Abstract
The polyadenosine RNA binding protein, polyadenylate-binding nuclear protein 1 (PABPN1), plays key roles in post-transcriptional processing of RNA. Although PABPN1 is ubiquitously expressed and presumably contributes to control of gene expression in all tissues, mutation of the PABPN1 gene causes the disease Oculopharyngeal Muscular Dystrophy (OPMD), in which a limited set of skeletal muscles are effected. A major goal in the field of OPMD research is to understand why mutation of a ubiquitously expressed gene leads to a muscle-specific disease. PABPN1 plays a well-documented role in controlling the poly(A) tail length of RNA transcripts but new functions are emerging through studies that exploit a variety of unbiased screens as well as model organisms. This review addresses: (1) the molecular function of PABPN1 incorporating recent findings that reveal novel cellular functions for PABPN1 and (2) the approaches that are being used to understand the molecular defects that stem from expression of mutant PABPN1. The long-term goal in this field of research is to understand the key molecular functions of PABPN1 in muscle as well as the mechanisms that underlie the pathological consequences of mutant PABPN1. Armed with this information, researchers can seek to develop therapeutic approaches to enhance the quality of life for patients afflicted with OPMD.
Introduction
The function and fate of all cells is determined by their gene expression profile. While gene transcription is critical, post-transcriptional events also influence both spatial and temporal control of gene expression. Nascent messenger RNAs undergo extensive modification including capping, splicing and 3′-end processing. The 3′-end processing of transcripts involves a site-specific endonucleolytic cleavage followed by the addition of adenosine residues to the cleaved product, generating a polyadenosine or poly(A) tail. These tightly coupled and highly regulated events require the assembly and function of multiple protein complexes to yield a correctly cleaved and polyadenylated 3′-end. The poly(A) tails generated on mRNA transcripts are critical for regulating the fate of RNAs through modulating post-transcriptional processes including transcript stability, nuclear export and translation [1, 2] largely through interactions with poly(A) RNA binding proteins [3, 4]. These poly(A) binding proteins decorate poly(A) tails from the time of their synthesis in the nucleus until translation of the RNA in the cytoplasm. While a cytoplasmic poly(A) binding protein, PABPC1, mediates cytoplasmic effects on RNA stability and directly modulates translation [5], the nuclear poly(A) binding protein, PABPN1, plays multiple roles in modulating the fate of RNA transcripts [6–11]. Early biochemical characterization of PABPN1 revealed a key role in ensuring proper polyadenylation [6, 9], but emerging evidence supports additional cellular roles for PABPN1 [12, 13].
Understanding the function of PABPN1 specifically in skeletal muscle is particularly critical as mutation of the PABPN1 gene results in the muscle-specific disease, Oculopharyngeal Muscular Dystrophy (OPMD) [14]. OPMD usually presents in the fifth decade of life with its main symptoms being eyelid drooping and difficulty in swallowing due to weakness in the levator palpebrae superioris and pharyngeal muscles, respectively [15]. Depending on the specific patient population studied, additional weakness is noted in proximal limb, facial and extraocular muscles [16, 17]. Although life expectancy seems not to be shortened, frequent aspiration pneumonia, together with malnutrition due to difficulties in swallowing, significantly impair the quality of life of patients [18]. Autosomal dominant OPMD, which is by far the most prevalent form of the disease [14, 19, 20], results from a GCN expansion within the coding region of the PABPN1 gene that creates a short alanine expansion (10 alanines expanded to 12–17 alanines) in the N-terminal domain of PABPN1. How this modest change in a ubiquitously expressed protein that plays key roles in post-transcriptional regulation of gene expression leads to a muscle disease that impacts a specific subset of muscles remains a major question in the field of OPMD research. The lack of a comprehensive understanding of the function and regulation of PABPN1 is one of the underlying causes behind the unanswered questions regarding the pathogenesis and tissue-specificity of OPMD. In this review we will first consider the function of PABPN1 including information that has emerged from studies of nuclear poly(A) binding proteins in model organisms. We then consider how ubiquitous expression of mutant PABPN1 could give rise to a tissue-specific disease. Ultimately, understanding the function of PABPN1 particularly in skeletal muscle could yield insight into the molecular events that underlie muscle pathology in OPMD and suggest therapeutic strategies for this devastating disease.
PABPN1 Function
Over the years a number of approaches have been employed to analyze the function of PABPN1. Early studies revealed that PABPN1 modulates polyadenylation through enhancing the processivity of the poly(A) polymerase (PAP) [6, 9, 10]. However, a number of recent studies have revealed new functions of PABPN1. A detailed understanding of the cellular role of PABPN1, especially in the muscles affected in OPMD, will provide insight not only into the function of a key regulator of post-transcriptional RNA processing and hence, a general modulator of gene expression, but also into the cellular processes that could be altered in OPMD.
Domain structure of PABPN1
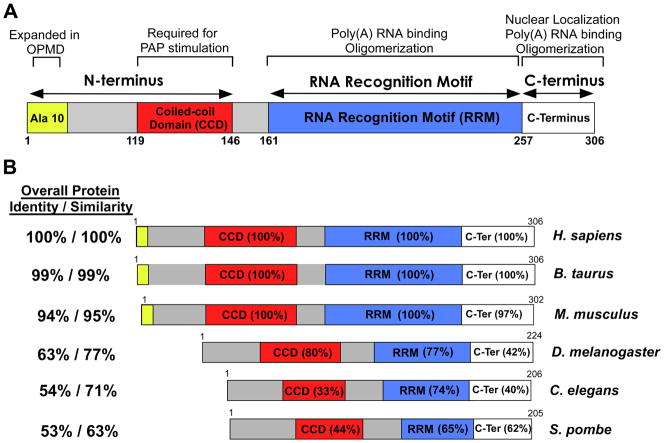
A first step toward considering protein function is identifying the key domains within that protein and understanding what property those domains might confer on the protein of interest. The domain structure of the human PABPN1 protein is shown in Figure 1A. Studies over the years have analyzed PABPN1 and related nuclear poly(A) binding proteins using a variety of different approaches ranging from biochemical studies with purified proteins to use of genetic model organisms. The domain structures of the proteins from different species that have been exploited to provide insight into the function of PABPN1, together with their amino acid identity compared to human PABPN1, are shown in Figure 1B. These proteins include bovine and mouse PABPN1, which are nearly identical to human PABPN1, as well as more distantly related proteins in genetic model organisms including Drosophila melanogaster, Caenorhabditis elegans, and the fission yeast, Schizosaccharomyces pombe (Fig. 1B).
Fig. 1. Domain structures and comparison of nuclear poly(A) binding proteins.
(A) Domain structure of human PABPN1. PABPN1 is made up of three major domains: 1) an N-terminal domain that contains a stretch of 10 alanines that is expanded to between 12 and 17 alanines in autosomal dominant OPMD and a coiled-coiled region required for binding to the poly(A) polymerase (PAP); 2) a single RNA Recognition Motif (RRM) that is required for poly(A) RNA binding and also contributes to oligomerization; and 3) a C-terminal region that contains a nuclear localization signal and contributes to PABPN1 oligomerization. The function(s) attributed to each domain is (are) indicated at the top. (B) Alignment of domain structures of human PABPN1 with the various nuclear poly(A) binding proteins from different organisms that have been studied. Both bovine (B. taurus) and murine (M. musculus) PABPN1 are shown as are nuclear poly(A) binding proteins from D. melanogaster (PABP2), C. elegans (PABP-2), and S. pombe (Pab2). Of the proteins shown, only the human, bovine and mouse PABPN1 proteins contain the N-terminal stretch of alanines that is expanded in OPMD. The Overall Protein Identity/Similarity as compared to human PABPN1 is indicated to the left of each domain structure. The similarity between the domains of different nuclear poly(A) binding proteins and the corresponding domain of human PABPN1 is indicated as percent amino acid identity within each domain.
The PABPN1 protein can be divided into three major domains: an acidic N-terminus; a central ribonucleoprotein-type RNA binding motif (RRM); and a basic arginine-rich C-terminus (Fig. 1A). Within the N-terminal domain, the initiating methionine is followed immediately by a stretch of 10 alanines. In autosomal dominant OPMD, the most common form of the disease [14, 19, 20], this alanine stretch is expanded to 12–17 alanines [14]. The N-terminal domain also contains a coiled-coil region, which is essential for the interaction of PABPN1 with the poly(A) polymerase (PAP) and hence, for modulating polyadenylation [9].
PABPN1 contains a single RNA recognition motif (RRM), which mediates high affinity binding to poly(A) RNA [21]. Although no crystal structure is available for the intact PABPN1 protein, the RRM of PABPN1 has been crystalized at 2Å resolution [22]. While the RRM of PABPN1 is very different in sequence from most other known RRMs, including those found in PABPC1 (~20% similarity), it adopts a typical RRM fold, which consists of four-stranded antiparallel beta-sheets coupled with two perpendicular alpha-helices [22]. The only potential insight into how PABPN1 could recognize polyadenosine RNA comes from analysis of an embryonic cytoplasmic poly(A) binding protein in Xenopus laevis called embryonic poly(A) binding protein 2 (ePABP2) [23]. Although the function of this specialized poly(A) binding protein has not been fully characterized, ePABP2 has an RRM motif similar to that found in PABPN1. A solution structure for the ePABP2 RRM in complex with RNA [24] reveals some details of how this type of RRM could recognize polyadenosine RNA.
The C-terminal domain of PABPN1 contains no obvious motif but functional characterization reveals that this domain cooperates with the RRM to mediate RNA binding and also plays a role in the oligomerization of PABPN1 molecules [22, 25]. In addition, the C-terminal domain contains the nuclear localization signal (NLS) as well as arginine residues that are asymmetrically dimethylated [26, 27]. Methylation of these arginine residues has no effect on PABPN1 binding to RNA, but appears to modulate the nuclear accumulation of PABPN1 by weakening the interaction between PABPN1 and the PABPN1 nuclear import receptor, transportin [28, 29].
Mammalian PABPN1
Biochemical analyses
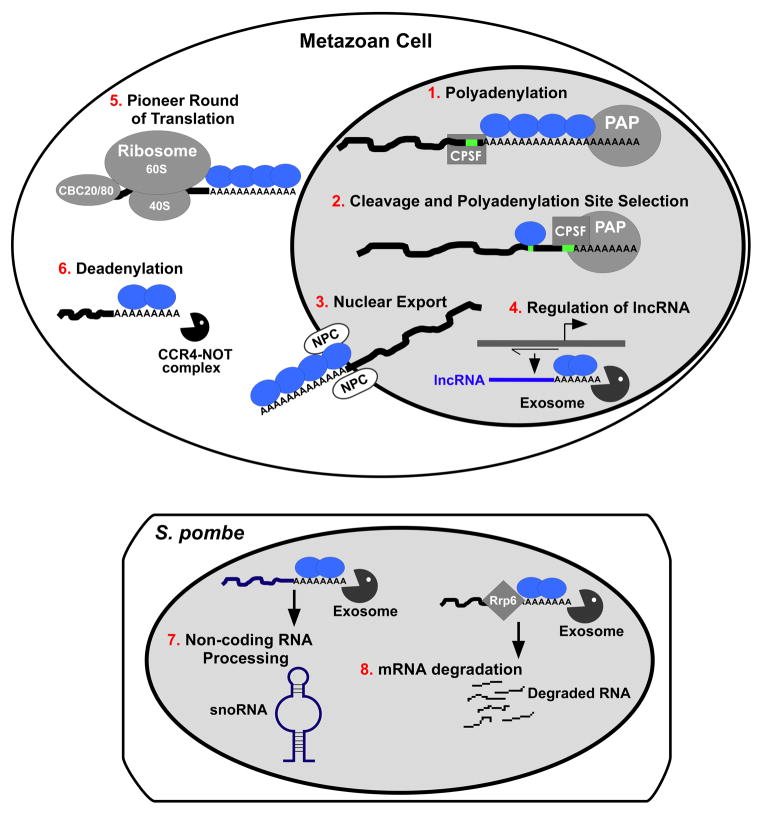
As illustrated in Figure 2, studies in mammalian cells implicate PABPN1 in a number of key post-transcriptional functions within the nucleus including control of poly(A) tail length (Fig. 2-1) and poly(A) RNA export (Fig. 2-3). More recently, PABPN1 has been implicated in poly(A) cleavage site selection (Fig. 2--2) and regulation of long non-coding RNAs (lncRNA) (Fig. 2-4) [11–13].
