winter diets along a large scale gradient of forage availability in southern Norway. Eur J Wildl Res. 2010;56:745–755. doi: 10.1007/s10344-010-0370-4. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 173 kb)


