Abstract
Neoadjuvant radiochemotherapy is the gold standard for locally advanced rectal cancer, but this strategy does not achieve benefit in all patients. Analysis of intracellular signal pathways in patients with locally advanced rectal cancer identified a phosphoproteomic profile that may cluster patients who do not respond to neoadjuvant chemoradiotherapy.
Background
Currently there is no reliable technique for predicting clinical or pathologic complete tumor response after radiochemotherapy (RCT) in patients with rectal cancer. We applied reverse phase protein microarray (RPMA) technology to find a signal pathway that may predict the response to preoperative treatment.
Patients and Methods
Fifteen rectal cancer samples were collected during preoperative RCT. Seven patients had a good response to preoperative therapy (Mandard grade I–II) and 8 patients had a poor response (Mandard grade III-V). Using laser capture microdissection (LCM) and RPMA analysis, we measured the phosphorylation level of nearly 80 end points and analyzed the signaling pathways.
Results
We identified 4 signaling proteins whose phosphorylation levels were significantly different (P < .05) between the good vs. poor responders; CHK2 and β-catenin were more highly phosphorylated in poor responders, whereas PDK1 and glycogen synthase kinase (GSK)-3α/β had lower phosphorylation levels in poor responders. Interestingly GSK-3α/β, β-catenin, and PDK1 are all present in the phosphatidylinositol-3-kinase (PI3K)-AKT signaling pathway.
Conclusions
Based on our results, we hypothesize that the activating state of the PI3K-AKT pathway can stratify patients who could benefit most from neoadjuvant treatment. Moreover, identification of theranostic targets has the potential to pinpoint new therapeutic strategies for the nonresponsive population.
Keywords: Predictive marker, Protein kinase, Rectal cancer, Neoadjuvant treatment
Introduction
Colon and rectal cancers accounted for about 1 million new cases in 2002 (9.4% of the world total) and colorectal cancers rank fourth in frequency in men and third in frequency in women. Survival estimates at 5 years are 65% in North America and 54% in western Europe. Since patients with locally advanced disease have a high risk of local recurrence,1,2 neoadjuvant radiochemotherapy (RCT) is increasingly accepted as the gold standard for patients with stage T3 or T4 and/or stage N1 to N2 rectal tumors. Neoadjuvant therapy has been shown to significantly reduce the risk of local recurrence compared with surgery alone and also when compared with adjuvant RCT.3 Moreover, this approach allows an increase of more than 20% in the rate of sphincter-saving surgery.3
Only a portion of patients will respond to RCT, with a pathologic complete response of 6% to 26% and pathologic downstaging in 40% to 60% of patients; moreover, clinical complete response is associated with pathologic complete response in 30% of cases.4 However, the majority of patients who underwent neoadjuvant RCT experienced toxicity from chemotherapy or radiotherapy; up to 85% of patients will experience grade 0 to grade 2 toxicity and up to 15% of patients will have grade 3 to 4 toxicity.5,6 Some authors claim that preoperative treatment increases the postoperative complications, especially anastomotic leakage.7,8 The possibility of identifying which patients are resistant to RCT or which are likely to benefit from the treatment would confer major optimization on current treatment regimens. More than 1200 articles can be found on PubMed concerning the possible utility of several markers to predict which cancer patients will respond to neoadjuvant therapy; unfortunately, at present none of these can be translated to a clinical setting.9
Reverse phase protein array (RPMA) has recently been developed to map the state of key signal transduction pathways from human specimens by looking at dozens of kinases at once through multiplexed phosphospecific antibody analysis.10–14 More than 100 protein kinases are implicated in human cancers: activation and inhibition of protein kinases by phosphorylation is a central element in cancer and cancer therapeutics. A deregulation of the signaling pathways leads to alterations in cell growth, apoptosis, proliferation, migration, and adhesion.
The aim of this study was to use multiplexed RPMA-based functional protein pathway mapping to identify the pathway biomarkers that could be predictive of response to RCT neoadjuvant treatment.
Patients and Methods
Patients
The patient population consisted of 15 patients (13 men and 2 women; mean age 59 years [range, 39–72 years]) with primary adenocarcinoma of the midrectum (7–11 cm from the anal verge) or lower rectum (4–7 cm from the anal verge) who received neoadjuvant chemoradiotherapy and then underwent surgical resection between 2002 and 2007 at the Clinica Chirurgica II, Department of Oncological and Surgical Science, University of Padova and at the Department of Surgical Oncology, CRO-IRCCS National Cancer Institute of Aviano, Italy.