Fig. 2. Schematic of PABPN1 functions identified in metazoan cells and fission yeast.
In metazoan cells (top), PABPN1 (represented as blue circles) is implicated in functions within both the nucleus and the cytoplasm. Nuclear functions linked to PABPN1 include: (1) binding to the poly(A) tail of nascent mRNA transcripts to stimulate polyadenylation by the poly(A) polymerase (PAP) [9] in conjunction with the cleavage and polyadenylation specificity factor (CPSF); (2) modulating alternative polyadenylation through binding to and masking “weak” proximal cleavage/polyadenylation sites allowing the cleavage and polyadenylation machinery to assemble at distal “stronger” sites [12]; (3) following polyadenylation, accompanying the transcript to the nuclear pore complex (NPC) and directly or indirectly aiding in nuclear RNA export [11, 66]; (4) regulating the steady-state level of long non-coding RNAs (lncRNA) by mediating their turnover via the nuclear exosome [13]. In the cytoplasm, functions linked to PABPN1 include: (5) contributing to a pioneer round of translation that is vital for mRNA quality control [5] where the transcript is associated with the nuclear cap binding complex (CBC) consisting of CBC20 and CBC80 as well as the ribosome after which PABPN1 is replaced by cytoplasmic PABPC1; and (6) as identified only in Drosophila thus far, cytoplasmic deadenylation of transcripts via the CCR4-NOT complex [41]. In the fission yeast S. pombe (bottom), the nuclear poly(A) binding protein, Pab2, is involved in (7) processing of small nucleolar RNAs [46] and (8) turnover of meiotic transcripts via the nuclear exosome [49].
Early biochemical experiments defined a role for mammalian PABPN1 in stimulating polyadenylation by the poly (A) polymerase (PAP) (Fig. 2-1) [6, 9]. Assays using purified mammalian proteins demonstrate that PABPN1 binds the growing mRNA tail after the distributive addition of ~10 adenosines by PAP. PABPN1 physically interacts with PAP resulting in a marked increase in the affinity of PAP for RNA without a change in PAP catalytic activity [9] resulting in a stimulation of the processivity of PAP. Structure-function studies have revealed important insights into how PABPN1 stimulates processive polyadenylation. RNA binding mutants of PABPN1, including those with amino acid changes in the RRM (Y175A or F215A) that decrease RNA binding, are unable to increase PAP affinity for RNA and as a result cannot stimulate polyadenylation, which provides evidence that PABPN1 tethers PAP to RNA as a mode of regulating poly(A) tail length [10]. Further studies provide evidence that the N-terminal domain of PABPN1 including the coiled-coil region interacts with PAP. Amino acid changes in this domain, especially at leucine 136, eliminate the capacity of PABPN1 to stimulate processive polyadenylation by PAP without altering the affinity of PABPN1 for poly(A) RNA [9]. Taken together, such studies support a model where PABPN1 binds to poly(A) RNA through the single RRM with a contribution from the C-terminal domain, while interaction with the poly(A) polymerase (PAP) occurs via the coiled-coil domain in the N-terminal region of PABPN1 (Fig. 1A). Both these interactions are consistent with an in vivo role for PABPN1 in stimulating polyadenylation of nascent transcripts by PAP. However, given new functions emerging for PABPN1 as described below and summarized in Figure 2, continued structure-function analyses will be required to understand the molecular interactions that are critical to the function of PABPN1.
In addition to directly interacting with PAP to modulate polyadenylation, PABPN1 also may indirectly modulate poly(A) tail length through facilitating an ongoing interaction between PAP and the cleavage and specificity factor (CPSF) [10]. This “molecular ruler” model posits that PABPN1 effectively counts the number of adenosines incorporated into the mRNA tail and terminates processive polyadenylation when a threshold size is reached [10]. One possible molecular explanation consistent with this model is that PABPN1 coats the elongating poly(A) tail, forming a spherical particle that allows the RNA to fold back and maintain an interaction between CPSF and PAP. On reaching a threshold length of ~250 adenosines, the structure of this polyadenylation complex is disrupted, and PABPN1 can no longer facilitate the interaction between CPSF and PAP terminating the processive phase of polyadenylation [10]. Support for this model comes from electron microscopy and scanning force microscopy images using synthetic poly(A) RNA and purified PABPN1 which visualized spherical particles that form in the presence of both PABPN1 and poly(A) RNA [30].
Taken together, these biochemical studies have provided important insight into how PABPN1 can modulate poly(A) tail length, first enhancing processivity of PAP and then, following addition of a certain number of As, decreasing PAP processivity. Thus PABPN1 plays a key role in post-transcription regulation of gene expression. However, additional studies are required to understand the details of PABPN1 function in the cell nucleus. Several recent studies have revealed new roles for PABPN1 that now must be integrated with our understanding of the role that PABPN1 plays in modulating polyadenylation.
Cellular analyses of PABPN1 function
Although the biochemical work done to characterize PABPN1 is extremely informative, characterization of PABPN1 function in muscle cells, the cell type affected in OPMD, is essential. Depleting PABPN1 in primary muscle cells in culture using siRNA results in decreased cell proliferation and defects in differentiation [11]. Consistent with the characterized role of PABPN1 in polyadenylation, a significant decrease in poly(A) tail length of bulk RNA as well as poly(A) tail length of specific transcripts examined was also detected (Fig. 2-1). An additional consequence noted was the nuclear accumulation of poly(A) RNA suggesting a defect in poly(A) RNA export from the nucleus (Fig. 2-3) [11]. This finding could indicate that PABPN1 plays a direct role in the export of poly(A) RNA from the nucleus, a function that could be supported by the previous finding that PABPN1 shuttles between the nucleus and the cytoplasm [8]. Alternatively, defects in polyadenylation could impair upstream RNA processing steps required for efficient poly(A) RNA export leading to nuclear accumulation as an indirect consequence of decreased PABPN1 function. Importantly, the defects in proliferation observed in primary muscle cells were also detected in a fibroblast cell line [11], highlighting the critical importance of understanding what makes muscle susceptible to expression of mutant PABPN1. Further studies will be required to understand the direct roles of PABPN1 in RNA biogenesis as well as the precise roles in muscle.
As a complement to examining the conventional role of PABPN1 in poly(A) tail length control, a recent unbiased RNAi-based candidate screen for regulators of alternative polyadenylation in the HeLa cancer cell line identified PABPN1 as the top hit among the RNA binding proteins examined [12]. Alternative polyadenylation plays a key and increasingly appreciated role in modulating gene expression as any change in the length of the 3′-UTR of a transcript can alter interactions with RNA binding proteins as well as microRNAs (miRNAs) significantly impacting transcript stability and translation [31]. Further analysis of the role of PABPN1 in alternative polyadenylation uncovered a link between PABPN1 and alternative polyadenylation/cleavage site selection in muscle cells and also identified changes in polyadenylation site selection in the muscle tissue of an OPMD mouse model (Fig 2--2) [12, 32]. Depletion of PABPN1 results in a significant shift in cleavage/polyadenylation site selection from distal sites to more proximal sites [12, 32]. The selection of a proximal polyadenylation site alters the post-transcriptional control of transcripts by abrogating the susceptibility to miRNAs that have binding sites downstream of the proximal polyadenylation signal [12]. The importance of alternative polyadenylation has recently been demonstrated in regulation of muscle stem cells through modulating miRNA-mediated regulation of the transcript encoding a key myogenic regulator [33]. A model has been proposed in which PABPN1 binds and masks proximal “weak” polyadenylation sites, enhancing the use of distal canonical polyadenylation sites [12]. Alternative polyadenylation is critical for regulation of gene expression [34, 35] and misregulation of transcripts critical for the maintenance and regeneration of skeletal muscles could contribute to OPMD pathogenesis.
In addition to modulating post-transcriptional regulation of messenger RNAs, PABPN1 has recently been implicated in the processing of long non-coding RNAs (lncRNAs) (Fig 2-4) [13]. Non-coding RNAs are increasingly being linked to the regulation of gene expression in a wide variety of tissues and in human disease [36–38]. With the goal of identifying genes regulated by PABPN1, Bachand and coworkers performed a global analysis of gene expression in a HeLa cell line depleted of PABPN1 [13]. No change in bulk poly (A) tail length was reported when PABPN1 was depleted in this cell type [13, 39] lending support to the idea that PABPN1 may have distinct roles in different cell types. Conclusions drawn from studies using different cell types can be influenced by the cell-specific spectrum of transcripts affected or examined. In this particular study, depletion of PABPN1 in HeLa cells resulted in a significant increase in the steady-state level of about 13% of the known lncRNAs, which is significantly higher when compared to altered expression of only approximately 4% of protein coding genes in these cells [13]. The lncRNAs that accumulate upon PABPN1 depletion are predominantly located close to transcription start sites of downstream genes and are almost exclusively transcribed in an orientation antisense to those downstream genes suggesting that the lncRNAs affected by PABPN1 could modulate expression of neighboring transcripts. PABPN1-mediated negative regulation of specific lncRNAs is dependent upon the activity of the evolutionarily conserved nuclear exosome, a complex of 3′-5′ riboexonucleases responsible for processing and degradation of much of cellular RNA [13, 40]. Misregulation of specific PABPN1-dependent lncRNAs necessary for normal muscle function could contribute to the pathology in OPMD.
Roles for PABPN1 have also been identified in the cytoplasm (Fig. 2). One study implicated PABPN1 in the first round of translation of transcripts in the cytoplasm, termed the pioneer round of translation (Fig 2-5) [5]. This study presented the idea that PABPN1 accompanies transcripts from the nucleus to the cytoplasm and is then replaced by the conventional cytoplasmic PABPC1 following the first or pioneer round of translation. As the pioneer round of translation provides the substrate for a form of RNA quality control termed nonsense-mediated decay, where mRNAs with premature stop codon are targeted for degradation [5], this role would implicate PABPN1 in this form of RNA quality control. Another cytoplasmic function that has thus far only been defined in the cytoplasm of Drosophila oocytes is deadenylation via the CCR4-NOT complex (Fig 2-6) providing a mechanism to regulate translation of specific transcripts in the cytoplasm of these oocytes [41]. Further studies will be required to refine our understanding of cytoplasmic PABPN1 functions.
Unbiased approaches have expanded the functional repertoire of PABPN1 from stimulating canonical polyadenylation of messenger RNAs to previously unknown roles in alternative polyadenylation and regulation of non-coding RNAs highlighting the need to fully characterize the spectrum of PABPN1 functions. Many of the proteins involved in polyadenylation and other steps of 3′-end processing are evolutionarily conserved [42, 43]. Therefore, one approach to identify functions of the nuclear poly(A) binding proteins including PABPN1 is to exploit information gained from studies of these proteins in genetically tractable model organisms.
Nuclear Poly(A) Binding Proteins: What Can We Learn from Model Organisms?
As shown in Figure 1B, S. pombe, C. elegans, and D. melanogaster each express a nuclear-localized poly(A) binding protein that shares the PABPN1 domain structure. Molecular studies in these genetically tractable model organisms have provided insight into the function and regulation of nuclear poly(A) binding proteins. Some of the new functions that have been uncovered from these model systems might be relevant to PABPN1 function in muscle cells and could provide clues to the pathogenesis of OPMD. However, the specialized roles of these proteins may have changed or been adopted by other nuclear poly(A) binding proteins over the course of evolution. Thus, the insight gained should be considered with the caveat that the respective poly(A) binding proteins from these organisms have not yet been demonstrated to be true functional orthologs of PABPN1.