Pretreatment Staging
Pretreatment staging consisted of proctoscopy, transrectal ultra-sonography (TRUS), pelvic computed tomography (CT), and magnetic resonance imaging (MRI). Tissue sampling was done before preoperative therapy during colonoscopy examination at the time of diagnosis and tissues were immediately snap frozen and stored in liquid nitrogen until used. Five tumors were located in the midrectum and 10 in the lower rectum. Eleven patients were classified as having N-positive disease (stage III): 1 patient had T2 disease, 6 patients had T3 disease, and 4 patients had T4 disease. Four patients were classified as having N-negative disease: 1 patient had stage I, T2 disease and 3 patients had stage II, T3 disease (Table 1).
Table 1.
Clinicopathologic Characteristics of the 15 Patients Who Underwent Preoperative Radiochemotherapy and Surgery for Midrectum and Lower Rectum Cancer
| Patients | Age (y) | Radiotherapy (Gy) | Computed Tomography |
cTNM | pTNM | Stage | Stage | Mandard |
|---|---|---|---|---|---|---|---|---|
| Preoperative | Preoperative | Preoperative | Preoperative | Postoperative | Score | |||
| 1 | 52 | 5040 | 5-FU+ oxa | T4N1M0 | T2N0M1 | III | IV | 4 |
| 2 | 46 | 5040 | 5-FU | T4N1M0 | T3N1M0 | III | III | 2 |
| 3 | 55 | 5040 | 5-FU | T4N1M0 | T3N0M0 | III | II | 3 |
| 4 | 64 | 5040 | 5-FU | T3N1M0 | T3N0M0 | III | II | 3 |
| 5 | 72 | 5040 | 5-FU | T3N1M0 | T0N0M0 | III | 0 | 1 |
| 6 | 66 | 5040 | 5-FU | T3N1M0 | T2N0M0 | III | II | 5 |
| 7 | 51 | 5040 | 5-FU | T4N1M0 | T2N0M0 | III | II | 3 |
| 8 | 61 | 3060 | Cape+oxa | T2N1M0 | T1N0M0 | III | I | 4 |
| 9 | 68 | 5040 | 5-FU+oxa | T3N1M0 | T0N0M0 | III | 0 | 1 |
| 10 | 64 | 5500 | Cape | T3N1M0 | T2N0M0 | III | I | 3 |
| 11 | 39 | 5040 | Cape+oxa | T3N0M0 | T3N0M0 | II | II | 3 |
| 12 | 63 | 5040 | Cape+oxa | T3N0M0 | T2N0M0 | II | I | 2 |
| 13 | 59 | 5040 | Cape+oxa | T3N1M0 | T0N0M0 | III | 0 | 1 |
| 14 | 63 | 5040 | Cape | T2N0M0 | T0N0M1 | I | IV | 1 |
| 15 | 61 | 5040 | Cape | T3N0M0 | T1N0M0 | II | I | 2 |
Abbreviations: 5-FU = 5-fluorouracil; Oxa = oxaliplatin; Cape = capecitabine.
Neoadjuvant Regimen
External-beam radiotherapy was delivered in fractions of 1.8 Gy/d by using the 3-field or box technique: 13 patients received a total of 50.4 Gy, 1 received a total of 55.0 Gy, and 1 received only 30.6 Gy because of toxicity.
Patients received chemotherapy throughout the course of radiotherapy: 5 patients received oral capecitabine plus intravenous oxaliplatin, 2 patients received oral capecitabine, 2 patients received intravenous 5-fluorouracil (5-FU) plus intravenous oxaliplatin, and 6 patients received intravenous 5-FU (Table 1).
Adverse effects were graded according to the National Cancer Institute Common Toxicity Criteria, version 3.0. During chemotherapy the following adverse events were observed: 1 (6.6%) patient had grade 3 to 4 gastrointestinal toxicity and 6 (40%) patients had grade 1 to 2 gastrointestinal toxicity, 4 (26.6%) patients had grade 1 to 2 hematologic toxicity, 3 (20%) patients had grade 1 to 2 neuropathy, 1 (6.6%) patient had grade 3 to 4 skin toxicity, and 5 (33.3%) patients had grade 1 to 2 skin toxicity.