Pab2 in Schizosaccharomyces pombe
While no apparent ortholog of PABPN1 is found in Saccharomyces cerevisiae, a nuclear poly(A) binding protein from the fission yeast Schizosaccharomyces pombe, Pab2, has been studied extensively [44–49]. As shown in Figure 1B, Pab2 shares 53% identity and 63% similarity with human PABPN1, with a similar coiled-coil domain, RRM and an arginine rich C-terminus that contains a nuclear localization signal [47]. The pab2 gene is not essential in S. pombe but deletion of pab2 (Δpab2) results in cold-sensitive cell growth. In contrast to results in muscle cells, deletion of pab2 causes hyperadenylation of RNAs [45] perhaps suggesting differential regulation of specific RNA targets. Pab2, which is co-transcriptionally recruited to specific transcripts prior to initiation of polyadenylation regulates both the biogenesis and stability of these target transcripts [44]. Additional studies demonstrate that Pab2 interacts with components of the nuclear exosome (Rrp6 and Dis3) and plays a role in the synthesis of small non-coding nucleolar RNAs (snoRNA) (Fig. 2-7). Genome-wide transcriptome analysis of Δpab2 cells shows that the steady-state level of the majority of transcripts do not change significantly upon loss of Pab2 [46]; however, many snoRNAs are upregulated in the absence of Pab2. These snoRNAs have poly(A) tails much longer (~300 adenosines) than in wildtype cells, which could explain the hyperadenylation observed in Δpab2 cells [45, 46]. Pab2 also plays a role in turnover of inefficiently spliced mRNA transcripts [48]. In a further link to control of RNA levels, the interaction of Pab2 with the nuclear exosome is vital for the negative regulation of meiotic transcripts during mitosis in S. pombe (Fig. 2-8). Pab2, in conjunction with another protein, Mmi1, and the nuclear exosome, directs polyadenylation-mediated degradation of meiotic transcripts in mitotic yeast [49, 50]. The association of Pab2 with the nuclear exosome to modulate levels of non-coding RNAs as well as inappropriately-expressed meiotic transcripts is reminiscent of recent data implicating mammalian PABPN1 in regulating lncRNAs [13]. This common function in targeting RNAs for destruction could reveal a general role for Pab2/PABPN1 in ensuring that aberrant or inappropriate RNAs are removed from the cell imparting quality control on the cellular RNA pool.
PABP-2 in Caenorhabditis elegans
PABP-2, the PABPN1-related nuclear poly(A) binding protein in C. elegans (Fig. 1B), was originally identified in a reverse screen for suppressors of the loss of function lethality of let-7, a highly conserved developmentally essential miRNA [51]. Depletion of PABP-2 by RNA interference rescues the phenotypes associated with loss of let-7 function [52]. The precise mechanism underlying this suppression is not well understood since PABP-2 depletion does not change the levels of let-7 or downstream targets [52]. Subsequent studies of PABP-2 function revealed that worms deleted for PABP-2 die in utero or at an early larval stage, indicating a critical developmental role for PABP-2 in larval development [52]. PABP-2 expression is temporally regulated, with protein levels decreasing significantly at later stages of larval development. Loss of function of PABP-2 also causes premature differentiation of the stem cell-like seam cells that are critical for larval development. C. elegans PABP-2 lacks the N-terminal region that carries the polyalanine stretch expanded in OPMD; however, the protein does have homology to human PABPN1 in the coiled-coiled domain, RRM and the C-terminus (Fig. 1B). Despite this homology, developmental defects are the only phenotype associated with PABP-2 loss of function, with no significant effect on steady state mRNA levels and/or global translation. Detailed characterization of the effects of PABP-2 depletion on poly(A) tail length, nuclear export and alternative polyadenylation would yield information about whether this protein modulates post-transcriptional processing in ways similar to those described for other nuclear poly(A) binding proteins. In addition, understanding the nature of the genetic suppression of let-7 may provide clues into the function of PABP-2 in C. elegans and thus yield insight into whether information that emerges from studies in this model could inform studies of human PABPN1.
PABP2 in Drosophila melanogaster
The Drosophila melanogaster nuclear poly(A) binding protein, PABP2, shows considerable similarity to human PABPN1 (Figure 1B). Like PABPN1, PABP2 contains a coiled-coiled domain, a single RRM, and a C-terminal domain containing multiple arginine residues. Consistent with biochemical studies of mammalian PABPN1, PABP2 can stimulate polyadenylation by PAP as assessed with purified recombinant proteins [53]. PABP2 is a primarily nuclear protein that is expressed throughout the life cycle of flies and is essential for viability [41, 53]. Flies lacking PABP2 (Pabp255, a null allele) die in late embryonic or early larval stages of development. Interestingly, a mutant (I61S) form of PABP2 (analogous to the RNA binding mutant, L136S in human PABPN1 [9]), which does not interact with or stimulate PAP, cannot rescue the lethality of the null allele. This result indicates that the interaction of PABP2 with PAP is required for the essential function of PABP2 in flies. There is a significant shortening of poly(A) tails of both specific mRNA transcripts and bulk poly(A) RNA in Pabp2 null larvae, indicating that PABP2 stimulates polyadenylation in flies [41]. In addition to this nuclear role in polyadenylation, PABP2 also has an important role in the cytoplasm of Drosophila oocytes and early embryos that appears to be in opposition to its nuclear function (Fig. 2-6). PABP2 along with the Drosophila PABPC1 homolog and the deadenylase, CCR4, mediate the trimming of poly(A) tails of developmentally critical transcripts [41]. These distinct roles of PABP2 in Drosophila lend credence to the idea that nuclear poly(A) binding proteins, including PABPN1, could have distinct effects on different RNA targets and also highlight the idea that the function of PABPN1 could differ depending on cellular compartment.
PABPN1 and Muscle Disease
OPMD is caused by an expansion of a short (GCN) trinucleotide repeat in the coding sequence of PABPN1 (GCG6 expanded to GCG8–13). In autosomal dominant OPMD, the stretch of 10 alanines at the N-terminus of PABPN1 (Fig. 1A) is expanded to 12–17 alanines [14]. While GCG6 is the normal repeat length, a small percentage of the population carries a GCG7 polymorphism [14]. Homozygosity for the GCG7 allele results in a rare autosomal recessive form of OPMD [14]. Patients that are compound heterozygotes for a GCG7 allele and a GCG9 expansion develop more severe disease phenotypes than patients that have only a single GCG9 expansion. These observations suggest that the GCG7 polymorphism can act as a modifier of autosomal dominant OPMD [14].
A hallmark of the autosomal dominant form of OPMD is the presence of nuclear aggregates in muscle tissue [54] similar to other polyalanine and polyglutamine diseases [55–57]. A number of approaches have been used over the years to try and understand how a rather modest change in PABPN1, addition of a few alanines, can lead to the dystrophic phenotype that occurs in OPMD. In autosomal dominant OPMD, one mutant allele of PABPN1 replaces one normal allele of PABPN1. Thus, disease pathology could result from loss of one normal allele, gain of a mutant allele or the combination of both events. Below we consider the approaches used to study the impact of mutant PABPN1 on cellular function followed by the current hypotheses proposed to explain how mutant PABPN1 causes pathology in cells with special emphasis on muscle cells as this is the tissue impacted in OPMD.
Approaches used to study effects of mutant PABPN1
Biochemical studies
A few studies have been performed to directly examine the function of alanine-expanded PABPN1. Biochemical studies with recombinant proteins reveal that alanine-expanded PABPN1 function can enhance the processivity of PAP in a manner comparable to wildtype PABPN1 (E. Wahle, personal communication) [3]. Thus, at least in the context of purified proteins, mutant PABPN1 is not impaired in one key function attributed to the protein. More molecular details will need to emerge before alanine-expanded PABPN1 can be biochemically assessed in other cellular processes such as alternative polyadenylation and RNA quality control.
In vitro studies have also been performed to examine the propensity of mutant and wildtype PABPN1 to form aggregates. Work using purified proteins demonstrates that the N-termini of both wildtype and alanine-expanded PABPN1 form fibrils similar to the nuclear aggregates characteristic of OPMD [58]. Purified fragments of the N-terminal domain of PABPN1 carrying an expanded stretch of 17 alanines show a greater propensity to form fibrils that are more resistant to solubilization as compared to fragments with 10 alanines [59]. However, subsequent work using full-length PABPN1 revealed that fibril formation is independent of the alanine tract and that the C-terminal domain confers aggregation [60] as both the wildtype and alanine-expanded PABPN1 proteins could form aggregates and removing the N-terminal stretch of alanines did not prevent aggregation [60]. Thus, these in vitro studies do not support a model where alanine-expanded PABPN1 is more prone to aggregation, at least in vitro, than the wildtype protein. Clearly, further studies as well as other complementary approaches are required to understand how expression of a modestly altered PABPN1 protein leads to muscle defects in OPMD.
Cell culture studies
Cell culture models for OPMD have been established by overexpressing alanine-expanded PABPN1 in a variety of cell types [25, 61–65]. Although many of these studies have employed non-muscle cell types, such approaches have provided insight into some of the proteins that may be present in the nuclear aggregates found in OPMD as well as identified cellular pathways that may be altered upon expression of mutant PABPN1.
Overexpression of alanine-expanded PABPN1 in multiple cell types results in the formation of nuclear aggregates reminiscent of those found in OPMD muscle tissue. These aggregates contain poly(A) RNA, components of the ubiquitin-proteasome machinery and RNA binding proteins including PABPN1 [54, 63, 65–69]. Time-lapse microscopy performed in HeLa cells overexpressing wildtype or alanine-expanded PABPN1 shows that PABPN1 can freely move into and out of the aggregates [61]. Similar time-lapse experiments performed in the human A549 cancer cell line overexpressing alanine-expanded PABPN1 demonstrate that the characteristic nuclear aggregates can disassemble during mitosis and are thus not irreversible [70]. The aggregates formed by alanine-expanded PABPN1 are larger, greater in number and more resistant to salt extraction than those formed by the wildtype protein [25, 62] suggesting that mutant PABPN1 does have properties that could contribute to an increase in dwell time of macromolecules within the aggregates.
These cell culture studies suggest that decreased PABPN1 function within the nucleus may be a critical component of cellular dysfunction. Targeting alanine-expanded PABPN1 to the cytoplasm by either attaching a nuclear export signal or deleting the nuclear localization signal prevents formation of aggregates in HeLa cells [71]. Formation of nuclear aggregates in HeLa cells or mouse myogenic C2 cells is also abrogated by mutations in domains of alanine-expanded PABPN1 that are required for interaction with RNA or the poly(A) polymerase [61]. Taken together, these data argue that the deleterious effects of mutant PABPN1 require not only the nuclear environment but also maintenance of the functional interactions of PABPN1, which suggests that the expanded alanine tract is not sufficient to trigger aggregation in the absence of other key interactions.
Although apoptosis has not been reported in muscles of OPMD patients, markers of apoptosis are significantly increased in some of these cell culture studies [69, 72, 73]. Experiments performed in HeLa and human embryonic kidney (HEK 293) cells identify the p53 pathway as a key mediator of apoptosis resulting from expression of alanine-expanded PABPN1 [72], which is similar to the mechanism of neuronal apoptosis linked to some polyglutamine expansion diseases [74, 75]. Expressing both wildtype and alanine-expanded PABPN1 simultaneously in monkey fibroblast cells (COS) decreased the expression of apoptotic markers when compared to cells expressing alanine-expanded PABPN1 alone. This anti-apoptotic activity of wildtype PABPN1 does not correlate with a decrease in the formation of nuclear aggregates, suggesting that the aggregates are not solely responsible for cellular dysfunction [76].
Although OPMD affects skeletal muscle, surprisingly little work has been done to examine muscle-specific functions of PABPN1. Defining the muscle-specific roles of PABPN1 is critical for understanding the pathogenesis of OPMD. Although mutant PABPN1 overexpression studies have predominantly been performed in cell culture systems unrelated to muscle [25, 67, 69, 73], some studies have examined myogenic cell lines using this approach. Overexpression of alanine-expanded PABPN1 in the C2C12 mouse muscle cell line resulted in decreased protein levels of muscle-specific proteins such as muscle creatine kinase, MyoD and myogenin [64]. In addition to the altered expression of muscle-specific proteins, the nuclear aggregates showed sequestration of myogenic transcription factors, Pax3/7 and Myf5, which are vital for muscle differentiation [64, 77, 78]. Overexpression of wildtype or alanine-expanded PABPN1 at relatively low levels in myoblasts did not result in altered expression of muscle-specific proteins [79]. However, these cell lines did show a marked dysregulation of gene expression similar to that observed in skeletal muscles of an OPMD mouse model and in OPMD patients [80, 81]. Some of the genes affected were in the ubiquitin-proteasome pathway, which is significantly impacted upon expression of mutant PABPN1 in vitro as well as in OPMD animal models and in OPMD patients [80]. This study also revealed that mutant PABPN1 forms nuclear aggregates faster than the wildtype protein in these cell lines, which could be due to the decreased ubiquitination and proteasomal clearance of mutant PABPN1 [79]. Together these findings led to speculation that the sequestration of critical muscle-specific proteins in aggregates could lead to functional depletion triggering a compensatory upregulation in the expression of the corresponding genes.
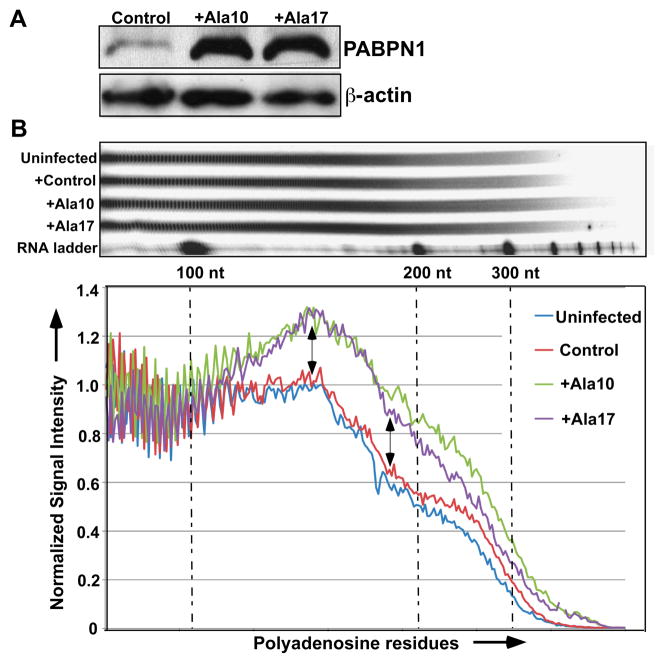
Some caveats need to be considered when interpreting the data from these cell culture studies. Historically these experiments have been performed using non-myogenic cell lines though recent studies have begun to focus on understanding the pathology of mutant PABPN1 in muscle cell lines [61, 64, 79]. In addition, all these models rely on various levels of overexpression of alanine-expanded PABPN1 in cells that retain the normal copy number of wildtype PABPN1. Thus, this scenario does not recapitulate the genotype of autosomal dominant OPMD patients. In fact, overexpression of wildtype PABPN1 can alter polyadenylation as shown in Figure 3B where similar effects on poly(A) tail length are detected in primary mouse muscle cells overexpressing either wildtype or mutant PABPN1. Thus, care needs to be taken in such models to ensure that changes detected result from the presence of mutant PABPN1 and not merely an increase in total PABPN1 protein levels. Comparing different studies is further complicated as the level of overexpression of mutant PABPN1 likely differs markedly from study to study.
Fig. 3. Overexpression of wildtype or mutant PABPN1 similarly impacts poly(A) tail length in murine myoblasts.
Pure cultures of primary mouse myoblasts from limb muscles were infected with retrovirus expressing wildtype (Ala 10) or expanded (Ala 17) murine PABPN1. As a control, cells were infected with retrovirus carrying an empty vector (Control). (A) Protein extracts were analyzed by immunoblotting to confirm expression of PABPN1. β-actin serves as a loading control. Protein extracts from myoblasts infected with retrovirus carrying an empty vector show the level of endogenous PABPN1 in these cells. (B) Representative distribution and graphical representation of bulk poly(A) tails from murine myoblasts expressing wildtype (Ala 10) or expanded (Ala 17) PABPN1. Total RNA was labeled with [32P] cytidine 3′, 5′-bis(phosphate) using T4 RNA ligase and digested with RNase A/T1. Samples were resolved by electrophoresis in denaturing polyacrylamide gels and exposed to radiographic film. The resulting audioradiogram was scanned to generate the graphical representation shown below which plots the signal intensity relative to polyadenosine tract length. The positions of 100, 200, and 300 nucleotides (nt) are indicated. Cells overexpressing either wildtype (Ala10, green) or expanded (Ala17, purple) PABPN1 show longer bulk poly(A) tails when compared to uninfected myoblasts (blue) or myoblasts infected with control retrovirus (red).
While these in vitro models have identified some intriguing dysregulated pathways that could contribute to muscle-specific pathology in OPMD, results obtained must be translated to in vivo systems to assess the effects of mutant PABPN1 in the context of the whole organism. Various in vivo systems have been developed both to understand the molecular events that underlie disease phenotypes and also to identify possible therapeutic strategies.
Transgenic animal models
Animal models of OPMD have been established in C. elegans, D. melanogaster and mice (Table 1). While these models differ from one another in the details, all are standard transgenics and thus contain two normal alleles of PABPN1 and one mutant allele of alanine-expanded PABPN1 integrated randomly into the genome.
Table 1.
Comparison of transgenic animal models of OPMD
| C. elegans [79] | D. melanogaster [81] | Mouse Hino et al. [87] | Mouse Dion et al. [88] | Mouse Mankodi et al. [93] | Mouse Davies et al. [83] | |
|---|---|---|---|---|---|---|
| PABPN1 transgene | Ala13 (human) | Ala17 (bovine) | Ala13 (human) | Ala13 (human) | Ala16 (human) | Ala17 (bovine) |
| Copies of PABPN1 | 2 endogenous + transgene | 2 endogenous + transgene | 2 endogenous + transgene | 2 endogenous + transgene | 2 endogenous + transgene | 2 endogenous + transgene |
| Promoter driving transgene expression | Muscle-specific Minor myosin heavy chain (Pmyo-3) |
Muscle-specific Myosin heavy chain (Mhc-Gal4) |
Ubiquitous CMV early enhancer/chic ken beta actin (CAG) |
Ubiquitous Putative native PABPN1 |
Ubiquitous Mifepristone- inducible system |
Muscle-specific Human skeletal actin |
| PABPN1 nuclear aggregates | Yes (weak) | Yes | Yes | Yes (neurons only) | Yes | Yes |
| Muscles examined for histopathology | Body wall muscle cells | Indirect flight muscles | Limb, eyelid, pharyngeal | Eyelid, soleus | Limb, pharyngeal, diaphragm, paraspinals | Biceps |
| Muscle histology | Loss of muscle cells | Muscle degeneration Thin, disorganized myofibrils Apoptotic nuclei Loss of mitochondria |
Muscle degeneration Increased fibrosis Increased fat |
Abnormal increase in regenerated myofibers | Progressive non- necrotic myopathy | Apoptosis Abnormal increase in regenerated myofibers |
| Phenotype | Abnormal motility: Decreased amplitude of sinusoidal wave Increased body bends |
Abnormal wing position: Wings held up Wings held down Dysregulation of ubiquitin-proteasome pathway genes |
Muscle weakness Decreased survival Reduced body weight |
Muscle weakness Primarily peripheral nerve alterations |
Muscle weakness Decreased survival Cardiomyopathy |
Progressive muscle weakness Myofiber atrophy Dysregulation of ubiquitin-proteasome pathway genes |
| Age of onset* | 5 days (~25-day lifespan) | 2 days (~25-day lifespan) | 3 months (juvenile) | 12 months (adult) | 10–12 weeks after induction | 6 months (adult) |
Age at which functional muscle deficits were first documented.
OPMD models have been established and exploited in C. elegans and D. melanogaster using muscle-specific overexpression of alanine-expanded PABPN1 (Table 1) [82–85]. As the nuclear poly(A) binding proteins in C. elegans and D. melanogaster do not contain the alanine stretch that is expanded in OPMD (Fig. 1B), these models depend on expression of alanine-expanded human (C. elegans) or bovine (D. melanogaster) PABPN1. In both models, expression of mutant PABPN1 results in muscle degeneration that manifests as mobility defects in C. elegans and wing position abnormalities in D. melanogaster. These model organisms with obvious muscle phenotypes are valuable tools because they can be easily manipulated to address mechanistic questions and also used for genetic screening. In these genetically tractable organisms, these approaches are rapid and inexpensive providing an excellent discovery platform. Thus, both worm and fly models of OPMD have been used to analyze mechanistic aspects of mutant PABPN1 required to confer muscle pathology and to screen for modifiers of OPMD pathology in vivo that can be further validated in more complex model organisms.
The C. elegans model of OPMD provides insight into whether aggregate formation and muscle pathology are linked to one another. In fact, these studies reveal that the marked muscle and mobility defects that occur upon overexpression of alanine-expanded PABPN1 in this model do not directly correspond to the presence of nuclear aggregates [82]. The Drosophila model has been used to dissect the functions of mutant PABPN1 required to confer pathology [84]. These studies suggest that the RNA binding function of PABPN1 is essential for the development of muscle pathology as expression of alanine-expanded PABPN1 with mutations in the RRM domain does not cause defects in muscle function. Further results from this study argue that the alanine expansion in PABPN1 is not absolutely required to confer muscle defects as expression of wildtype bovine PABPN1 or bovine PABPN1 lacking the alanine tract altogether could induce wing position defects leading these authors to suggest that an increase in the overall levels of PABPN1 protein confers muscle phenotypes independent of the alanine expansion [84]. Results from these mechanistic studies suggest that factors other than polyalanine toxicity may contribute to the development of OPMD pathology.
In addition to providing mechanistic insight into the function of mutant PABPN1, these model organisms can be valuable tools for identifying potential therapeutic targets. For example, in the worm OPMD model, longevity and cell metabolism regulatory molecules modulate the effects of mutant PABPN1 expression [82, 83]. Genetic studies exploiting the Drosophila OPMD model identify molecular chaperones, anti-apoptotic molecules and anti-aggregation compounds as potential modifiers of muscle phenotypes [84–86]. Anti-aggregation compounds have also shown promise in cell culture and mouse models of OPMD [64, 73, 76, 87, 88] showing that results from Drosophila have the potential to be relevant in mammalian systems and possibly in therapeutic applications.
Although value exists in the use of these genetically tractable models of OPMD, there are also clear caveats that need to be considered. Skeletal muscle structure in both flies and worms is distinct from humans [89, 90]. Given that pathology in OPMD occurs only in a specific subset of skeletal muscles, there could be aspects of the physiology of these specific muscles that are critical to development of disease phenotypes that are not recapitulated in simple models. Evolutionary differences in signaling pathways and target proteins could influence the direct translational potential of these model organisms as screening tools.
Multiple transgenic mouse models of OPMD have also been established to understand the underlying pathologic mechanisms and test potential therapeutic compounds. These models recapitulate some of the phenotypes associated with the human disease. Details of these transgenic mouse models are summarized in Table 1. In most of these mouse models, transgenic expression of the alanine-expanded human PABPN1 occurs via a ubiquitous promoter. For example, Hino et al. employed the cytomegalovirus early enhancer element/chicken beta actin (CAG) promoter for transgenic expression of mutant PABPN1 and detected muscle degeneration that worsened with age and was most prominent in the eyelid and pharyngeal muscles [91]. However, the muscle pathology in these mice was accompanied by growth retardation and a shortened lifespan that are not features of the human disease. Transgenic expression of mutant PABPN1 using the putative PABPN1 promoter in a model established by Dion et al. resulted in a primarily neuronal phenotype characterized by the presence of nuclear aggegrates that were accompanied by coordination defects and a peripheral neuropathy [92]. Although reports of neurological involvement exist in confirmed cases of OPMD [93–96], this model is not representative of the muscle pathology associated with the disease. Most recently, ubiquitous expression of transgenic alanine-expanded PABPN1 using an inducible gene expression system generated by Mankodi et al. revealed progressive skeletal muscle atrophy and dilated cardiomyopathy [97]. Appearance of nuclear aggregates in the skeletal muscle, heart and brain of these mice preceded the development of myopathy. Although cardiomyopathy is not a feature of the human disease, this model of OPMD provides insight into the reversibility of the muscle pathology as turning off the expression of the mutant PABPN1 resulted in a resolution of the aggregates and a significant improvement in the muscle pathology [97].