Surgery
Surgery was planned for 6 to 8 weeks after completion of preoperative CRT. Eleven patients (73.4%) underwent low anterior resection with colorectal anastomosis, 2 (13.3%) patients had low anterior resection with coloanal anastomosis, and 2 (13.3%) patients underwent transanal excision.
Postoperative major complications occurred in 4 (26.6%) patients: 3 patients experienced anastomotic leak and 1 patient had a rectovaginal fistula.
Restaging
Restaging by using pelvic CT and MRI, TRUS, and flexible rectosigmoidoscopy with at least 8 biopsies of the tumor bed were planned on the fifth week after completion of RCT.
Histopathologic Staging
Staging was carried out according to the TNM classification. T staging was available in all patients, with the following distribution: T0, n = 4 (26.7%); T1, n = 2 (13.3%); T2, n = 5 (33.3%); and T3, n = 4 (26.7%). N staging was available in all patients, 14 (93.3%) of whom had N-negative disease and 1 (6.7%) of whom had N-positive disease.
One patient with a complete response on the rectal wall (pT0) and 1 patient with a partial response on the rectal wall (pT2 from cT4) and complete response on the mesorectal lymph node (pN0) had previously undetected liver metastases found at operation (Table 1).
The evaluation of the tumor response to neoadjuvant treatment was performed based on the Mandard classification.15
Reverse Phase Protein Microarray Analysis
RPMA analysis was used in combination with LCM, as described previously.10–14 LCM was used to obtain a pure tumor cell population from all the biopsy samples. Eight-micrometer sections were obtained from each tissue and approximately 15,000 tumor cells underwent microdissection using a Pix Cell II Laser Capture System (PXL-200; Arcturus Engineering, Mountain View, CA). Microdissected cells were lysed in 30 µL of lysis buffer containing a 1:1 mixture of 2× Tris-glycine SDS Sample buffer with B mercaptoethanol (Novex, Life Technologies, Grand Island, NY) and TPER Reagent (Pierce Protein Research Products, Thermo Fisher Scientific, Inc, Rockford, IL). After cell lysis, samples were boiled and arrayed on an Aushon 2470 arrayer (Aushon BioSystems Inc, Billerica, MA) equipped with 350-µm pins and printed in duplicate 5-point 1:2 dilutions onto nitrocellulose-coated slides (FAST slides, Whatman, GE Healthcare, Little Chalfont, Buckinghamshire, UK).
Staining was carried out on an automated slide stainer (Dako, Carpinteria, CA) using the catalyzed signal amplification system (Dako). After application of cellular lysates, the slides were treated for 15 minutes with Reblot (Chemicon, Temecula, CA) and then washed 3 times for 10 minutes each time with TBS washing buffer. After treatment the slides were blocked in I Block Protein Blocking solution (Applied Biosystems, Carlsbad, CA) under constant rocking. Endogenous biotin and avidin were blocked using the biotin blocking kit (Dako), followed by application of protein block (Dako), primary antibody, and secondary antibody. The specificity of each antibody was tested by rigorous validation by Western blot. Only antibodies that generate a single band at the correct molecular weight and are ligand inducible are acceptable for RPMA. We used 80 antibodies: 77 directed against phosphorylation sites of phosphoproteins and 3 against total phosphoproteins (endothelial growth factor receptor [EGFR], cErb2/HER2, SMAC [second mitochondria-derived activator of caspases], and DIABLO).
All protein values were normalized to total protein to account for differences in intensity solely from starting lysate concentration variance. The total amount of protein present in each sample was estimated through SYPRO Ruby Protein Blot Stain (Molecular Probes, Eugene, OR) according to the manufacturer’s instructions as previously described.12–14 Stained slides were scanned individually on a Umax PowerLook III scanner (Umax Technologies, Dallas, TX) at 600 dpi and saved as TIFF files in Photoshop 6.0 (Adobe, San Jose, CA). The TIFF images for antibody-stained slides and SYPRO-stained slides were analyzed with array analysis software designed for protein microarray analysis: version 2.X00 (Vigene Tech, North Billerica, MA). The software performed spot finding, local background subtraction, replicate averaging, and total protein normalization, producing a single value for each sample at each end point.