Only the mouse model established by Davies et al. utilized muscle-specific overexpression of alanine-expanded PABPN1. These mice show nuclear aggregates along with apoptotic markers and progressive muscular weakness [87]. Microarray studies on RNAs isolated from skeletal muscles of these mice revealed deregulation of genes coding for mRNA processing, ubiquitin-proteasome and muscle cell differentiation factors as compared to wildtype mice or control mice overexpressing wildtype PABPN1 [80]. Progressive muscle atrophy was observed especially in fast glycolytic fibers, which contained more nuclear aggregates and were more severely affected as compared to oxidative muscle fibers [81]. This OPMD mouse model has been used most extensively for understanding disease pathogenesis and also as a screening tool for potential therapeutic interventions. Treatment of these transgenic animals with anti-aggregation drugs like cystamine, trehalose and doxycycline decreases nuclear aggregates, inhibits cell death and delays the development of muscle pathology [76, 87, 88]. Transgenic mice simultaneously expressing high levels of both wildtype and alanine-expanded PABPN1 in skeletal muscle show decreased apoptosis of muscle cells and improved muscle function compared to mice that overexpress alanine-expanded PABPN1 alone [98]. This decrease in muscle pathology is not accompanied by a decrease in nuclear aggregates. This mitigation of muscle phenotype without a corresponding resolution of aggregates argues that nuclear aggregates may not be directly involved in OPMD pathology in vivo.
Several caveats should be kept in mind when interpreting the data from these transgenic mouse models of OPMD. As with the in vitro cell culture models, the level of overexpression of mutant PABPN1 varies markedly among these models and in some cases is up to 30-fold higher than endogenous levels [87]. In addition, issues with the integration site of the transgene and lack of the proper 5′- and 3′-gene regulatory regions [92] confound analyses of these transgenic mice. Furthermore, transgenic expression of PABPN1 could cause different underlying molecular pathologies that lead to muscle disease in these overexpression models that are not found in OPMD patients. Importantly, none of these transgenic mouse models fully recapitulate the disease seen in OPMD patients. Thus, these mouse models do not accurately reflect either the genotype or the phenotype of patients afflicted with the disease. A mouse model with one allele each of wildtype and mutant PABPN1 would be representative of the genotype found in autosomal dominant OPMD and allow us to better understand the disease process.
OPMD patient cells and tissues
A limited number of experiments have been performed using muscle samples from autosomal dominant OPMD patients aimed at understanding the molecular changes that arise in the disease which may give clues to the underlying pathologic mechanisms. A major advantage in utilizing human OPMD samples is that cellular and molecular phenotypes result from endogenous levels of wild type and mutant PABPN1 as opposed to the overexpression models discussed above. Furthermore, patient tissues provide a comparison to evaluate the utility of various OPMD models assessing how well they recapitulate the molecular phenotypes present in the disease. One question that is uniquely poised to be answered using patient tissues is whether wildtype and mutant PABPN1 are present at equal levels in cells. Recently the steady-state level of the transcript corresponding to the alanine-expanded PABPN1 allele was shown to be ~1.5-fold higher than the steady-state level of the transcript corresponding to the wildtype PABPN1 allele in OPMD muscles [99]. This finding suggests that mutant PABPN1 expression is dysregulated. However, current antibodies cannot distinguish between the wildtype and mutant PABPN1 proteins to determine whether this increase in transcript level leads to corresponding changes in mutant protein levels.
Patient tissues are also useful to address whether key functions of PABPN1 or molecular pathways are altered in the disease. Currently only polyadenylation has been assessed using cultured, satellite cell-derived myoblasts isolated from deltoid muscles of OPMD patients [66]. This study did not reveal any differences in bulk poly(A) tail length in these patient myoblasts compared to control myoblasts. To conclusively determine whether polyadenylation or other emerging functions (Fig. 2) are altered, studies in affected patient tissues are required. Microarray studies have been valuable in elucidating molecular pathways, such as the ubiquitin proteasome system, that are altered in OPMD muscle tissue [80, 81]. Other approaches such as a cytokine antibody array reveal differential expression of factors involved in fibrosis in affected and nonaffected muscles in OPMD patients suggesting potential pathways to target for therapy [100]. Cell culture studies of OPMD satellite cell-derived myoblasts can assess whether these critical stem cells may be functionally impaired. Satellite cells isolated from affected cricopharyngeal muscles of OPMD patients gave rise to myoblasts in vitro that underwent premature proliferative arrest suggesting that the ability of OPMD myoblasts to proliferate and fuse with myofibers may be hindered in vivo [100]. Furthermore, a recent study showed an increased number of Pax7+ cells both in the satellite cell position and in the interstitium in cricopharyngeal muscle of OPMD patients as compared to control cricopharyngeal muscle [101]. These observations suggest dysregulation of satellite cells in this disease.
Although OPMD biopsy samples provide some clear advantages over the animal models described above several unique problems are inherent in the use of human biopsy material. These problems include limited quantities of tissues and the influences of genetic/environmental variation. Furthermore, the inability to sample multiple time points during disease progression in the same patients makes it hard to distinguish between molecular events that cause pathology from those that are secondary to the dystrophic changes in the muscles. Finally, one should consider whether results of a small biopsy taken from an area of muscle whose pathologic status is unknown is reflective of the muscle as a whole.
Molecular mechanisms by which mutant PABPN1 may cause disease
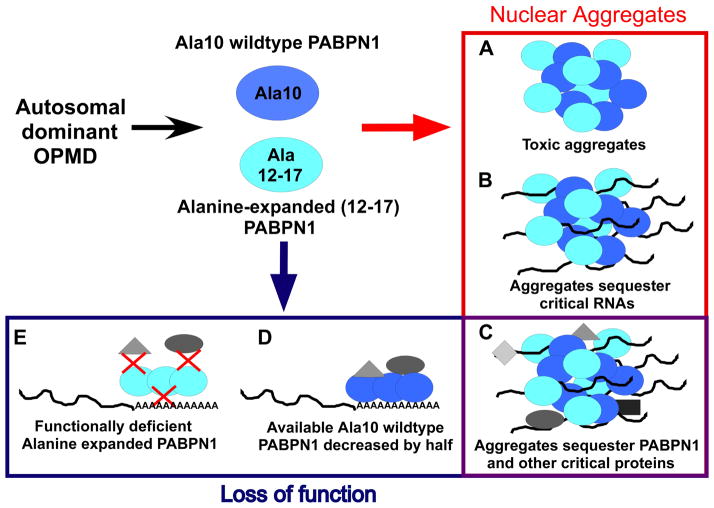
Experiments using the model systems described above have led to speculation regarding how mutant PABPN1 could impact cellular function. Multiple models currently exist to explain the mechanism by which alanine-expanded PABPN1 could contribute to pathology in OPMD (Figure 4). The challenge in the future will be to integrate the information gleaned from these different studies into a cohesive model to understand the muscle-specific nature of OPMD.
Fig. 4. Models for OPMD pathogenesis.
Currently two general models are used to explain how alanine-expanded PABPN1 confers muscle pathology in autosomal dominant OPMD where patients have one normal and one mutant allele of PABPN1. One model suggests that nuclear aggregates cause disease (right column, outlined in red). A second model suggests that loss of PABPN1 function (bottom row, outlined in blue) underlies pathology. Importantly, these models are not mutually exclusive and elements of both could contribute to muscle pathology including the indicated overlap in these models (bottom right, outlined in purple). In considering possible mechanisms by which nuclear aggregates could confer muscle pathology: A) Both wildtype and expanded PABPN1 are predisposed to aggregation [61], and these nuclear aggregates may directly lead to cellular toxicity and apoptosis; B) the aggregates could sequester poly(A) RNA; and (C) aggregates could sequester a variety of proteins including PABPN1 [63, 66, 68, 69, 114]. The sequestration of critical transcripts and proteins vital for cell function into these aggregates could be responsible for the muscle defects in OPMD through loss of function for PABPN1. Other possible contributions through loss of function include: D) patients with autosomal dominant OPMD have only one copy of wildtype PABPN1 as compared to unaffected individuals raising the possibility that defects observed in OPMD could be a direct result of haploinsufficiency of PABPN1; or E) as the function of alanine-expanded PABPN1 has not been extensively characterized in vivo, mutant PABPN1 might not perform all the cellular functions of wildtype PABPN1 with the same efficiency. This decreased functional capacity could be due to variability in the ability of mutant PABPN1 to interact with critical transcripts or protein partners essential for muscle cell function.
Nuclear aggregates
The filamentous nuclear aggregates found in OPMD are similar to aggregates associated with the pathogenesis of polyglutamine expansion disorders and some of the other polyalanine expansion diseases [55–57]. As with a number of other polyglutamine and polyalanine expansion diseases, much of the research in the OPMD field has focused on the role of these aggregates [55, 57, 64, 69, 71, 76, 88, 102, 103]. Although they are considered a pathological hallmark of the disease, nuclear aggregates are found in less than 20% of myofiber nuclei in muscle sections from OPMD patients [54, 101, 104]. Furthermore, nuclear aggregates containing wildtype PABPN1 have been found in hypothalamic neurons under physiologic conditions, which is in agreement with in vitro data demonstrating the propensity of the wildtype protein to aggregate [105]. A multitude of experiments that overexpressed mutant PABPN1 have made the case for a toxic gain-of-function model in which alanine-expanded PABPN1 leads to apoptosis resulting from the formation of toxic aggregates (Fig. 4-A). Evidence cited in favor of this model is the ability of anti-aggregation drugs to ameliorate the pathology caused by the overexpression of alanine-expanded PABPN1 in vitro and in vivo [76, 87, 88]. However, both wildtype and alanine-expanded PABPN1 proteins can form these aggregates and removing the N-terminal stretch of alanines does not prevent aggregation [61]. Multiple lines of evidence have also demonstrated that formation of these nuclear aggregates does not always correlate with detrimental effects of expressing mutant PABPN1 [82, 84, 98].
A variation on the gain-of-function model via formation of toxic nuclear aggregates is that alanine-expanded PABPN1 could act via a dominant-negative mechanism in which the mutant protein inhibits the function of the wildtype protein [106–108]. Although such an effect has been demonstrated in synpolydactyly, which is caused by a polyalanine expansion in the HoxD13 transcription factor [109], no experimental evidence exists suggesting such a dominant-negative effect occurs in OPMD.
In addition to having a gain-of-function toxic effect, nuclear aggregates could sequester RNAs (Fig. 4-B) or proteins including PABPN1 (Fig. 4-C) essential for proper cellular function. Recent data from muscle sections of OPMD patients and primary human myoblasts overexpressing mutant PABPN1 show that nuclear aggregates develop in close proximity to nuclear speckles, and progressively deplete the speckles of both PABPN1 and poly(A) RNA [110]. This disruption of nuclear speckles is likely to have an adverse effect on nascent mRNA processing and could account for the dysregulation of gene expression observed in OPMD. In autosomal dominant OPMD, the cells already lack one allele of wildtype PABPN1; any further depletion of this critical RNA processing factor could lead to cellular dysfunction and pathology could ensue. In this case, anti-aggregation compounds that ameliorate the deleterious effects of expressing mutant PABPN1 could release functional wildtype PABPN1 or critical RNA from the aggregates increasing the functional pool of these vital molecules.