Statistical Analysis
Statistical analyses were performed with SAS, version 9 software (SAS Institute, Cary, NC). The differences of end point intensities between the 2 groups were assessed. Initially, the distribution of variables was checked. If 2 groups of the variable followed the normal distribution, a 2-sample t test was performed. If the variances of 2 groups were equal, a 2-sample t test with a pooled variance procedure was used to compare the means of intensity between the 2 groups. Otherwise, a 2-sample t test without a pooled variance procedure was adopted. For nonnormally distributed variables, the Wilcoxon rank-sum test was used. All significance levels were set at P = .05. Unsupervised hierarchical clustering was performed using JMP analysis, (5.1 software; SAS Institute).
Results
Protein Signal Transduction Mapping
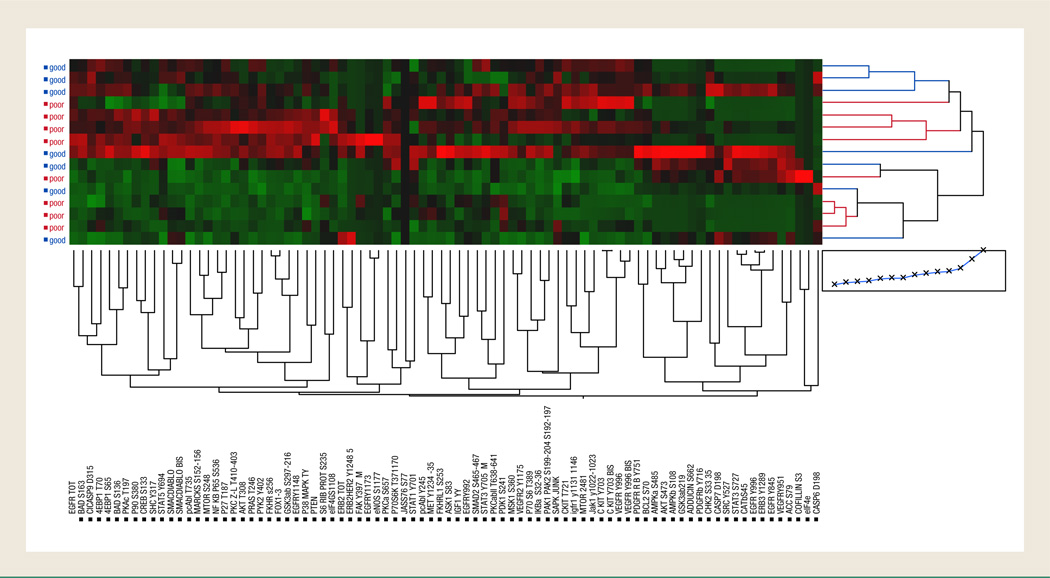
The RPMA data obtained from the 15 patient tumor specimens analyzed were divided into 2 groups according to the pathologic response of the patient; group A (good responders) included patients whose response was grade I and II (7 patients) and group B (poor responders) included patients with grade III, IV, or V (8 patients). These samples were analyzed for signaling pathway activation by RPMA using a series of 80 phosphospecific and total protein antibodies (Table 2) against key signaling molecules regulating cell prolif-eration, motility, apoptosis, and survival. Unsupervised 2-way hierarchical clustering analysis (Figure 1) using all the 80 signaling end points measured failed to correctly segregate the good responders from the poor responders, which indicates that global signaling is not significantly altered between the 2 cohorts. As seen in Figure 1, we noted a high degree of heterogeneity in the pathway portraits, with each patient’s tumor profile appearing as a unique molecular subtype.
Table 2.