Loss of PABPN1 function
An emerging concept in OPMD pathology is that expression of mutant PABPN1 results in a relative loss of PABPN1 function in cells. There is precedence for a loss of function hypothesis in other diseases that are associated with the formation of nuclear aggregates. In the case of both polyalanine and polyglutamine expansion disorders, mutations leading to loss of function of the respective proteins give rise to similar disease phenotypes, although only the expansions result in the formation of nuclear aggregates [56, 109, 111, 112]. As discussed above, one mechanism that could lead to loss of PABPN1 function is sequestration of PABPN1 in nuclear aggregates (Fig. 4-C), which would lead to functional depletion of PABPN1. This model includes elements of the nuclear aggregate and loss of function models thus unifying the two models. Another potential mechanism stems from the fact that in autosomal dominant OPMD, patients have one allele of each of the wildtype and mutant PABPN1; thus, the steady-state levels of wild type PABPN1 protein would be predicted to be 50% lower than in normal individuals (Fig. 4-D). Although both wildtype and alanine-expanded PABPN1 seem to have similar capacities to promote polyadenylation in vitro and in cultured cells [3, 66] (Fig. 3), the ability of mutant PABPN1 to execute the other functions of wildtype PABPN1 has not been analyzed in vitro or in vivo so an impaired function of mutant PABPN1 could contribute to muscle pathology (Fig. 4-E). Finally, proteomic analysis of C2C12 muscle cell lysates identified multiple proteins that differentially interacted with recombinant, purified wildtype or mutant PABPN1 [65]. Variability in protein interactions or altered affinity for critical protein partners of mutant Ataxin-1 has been previously demonstrated to contribute to disease pathogenesis in spinocerebellar ataxia 1, a polyglutamine expansion disorder [113]. The alanine expansion in PABPN1 may similarly prevent interaction of critical protein partners with PABPN1 thus leading to a loss of functional PABPN1 (Fig. 4-E).
Why is OPMD a muscle-specific disease?
Why expression of alanine-expanded PABPN1 leads to disease in skeletal muscle despite ubiquitous expression is unknown but may be related to unique characteristics of this tissue. Skeletal muscle is one of the few tissues that display extensive regenerative ability. This regenerative ability is due to the ability of a type of resident muscle stem cell called satellite cells to undergo myogenesis. In response to injury, satellite cells begin to proliferate and give rise to progeny called myoblasts. During muscle degeneration and regeneration, these myoblasts differentiate and fuse to form new myofibers, hence restoring normal tissue architecture. Defects in myoblast proliferation, differentiation and fusion coupled with dysregulation of satellite cells with aging [114, 115] could contribute to the pathology observed in OPMD. Indeed, satellite cells isolated from cricopharyngeal muscles of OPMD patients showed defects in cell proliferation in vitro [100] but whether poor cell proliferation results from direct effects of mutant PABPN1 on cellular function or is an indirect affect arising from chronic cycles of degeneration/regeneration in affected muscles and reflects exhaustion of the stem cell pool as in other dystrophies [116] is unknown.
A further conundrum in OPMD is why a limited subset of muscle groups are affected. PABPN1 function may have greater importance in certain muscle types than others, which may explain the very specific muscle pathology observed in OPMD. In particular, specific physiologic properties of craniofacial muscles such as the extraocular, tongue and pharyngeal muscles may make them particularly vulnerable to alanine expansion of PABPN1. Very limited molecular studies have been performed on pharyngeal muscles to date [100, 101], but intrinsic differences exist between craniofacial and limb muscles [101, 117, 118]. Furthermore, innate differences are also noted in the satellite cells of craniofacial and limb muscles [101, 119]. These differences may reflect the distinct embryonic origins of craniofacial muscles compared to limb muscles [120]. Intriguingly, the number of Pax7+ satellite cells is increased in normal human cricopharyngeal muscles compared to other muscles, with an even greater increase noted in cricopharyngeal muscles of OPMD patients in the absence of any signs of muscle regeneration at the time point analyzed [101]. Whether these changes in satellite cell number contribute to the pathogenesis of OPMD is unknown, but merits further investigation.
Concluding Remarks
Our knowledge about the cellular functions of PABPN1 has greatly expanded in recent years. Unfortunately far less is understood about how expression of alanine-expanded PABPN1 leads to disease much less a muscle-specific disease that affects a limited subset of skeletal muscles in the body starting in middle age. Further research on the various functions of PABPN1 especially in muscle cells is likely to uncover key information that will inform our understanding of the pathologic mechanisms at work in OPMD and provide potential opportunities for therapeutic intervention.
Acknowledgments
We are grateful to colleagues for sharing unpublished data. We thank Drs. F. Bachand and E. Wahle for comments and sharing information. Due to space limitations not all relevant citations could be included. We apologize to our colleagues for omission of any particular citations. AB and LHA are supported by Development Grants from the Muscular Dystrophy Foundation. AHC and GKP are supported by a collaborative NIH grant (AR061987).
Abbreviations
- CPSF
cleavage specificity factor
- lncRNA
long non-coding RNAs
- miRNA
micro RNA
- NLS
nuclear localization signal
- OPMD
Oculopharyngeal Muscular Dystrophy
- PABPN1
Poly(A) Binding Protein Nuclear 1
- PAP
Poly(A) polymerase
- poly(A) tail
polyadenosine tail
- RRM
ribonucleoprotein-type RNA binding motif
- RNAi
RNA interference
References
- 1.Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley interdisciplinary reviews. 2011;2:348–61. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- 2.Weill L, Belloc E, Bava FA, Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nature structural & molecular biology. 2012;19:577–85. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn U, Wahle E. Structure and function of poly(A) binding proteins. Biochimica et biophysica acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome biology. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–17. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 6.Wahle E. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell. 1991;66:759–68. doi: 10.1016/0092-8674(91)90119-j. [DOI] [PubMed] [Google Scholar]
- 7.Krause S, Fakan S, Weis K, Wahle E. Immunodetection of poly(A) binding protein II in the cell nucleus. Experimental cell research. 1994;214:75–82. doi: 10.1006/excr.1994.1235. [DOI] [PubMed] [Google Scholar]
- 8.Calado A, Kutay U, Kuhn U, Wahle E, Carmo-Fonseca M. Deciphering the cellular pathway for transport of poly(A)-binding protein II. RNA (New York, NY. 2000;6:245–56. doi: 10.1017/s1355838200991908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerwitz Y, Kuhn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, Schwarz E, Wahle E. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. The EMBO journal. 2003;22:3705–14. doi: 10.1093/emboj/cdg347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn U, Gundel M, Knoth A, Kerwitz Y, Rudel S, Wahle E. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. The Journal of biological chemistry. 2009;284:22803–14. doi: 10.1074/jbc.M109.018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apponi LH, Leung SW, Williams KR, Valentini SR, Corbett AH, Pavlath GK. Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Human molecular genetics. 2009;19:1058–65. doi: 10.1093/hmg/ddp569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, Rubinsztein DC, Agami R. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–53. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 13.Beaulieu YB, Kleinman CL, Landry-Voyer AM, Majewski J, Bachand F. Polyadenylation-dependent control of long noncoding RNA expression by the poly(a)-binding protein nuclear 1. PLoS genetics. 2012;8:e1003078. doi: 10.1371/journal.pgen.1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brais B, Bouchard JP, Xie YG, Rochefort DL, Chretien N, Tome FM, Lafreniere RG, Rommens JM, Uyama E, Nohira O, Blumen S, Korczyn AD, Heutink P, Mathieu J, Duranceau A, Codere F, Fardeau M, Rouleau GA. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nature genetics. 1998;18:164–7. doi: 10.1038/ng0298-164. [DOI] [PubMed] [Google Scholar]
- 15.Victor M, Hayes R, Adams RD. Oculopharyngeal muscular dystrophy. A familial disease of late life characterized by dysphagia and progressive ptosis of the evelids. The New England journal of medicine. 1962;267:1267–72. doi: 10.1056/NEJM196212202672501. [DOI] [PubMed] [Google Scholar]
- 16.Ruegg S, Lehky Hagen M, Hohl U, Kappos L, Fuhr P, Plasilov M, Muller H, Heinimann K. Oculopharyngeal muscular dystrophy - an under-diagnosed disorder? Swiss medical weekly. 2005;135:574–86. doi: 10.4414/smw.2005.11221. [DOI] [PubMed] [Google Scholar]
- 17.Van Der Sluijs BM, Hoefsloot LH, Padberg GW, Van Der Maarel SM, Van Engelen BG. Oculopharyngeal muscular dystrophy with limb girdle weakness as major complaint. Journal of neurology. 2003;250:1307–12. doi: 10.1007/s00415-003-0201-6. [DOI] [PubMed] [Google Scholar]
- 18.Messaed C, Dion PA, Abu-Baker A, Rochefort D, Laganiere J, Brais B, Rouleau GA. Soluble expanded PABPN1 promotes cell death in oculopharyngeal muscular dystrophy. Neurobiology of disease. 2007;26:546–57. doi: 10.1016/j.nbd.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Blumen SC, Nisipeanu P, Sadeh M, Asherov A, Blumen N, Wirguin Y, Khilkevich O, Carasso RL, Korczyn AD. Epidemiology and inheritance of oculopharyngeal muscular dystrophy in Israel. Neuromuscul Disord. 1997;7(Suppl 1):S38–40. doi: 10.1016/s0960-8966(97)00080-1. [DOI] [PubMed] [Google Scholar]
- 20.Bouchard JP, Brais B, Brunet D, Gould PV, Rouleau GA. Recent studies on oculopharyngeal muscular dystrophy in Quebec. Neuromuscul Disord. 1997;7(Suppl 1):S22–9. doi: 10.1016/s0960-8966(97)00077-1. [DOI] [PubMed] [Google Scholar]
- 21.Kuhn U, Nemeth A, Meyer S, Wahle E. The RNA binding domains of the nuclear poly(A)-binding protein. The Journal of biological chemistry. 2003;278:16916–25. doi: 10.1074/jbc.M209886200. [DOI] [PubMed] [Google Scholar]
- 22.Ge H, Zhou D, Tong S, Gao Y, Teng M, Niu L. Crystal structure and possible dimerization of the single RRM of human PABPN1. Proteins. 2008;71:1539–45. doi: 10.1002/prot.21973. [DOI] [PubMed] [Google Scholar]
- 23.Good PJ, Abler L, Herring D, Sheets MD. Xenopus embryonic poly(A) binding protein 2 (ePABP2) defines a new family of cytoplasmic poly(A) binding proteins expressed during the early stages of vertebrate development. Genesis. 2004;38:166–75. doi: 10.1002/gene.20015. [DOI] [PubMed] [Google Scholar]
- 24.Song J, McGivern JV, Nichols KW, Markley JL, Sheets MD. Structural basis for RNA recognition by a type II poly(A)-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15317–22. doi: 10.1073/pnas.0801274105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan X, Dion P, Laganiere J, Brais B, Rouleau GA. Oligomerization of polyalanine expanded PABPN1 facilitates nuclear protein aggregation that is associated with cell death. Human molecular genetics. 2001;10:2341–51. doi: 10.1093/hmg/10.21.2341. [DOI] [PubMed] [Google Scholar]
- 26.Smith JJ, Rucknagel KP, Schierhorn A, Tang J, Nemeth A, Linder M, Herschman HR, Wahle E. Unusual sites of arginine methylation in Poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. The Journal of biological chemistry. 1999;274:13229–34. doi: 10.1074/jbc.274.19.13229. [DOI] [PubMed] [Google Scholar]