End Points Whose Phosphorylation Levels Were Tested in the 15 Patients Using RPMA
| End Point | Company | Dilution | P Value |
|---|---|---|---|
| Acetyl-CoA Carboxylase (S79) | Cell signaling | 1:50 | 0.1 |
| 4E-BP1 (S65) | Cell signaling | 1:50 | 0.6 |
| 4E-BP1 (T70) | Cell signaling | 1:200 | 0.9 |
| Adducin (S662) | Upstate | 1:200 | 0.07 |
| Akt (S473) | Cell signaling | 1:100 | 0.1 |
| Akt (T308) | Cell signaling | 1:100 | 0.8 |
| AMPK Alpha (S485) | Cell signaling | 1:50 | 0.09 |
| AMPK Beta1 (S108) | Cell signaling | 1:50 | 0.1 |
| ASK1 (S83) | Cell signaling | 1:50 | 0.7 |
| Bad (S136) | Cell signaling | 1:200 | 0.6 |
| Bad (S163) | Cell signaling | 1:50 | 0.6 |
| BclII (S70) | Cell signaling | 1:200 | 0.2 |
| c-Kit (Y703) | Zymed | 1:200 | 0.9 |
| Caspase 6, Cleaved (D162) | Cell signaling | 1:50 | 0.07 |
| Caspase 7, Cleaved (D198) | Cell signaling | 1:100 | 0.07 |
| Catenin (beta) (S45) | Cell signaling | 1:50 | 0.05 |
| Chk2 (S33/35) | Cell signaling | 1:50 | 0.02 |
| c-Kit (T721) | Zymed | 1:200 | 0.3 |
| Caspase 9, Cleaved (D315) | Cell signaling | 1:50 | 0.45 |
| Cofilin (S3) | Cell signaling | 1:100 | 0.85 |
| CREB (S133) | Cell signaling | 1:200 | 0.4 |
| EGFR Total | Cell signaling | 1:100 | 0.8 |
| EGFR (Y845) | Cell signaling | 1:50 | 0.09 |
| EGFR (Y996) | Cell signaling | 1:100 | 0.04 |
| EGFR (Y1148) | Biosource | 1:100 | 0.3 |
| EGFR (Y1173) | Biosource | 1:100 | 0.3 |
| EGFR (Y992) | Cell signaling | 1:100 | 0.6 |
| eIF4E (S209) | Cell signaling | 1:50 | 0.2 |
| eIF4G (S1108) | Cell signaling | 1:1000 | 0.7 |