- 27.Fronz K, Otto S, Kolbel K, Kuhn U, Friedrich H, Schierhorn A, Beck-Sickinger AG, Ostareck-Lederer A, Wahle E. Promiscuous modification of the nuclear poly(A)-binding protein by multiple protein-arginine methyltransferases does not affect the aggregation behavior. The Journal of biological chemistry. 2008;283:20408–20. doi: 10.1074/jbc.M802329200. [DOI] [PubMed] [Google Scholar]
- 28.Fronz K, Guttinger S, Burkert K, Kuhn U, Stohr N, Schierhorn A, Wahle E. Arginine methylation of the nuclear poly(a) binding protein weakens the interaction with its nuclear import receptor, transportin. The Journal of biological chemistry. 2011;286:32986–94. doi: 10.1074/jbc.M111.273912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wahle E, Moritz B. Methylation of the nuclear poly(A) binding protein by type I protein arginine methyltransferases - how and why. Biological chemistry. 2013 doi: 10.1515/hsz-2013-0121. [DOI] [PubMed] [Google Scholar]
- 30.Keller RW, Kuhn U, Aragon M, Bornikova L, Wahle E, Bear DG. The nuclear poly(A) binding protein, PABP2, forms an oligomeric particle covering the length of the poly(A) tail. Journal of molecular biology. 2000;297:569–83. doi: 10.1006/jmbi.2000.3572. [DOI] [PubMed] [Google Scholar]
- 31.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Molecular cell. 2011;43:853–66. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Klerk E, Venema A, Anvar SY, Goeman JJ, Hu O, den Dunnen JT, van der Maarel SM, Raz V, t Hoen PA. Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic acids research. 2012 doi: 10.1093/nar/gks655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boutet SC, Cheung TH, Quach NL, Liu L, Prescott SL, Edalati A, Iori K, Rando TA. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell stem cell. 2012;10:327–36. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7028–33. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–84. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome research. 2008;18:1433–45. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharjee RB, Bag J. Depletion of nuclear poly(A) binding protein PABPN1 produces a compensatory response by cytoplasmic PABP4 and PABP5 in cultured human cells. PloS one. 2012;7:e53036. doi: 10.1371/journal.pone.0053036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–66. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 41.Benoit B, Mitou G, Chartier A, Temme C, Zaessinger S, Wahle E, Busseau I, Simonelig M. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Developmental cell. 2005;9:511–22. doi: 10.1016/j.devcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic acids research. 2010;38:2757–74. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemieux C, Bachand F. Cotranscriptional recruitment of the nuclear poly(A)-binding protein Pab2 to nascent transcripts and association with translating mRNPs. Nucleic acids research. 2009;37:3418–30. doi: 10.1093/nar/gkp207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perreault A, Lemieux C, Bachand F. Regulation of the nuclear poly(A)-binding protein by arginine methylation in fission yeast. The Journal of biological chemistry. 2007;282:7552–62. doi: 10.1074/jbc.M610512200. [DOI] [PubMed] [Google Scholar]
- 46.Lemay JF, D’Amours A, Lemieux C, Lackner DH, St-Sauveur VG, Bahler J, Bachand F. The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Molecular cell. 2010;37:34–45. doi: 10.1016/j.molcel.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 47.Mallet PL, Bachand F. A Proline-Tyrosine Nuclear Localization Signal (Py-Nls) Is Required For The Nuclear Import Of Fission Yeast Pab2, But Not Of Human Pabpn1. Traffic (Copenhagen, Denmark) 2012 doi: 10.1111/tra.12036. [DOI] [PubMed] [Google Scholar]
- 48.Lemieux C, Marguerat S, Lafontaine J, Barbezier N, Bahler J, Bachand F. A Pre-mRNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)-binding protein. Molecular cell. 2011;44:108–19. doi: 10.1016/j.molcel.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 49.St-Andre O, Lemieux C, Perreault A, Lackner DH, Bahler J, Bachand F. Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. The Journal of biological chemistry. 2010;285:27859–68. doi: 10.1074/jbc.M110.150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. The EMBO journal. 2010;29:2173–81. doi: 10.1038/emboj.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 52.Hurschler BA, Harris DT, Grosshans H. The type II poly(A)-binding protein PABP-2 genetically interacts with the let-7 miRNA and elicits heterochronic phenotypes in Caenorhabditis elegans. Nucleic acids research. 2011;39:5647–57. doi: 10.1093/nar/gkr145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benoit B, Nemeth A, Aulner N, Kuhn U, Simonelig M, Wahle E, Bourbon HM. The Drosophila poly(A)-binding protein II is ubiquitous throughout Drosophila development and has the same function in mRNA polyadenylation as its bovine homolog in vitro. Nucleic acids research. 1999;27:3771–8. doi: 10.1093/nar/27.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tome FM, Fardeau M. Nuclear inclusions in oculopharyngeal dystrophy. Acta neuropathologica. 1980;49:85–7. doi: 10.1007/BF00692226. [DOI] [PubMed] [Google Scholar]
- 55.Zoghbi HY, Orr HT. Polyglutamine diseases: protein cleavage and aggregation. Current opinion in neurobiology. 1999;9:566–70. doi: 10.1016/S0959-4388(99)00013-6. [DOI] [PubMed] [Google Scholar]
- 56.Nasrallah IM, Minarcik JC, Golden JA. A polyalanine tract expansion in Arx forms intranuclear inclusions and results in increased cell death. The Journal of cell biology. 2004;167:411–6. doi: 10.1083/jcb.200408091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caburet S, Demarez A, Moumne L, Fellous M, De Baere E, Veitia RA. A recurrent polyalanine expansion in the transcription factor FOXL2 induces extensive nuclear and cytoplasmic protein aggregation. Journal of medical genetics. 2004;41:932–6. doi: 10.1136/jmg.2004.024356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheuermann T, Schulz B, Blume A, Wahle E, Rudolph R, Schwarz E. Trinucleotide expansions leading to an extended poly-L-alanine segment in the poly (A) binding protein PABPN1 cause fibril formation. Protein Sci. 2003;12:2685–92. doi: 10.1110/ps.03214703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lodderstedt G, Hess S, Hause G, Scheuermann T, Scheibel T, Schwarz E. Effect of oculopharyngeal muscular dystrophy-associated extension of seven alanines on the fibrillation properties of the N-terminal domain of PABPN1. The FEBS journal. 2007;274:346–55. doi: 10.1111/j.1742-4658.2006.05595.x. [DOI] [PubMed] [Google Scholar]
- 60.Winter R, Kuhn U, Hause G, Schwarz E. Polyalanine-independent conformational conversion of nuclear poly(A)-binding protein 1 (PABPN1) The Journal of biological chemistry. 2012;287:22662–71. doi: 10.1074/jbc.M112.362327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tavanez JP, Calado P, Braga J, Lafarga M, Carmo-Fonseca M. In vivo aggregation properties of the nuclear poly(A)-binding protein PABPN1. RNA (New York, NY. 2005;11:752–62. doi: 10.1261/rna.7217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shanmugam V, Dion P, Rochefort D, Laganiere J, Brais B, Rouleau GA. PABP2 polyalanine tract expansion causes intranuclear inclusions in oculopharyngeal muscular dystrophy. Annals of neurology. 2000;48:798–802. [PubMed] [Google Scholar]
- 63.Abu-Baker A, Messaed C, Laganiere J, Gaspar C, Brais B, Rouleau GA. Involvement of the ubiquitin-proteasome pathway and molecular chaperones in oculopharyngeal muscular dystrophy. Human molecular genetics. 2003;12:2609–23. doi: 10.1093/hmg/ddg293. [DOI] [PubMed] [Google Scholar]
- 64.Wang Q, Bag J. Ectopic expression of a polyalanine expansion mutant of poly(A)-binding protein N1 in muscle cells in culture inhibits myogenesis. Biochemical and biophysical research communications. 2006;340:815–22. doi: 10.1016/j.bbrc.2005.12.078. [DOI] [PubMed] [Google Scholar]
- 65.Tavanez JP, Bengoechea R, Berciano MT, Lafarga M, Carmo-Fonseca M, Enguita FJ. Hsp70 chaperones and type I PRMTs are sequestered at intranuclear inclusions caused by polyalanine expansions in PABPN1. PloS one. 2009;4:e6418. doi: 10.1371/journal.pone.0006418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calado A, Tome FM, Brais B, Rouleau GA, Kuhn U, Wahle E, Carmo-Fonseca M. Nuclear inclusions in oculopharyngeal muscular dystrophy consist of poly(A) binding protein 2 aggregates which sequester poly(A) RNA. Human molecular genetics. 2000;9:2321–8. doi: 10.1093/oxfordjournals.hmg.a018924. [DOI] [PubMed] [Google Scholar]
- 67.Corbeil-Girard LP, Klein AF, Sasseville AM, Lavoie H, Dicaire MJ, Saint-Denis A, Page M, Duranceau A, Codere F, Bouchard JP, Karpati G, Rouleau GA, Massie B, Langelier Y, Brais B. PABPN1 overexpression leads to upregulation of genes encoding nuclear proteins that are sequestered in oculopharyngeal muscular dystrophy nuclear inclusions. Neurobiology of disease. 2005;18:551–67. doi: 10.1016/j.nbd.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 68.Fan X, Messaed C, Dion P, Laganiere J, Brais B, Karpati G, Rouleau GA. HnRNP A1 and A/B interaction with PABPN1 in oculopharyngeal muscular dystrophy. The Canadian journal of neurological sciences. 2003;30:244–51. doi: 10.1017/s0317167100002675. [DOI] [PubMed] [Google Scholar]
- 69.Bao YP, Cook LJ, O’Donovan D, Uyama E, Rubinsztein DC. Mammalian, yeast, bacterial, and chemical chaperones reduce aggregate formation and death in a cell model of oculopharyngeal muscular dystrophy. The Journal of biological chemistry. 2002;277:12263–9. doi: 10.1074/jbc.M109633200. [DOI] [PubMed] [Google Scholar]
- 70.Marie-Josee Sasseville A, Caron AW, Bourget L, Klein AF, Dicaire MJ, Rouleau GA, Massie B, Langelier Y, Brais B. The dynamism of PABPN1 nuclear inclusions during the cell cycle. Neurobiology of disease. 2006;23:621–9. doi: 10.1016/j.nbd.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 71.Abu-Baker A, Laganiere S, Fan X, Laganiere J, Brais B, Rouleau GA. Cytoplasmic targeting of mutant poly(A)-binding protein nuclear 1 suppresses protein aggregation and toxicity in oculopharyngeal muscular dystrophy. Traffic (Copenhagen, Denmark) 2005;6:766–79. doi: 10.1111/j.1600-0854.2005.00315.x. [DOI] [PubMed] [Google Scholar]
- 72.Bhattacharjee RB, Zannat T, Bag J. Expression of the poly alanine expansion mutatant of nuclear Poly (A) binding protein induces apoptosis via the p53 pathway. Cell biology international. 2012 doi: 10.1042/CBI20110348. [DOI] [PubMed] [Google Scholar]
- 73.Bao YP, Sarkar S, Uyama E, Rubinsztein DC. Congo red, doxycycline, and HSP70 overexpression reduce aggregate formation and cell death in cell models of oculopharyngeal muscular dystrophy. Journal of medical genetics. 2004;41:47–51. doi: 10.1136/jmg.2003.014548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chou AH, Lin AC, Hong KY, Hu SH, Chen YL, Chen JY, Wang HL. p53 activation mediates polyglutamine-expanded ataxin-3 upregulation of Bax expression in cerebellar and pontine nuclei neurons. Neurochemistry international. 58:145–52. doi: 10.1016/j.neuint.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 75.Tsoi H, Lau TC, Tsang SY, Lau KF, Chan HY. CAG expansion induces nucleolar stress in polyglutamine diseases. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13428–33. doi: 10.1073/pnas.1204089109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davies JE, Sarkar S, Rubinsztein DC. Trehalose reduces aggregate formation and delays pathology in a transgenic mouse model of oculopharyngeal muscular dystrophy. Human molecular genetics. 2006;15:23–31. doi: 10.1093/hmg/ddi422. [DOI] [PubMed] [Google Scholar]
- 77.Megeney LA, Rudnicki MA. Determination versus differentiation and the MyoD family of transcription factors. Biochemistry and cell biology = Biochimie et biologie cellulaire. 1995;73:723–32. doi: 10.1139/o95-080. [DOI] [PubMed] [Google Scholar]