| eNOS (S1177) | Cell signaling | 1:50 | 0.42 |
| cErb2/HER2 Total | Cell signaling | 1:100 | 0.8 |
| ErbB2/HER2 (Y1248) | Upstate | 1:1000 | 0.8 |
| ErbB3/HER3 (Y1289) (21D3) | Cell signaling | 1:100 | 0.07 |
| Fak (Y397) (18) | BD | 1:50 | 0.4 |
| FKHR (S256) | Cell signaling | 1:100 | 0.6 |
| FKHRL1 (S253) FOXO1 | Upstate | 1:1000 | 0.6 |
|
FKHR (T24)/FKHRL1 (T32) FOXO1-O3a |
Cell signaling | 1:200 | 0.9 |
| GSK-3α/β (Y279/216) | Biosourse | 1:500 | 0.5 |
| GSK-3α/β (S21/9) | Cell signaling | 1:100 | 0.04 |
|
IGF-1R (Y1135/36)/IR (Y1150/51) |
Cell signaling | 1:500 | 0.8 |
|
IGF-1 Rec (Y1131)/insulin Rec (Y1146) |
Cell signaling | 1:500 | 0.6 |
| IκB-Alpha (S32/36) | Cell signaling | 1:100 | 0.6 |
| Jak1 (Y1022/1023) | Cell signaling | 1:50 | 0.1 |
| MARCKS (S152/156) | Cell signaling | 1:100 | 0.7 |
| Met (Y1234/1235) | Cell signaling | 1:200 | 0.3 |
| MSK-1 (S360) | Cell signaling | 1:50 | 0.5 |
| MTOR (S2481) | Cell signaling | 1:100 | 0.6 |
| MTOR (S2448) | Cell signaling | 1:100 | 0.8 |
| NF-κB p65 (S536) | Cell signaling | 1:50 | 0.8 |
| p27 (T187) | Zymed | 1:200 | 0.8 |
| p38 MAP Kinase (T180/Y182) | Cell signaling | 1:50 | 1 |
| p70 S6 Kinase (T389) | Cell signaling | 1:20 | 0.6 |
| p70 S6 Kinase (S371) | Cell signaling | 1:50 | 0.8 |
| p90RSK (S380) | Cell signaling | 1:200 | 0.8 |
|
PAK1 (S199/204)/PAK2 (S192/197) |
Cell signaling | 1:50 | 0.95 |
| c-Abl (T735) | Cell signaling | 1:50 | 0.7 |
| c-Abl (Y245) | Cell signaling | 1:50 | 0.4 |
| PDGF Receptor Beta (Y751) | Cell signaling | 1:50 | 0.1 |