- 78.Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. The Journal of cell biology. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raz V, Routledge S, Venema A, Buijze H, van der Wal E, Anvar S, Straasheijm KR, Klooster R, Antoniou M, van der Maarel SM. Modeling oculopharyngeal muscular dystrophy in myotube cultures reveals reduced accumulation of soluble mutant PABPN1 protein. The American journal of pathology. 2011;179:1988–2000. doi: 10.1016/j.ajpath.2011.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anvar SY, t Hoen PA, Venema A, van der Sluijs B, van Engelen B, Snoeck M, Vissing J, Trollet C, Dickson G, Chartier A, Simonelig M, van Ommen GJ, van der Maarel SM, Raz V. Deregulation of the ubiquitin-proteasome system is the predominant molecular pathology in OPMD animal models and patients. Skeletal muscle. 2011;1:15. doi: 10.1186/2044-5040-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Trollet C, Anvar SY, Venema A, Hargreaves IP, Foster K, Vignaud A, Ferry A, Negroni E, Hourde C, Baraibar MA, t Hoen PA, Davies JE, Rubinsztein DC, Heales SJ, Mouly V, van der Maarel SM, Butler-Browne G, Raz V, Dickson G. Molecular and phenotypic characterization of a mouse model of oculopharyngeal muscular dystrophy reveals severe muscular atrophy restricted to fast glycolytic fibres. Human molecular genetics. 2010;19:2191–207. doi: 10.1093/hmg/ddq098. [DOI] [PubMed] [Google Scholar]
- 82.Catoire H, Pasco MY, Abu-Baker A, Holbert S, Tourette C, Brais B, Rouleau GA, Parker JA, Neri C. Sirtuin inhibition protects from the polyalanine muscular dystrophy protein PABPN1. Human molecular genetics. 2008;17:2108–17. doi: 10.1093/hmg/ddn109. [DOI] [PubMed] [Google Scholar]
- 83.Pasco MY, Catoire H, Parker JA, Brais B, Rouleau GA, Neri C. Cross-talk between canonical Wnt signaling and the sirtuin-FoxO longevity pathway to protect against muscular pathology induced by mutant PABPN1 expression in C. elegans. Neurobiology of disease. 2010;38:425–33. doi: 10.1016/j.nbd.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 84.Chartier A, Benoit B, Simonelig M. A Drosophila model of oculopharyngeal muscular dystrophy reveals intrinsic toxicity of PABPN1. The EMBO journal. 2006;25:2253–62. doi: 10.1038/sj.emboj.7601117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chartier A, Raz V, Sterrenburg E, Verrips CT, van der Maarel SM, Simonelig M. Prevention of oculopharyngeal muscular dystrophy by muscular expression of Llama single-chain intrabodies in vivo. Human molecular genetics. 2009;18:1849–59. doi: 10.1093/hmg/ddp101. [DOI] [PubMed] [Google Scholar]
- 86.Barbezier N, Chartier A, Bidet Y, Buttstedt A, Voisset C, Galons H, Blondel M, Schwarz E, Simonelig M. Antiprion drugs 6-aminophenanthridine and guanabenz reduce PABPN1 toxicity and aggregation in oculopharyngeal muscular dystrophy. EMBO molecular medicine. 2011;3:35–49. doi: 10.1002/emmm.201000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Davies JE, Wang L, Garcia-Oroz L, Cook LJ, Vacher C, O’Donovan DG, Rubinsztein DC. Doxycycline attenuates and delays toxicity of the oculopharyngeal muscular dystrophy mutation in transgenic mice. Nature medicine. 2005;11:672–7. doi: 10.1038/nm1242. [DOI] [PubMed] [Google Scholar]
- 88.Davies JE, Rose C, Sarkar S, Rubinsztein DC. Cystamine suppresses polyalanine toxicity in a mouse model of oculopharyngeal muscular dystrophy. Science translational medicine. 2010;2:34ra40. doi: 10.1126/scitranslmed.3000723. [DOI] [PubMed] [Google Scholar]
- 89.Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development (Cambridge, England) 2012;139:641–56. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Riddle DL, Blumenthal T, Meyer BJ, Priess JR. Introduction to C elegans. 1997. [PubMed] [Google Scholar]
- 91.Hino H, Araki K, Uyama E, Takeya M, Araki M, Yoshinobu K, Miike K, Kawazoe Y, Maeda Y, Uchino M, Yamamura K. Myopathy phenotype in transgenic mice expressing mutated PABPN1 as a model of oculopharyngeal muscular dystrophy. Human molecular genetics. 2004;13:181–90. doi: 10.1093/hmg/ddh017. [DOI] [PubMed] [Google Scholar]
- 92.Dion P, Shanmugam V, Gaspar C, Messaed C, Meijer I, Toulouse A, Laganiere J, Roussel J, Rochefort D, Laganiere S, Allen C, Karpati G, Bouchard JP, Brais B, Rouleau GA. Transgenic expression of an expanded (GCG)13 repeat PABPN1 leads to weakness and coordination defects in mice. Neurobiology of disease. 2005;18:528–36. doi: 10.1016/j.nbd.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 93.Boukriche Y, Maisonobe T, Masson C. Neurogenic involvement in a case of oculopharyngeal muscular dystrophy. Muscle & nerve. 2002;25:98–101. doi: 10.1002/mus.1213. [DOI] [PubMed] [Google Scholar]
- 94.Schober R, Kress W, Grahmann F, Kellermann S, Baum P, Gunzel S, Wagner A. Unusual triplet expansion associated with neurogenic changes in a family with oculopharyngeal muscular dystrophy. Neuropathology. 2001;21:45–52. doi: 10.1046/j.1440-1789.2001.00374.x. [DOI] [PubMed] [Google Scholar]
- 95.Blumen SC, Bouchard JP, Brais B, Carasso RL, Paleacu D, Drory VE, Chantal S, Blumen N, Braverman I. Cognitive impairment and reduced life span of oculopharyngeal muscular dystrophy homozygotes. Neurology. 2009;73:596–601. doi: 10.1212/WNL.0b013e3181b388a3. [DOI] [PubMed] [Google Scholar]
- 96.Dubbioso R, Moretta P, Manganelli F, Fiorillo C, Iodice R, Trojano L, Santoro L. Executive functions are impaired in heterozygote patients with oculopharyngeal muscular dystrophy. Journal of neurology. 2012;259:833–7. doi: 10.1007/s00415-011-6255-y. [DOI] [PubMed] [Google Scholar]
- 97.Mankodi A, Wheeler TM, Shetty R, Salceies KM, Becher MW, Thornton CA. Progressive myopathy in an inducible mouse model of oculopharyngeal muscular dystrophy. Neurobiology of disease. 2012;45:539–46. doi: 10.1016/j.nbd.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Davies JE, Sarkar S, Rubinsztein DC. Wild-type PABPN1 is anti-apoptotic and reduces toxicity of the oculopharyngeal muscular dystrophy mutation. Human molecular genetics. 2008;17:1097–108. doi: 10.1093/hmg/ddm382. [DOI] [PubMed] [Google Scholar]
- 99.Schroder JM, Klossok T, Weis J. Oculopharyngeal muscle dystrophy: fine structure and mRNA expression levels of PABPN1. Clinical neuropathology. 2011;30:94–103. doi: 10.5414/npp30094. [DOI] [PubMed] [Google Scholar]
- 100.Perie S, Mamchaoui K, Mouly V, Blot S, Bouazza B, Thornell LE, St Guily JL, Butler-Browne G. Premature proliferative arrest of cricopharyngeal myoblasts in oculo-pharyngeal muscular dystrophy: Therapeutic perspectives of autologous myoblast transplantation. Neuromuscul Disord. 2006;16:770–81. doi: 10.1016/j.nmd.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 101.Gidaro T, Negroni E, Perie S, Mirabella M, Laine J, Lacau St Guily J, Butler-Browne G, Mouly V, Trollet C. Atrophy, Fibrosis, and Increased PAX7-Positive Cells in Pharyngeal Muscles of Oculopharyngeal Muscular Dystrophy Patients. Journal of neuropathology and experimental neurology. 2013;72:234–243. doi: 10.1097/NEN.0b013e3182854c07. [DOI] [PubMed] [Google Scholar]
- 102.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nature medicine. 2004;10(Suppl):S10–7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 103.Di Zanni E, Bachetti T, Parodi S, Bocca P, Prigione I, Di Lascio S, Fornasari D, Ravazzolo R, Ceccherini I. In vitro drug treatments reduce the deleterious effects of aggregates containing polyAla expanded PHOX2B proteins. Neurobiology of disease. 2012;45:508–18. doi: 10.1016/j.nbd.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 104.Uyama E, Tsukahara T, Goto K, Kurano Y, Ogawa M, Kim YJ, Uchino M, Arahata K. Nuclear accumulation of expanded PABP2 gene product in oculopharyngeal muscular dystrophy. Muscle & nerve. 2000;23:1549–54. doi: 10.1002/1097-4598(200010)23:10<1549::aid-mus11>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 105.Berciano MT, Villagra NT, Ojeda JL, Navascues J, Gomes A, Lafarga M, Carmo-Fonseca M. Oculopharyngeal muscular dystrophy-like nuclear inclusions are present in normal magnocellular neurosecretory neurons of the hypothalamus. Human molecular genetics. 2004;13:829–38. doi: 10.1093/hmg/ddh101. [DOI] [PubMed] [Google Scholar]
- 106.Huang CC, Faber PW, Persichetti F, Mittal V, Vonsattel JP, MacDonald ME, Gusella JF. Amyloid formation by mutant huntingtin: threshold, progressivity and recruitment of normal polyglutamine proteins. Somatic cell and molecular genetics. 1998;24:217–33. doi: 10.1023/b:scam.0000007124.19463.e5. [DOI] [PubMed] [Google Scholar]
- 107.Preisinger E, Jordan BM, Kazantsev A, Housman D. Evidence for a recruitment and sequestration mechanism in Huntington’s disease. Philosophical transactions of the Royal Society of London. 1999;354:1029–34. doi: 10.1098/rstb.1999.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Human molecular genetics. 2000;9:2799–809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 109.Bruneau S, Johnson KR, Yamamoto M, Kuroiwa A, Duboule D. The mouse Hoxd13(spdh) mutation, a polyalanine expansion similar to human type II synpolydactyly (SPD), disrupts the function but not the expression of other Hoxd genes. Developmental biology. 2001;237:345–53. doi: 10.1006/dbio.2001.0382. [DOI] [PubMed] [Google Scholar]
- 110.Bengoechea R, Tapia O, Casafont I, Berciano J, Lafarga M, Berciano MT. Nuclear speckles are involved in nuclear aggregation of PABPN1 and in the pathophysiology of oculopharyngeal muscular dystrophy. Neurobiology of disease. 2012;46:118–29. doi: 10.1016/j.nbd.2011.12.052. [DOI] [PubMed] [Google Scholar]
- 111.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 112.Stromme P, Mangelsdorf ME, Shaw MA, Lower KM, Lewis SM, Bruyere H, Lutcherath V, Gedeon AK, Wallace RH, Scheffer IE, Turner G, Partington M, Frints SG, Fryns JP, Sutherland GR, Mulley JC, Gecz J. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nature genetics. 2002;30:441–5. doi: 10.1038/ng862. [DOI] [PubMed] [Google Scholar]
- 113.Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–8. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Renault V, Thornell LE, Eriksson PO, Butler-Browne G, Mouly V. Regenerative potential of human skeletal muscle during aging. Aging cell. 2002;1:132–9. doi: 10.1046/j.1474-9728.2002.00017.x. [DOI] [PubMed] [Google Scholar]
- 115.Conboy IM, Rando TA. Aging, stem cells and tissue regeneration: lessons from muscle. Cell cycle (Georgetown, Tex. 2005;4:407–10. doi: 10.4161/cc.4.3.1518. [DOI] [PubMed] [Google Scholar]
- 116.Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, Shkreli M, Delp S, Pomerantz JH, Artandi SE, Blau HM. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143:1059–71. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fischer MD, Budak MT, Bakay M, Gorospe JR, Kjellgren D, Pedrosa-Domellof F, Hoffman EP, Khurana TS. Definition of the unique human extraocular muscle allotype by expression profiling. Physiological genomics. 2005;22:283–91. doi: 10.1152/physiolgenomics.00158.2004. [DOI] [PubMed] [Google Scholar]
- 118.Porter JD, Israel S, Gong B, Merriam AP, Feuerman J, Khanna S, Kaminski HJ. Distinctive morphological and gene/protein expression signatures during myogenesis in novel cell lines from extraocular and hindlimb muscle. Physiological genomics. 2006;24:264–75. doi: 10.1152/physiolgenomics.00234.2004. [DOI] [PubMed] [Google Scholar]
- 119.Pavlath GK, Thaloor D, Rando TA, Cheong M, English AW, Zheng B. Heterogeneity among muscle precursor cells in adult skeletal muscles with differing regenerative capacities. Developmental dynamics : an official publication of the American Association of Anatomists. 1998;212:495–508. doi: 10.1002/(SICI)1097-0177(199808)212:4<495::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 120.Sambasivan R, Kuratani S, Tajbakhsh S. An eye on the head: the development and evolution of craniofacial muscles. Development (Cambridge, England) 2011;138:2401–15. doi: 10.1242/dev.040972. [DOI] [PubMed] [Google Scholar]