| PDGF Receptor Beta (Y716) | Upstate | 1:200 | 0.3 |
| PDK1 (S241) | Cell signaling | 1:200 | 0.04 |
| PKA C (T197) | Cell signaling | 1:200 | 0.45 |
| PKC Zeta/Lambda (T410/403) | Cell signaling | 1:50 | 0.8 |
| PKC Alpha (S657) | Upstate | 1:1000 | 0.2 |
| PKC Alpha/Beta II (T638/641) | Cell signaling | 1:50 | 0.9 |
| PRAS40 (T246) | Biosourse | 1:1000 | 0.6 |
| PTEN (S380) | Cell signaling | 1:500 | 0.6 |
| Pyk2 (Y402) | Cell signaling | 1:200 | 0.8 |
| S6 Ribosomal Protein (S235/236) (2F9) | Cell signaling | 1:200 | 0.5 |
| SAPK JNK (T183/Y185) | Cell signaling | 1:200 | 0.9 |
| Shc (Y317) | Upstate | 1:200 | 0.6 |
| SMAC DIABLO Total | Cell signaling | 1:1000 | 0.7 |
| SMAD2 (S465/467) | Cell signaling | 1:250 | 0.5 |
| Src (Y527) | Cell signaling | 1:500 | 0.07 |
| STAT1 (Y701) | Upstate | 1:200 | 0.8 |
| STAT3 (S727) | Cell signaling | 1:200 | 0.07 |
| STAT3 (Y705) | Upstate | 1:100 | 1 |
| STAT5 (Y694) | Cell signaling | 1:50 | 1 |
| VEGFR 2 (Y996) | Cell signaling | 1:50 | 1 |
| VEGFR 2 (Y1175) | Cell signaling | 1:50 | 0.3 |
| VEGFR 2 (Y951) | Cell signaling | 1:200 | 0.32 |
Figure 1.
Unsupervised Hierarchical 2-Way Clustering Analysis of the 15 Patients and the 80 End Points; There is No Clear Segregation Between the Responder and Nonresponder Groups of Patients According to the Levels of Phosphorylation of the 80 Protein Kinases
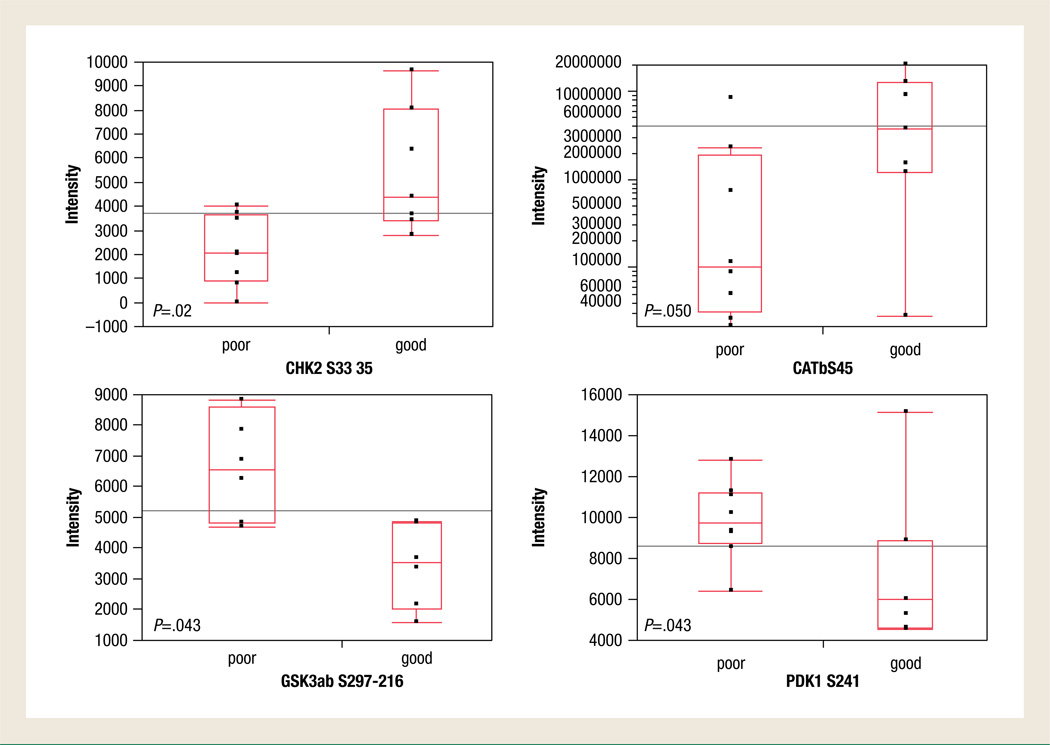
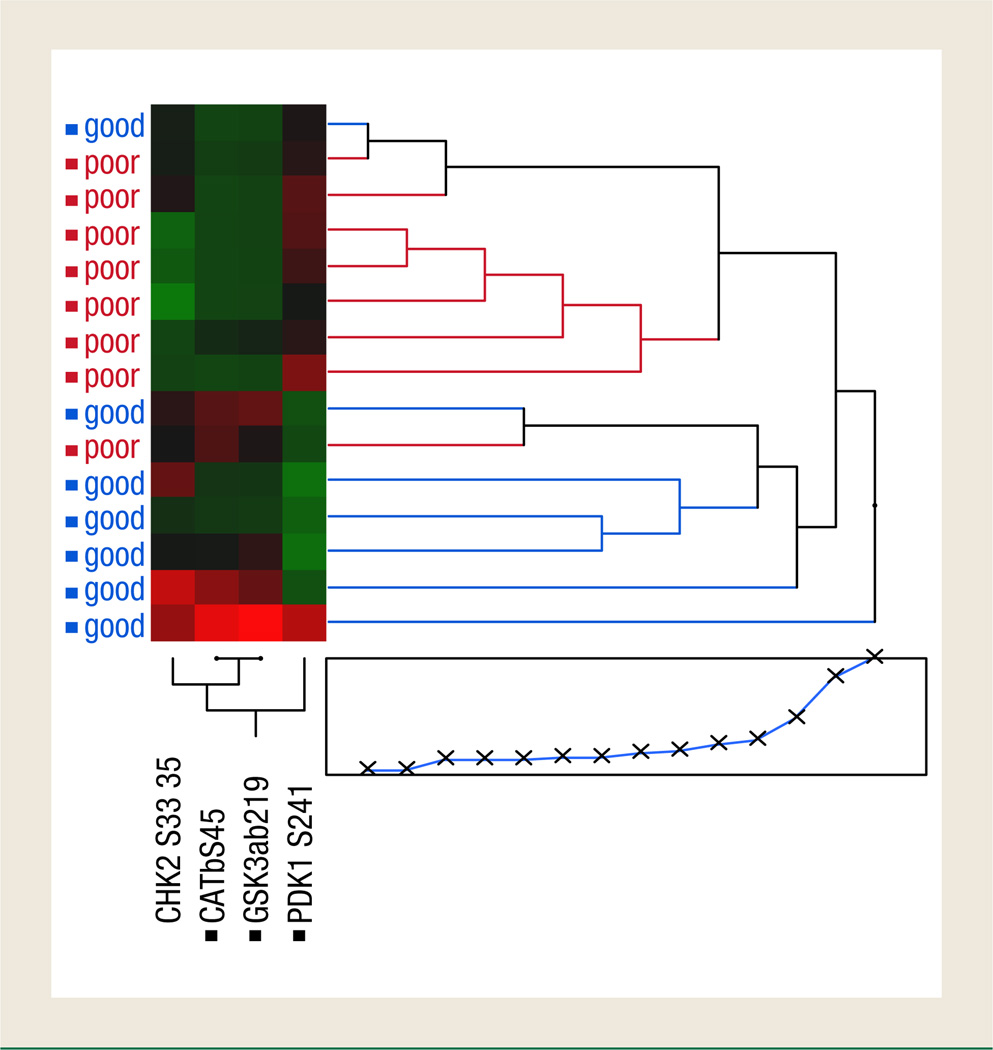
To determine if any specific protein signaling activation was different between good and poor responders, we used supervised statistical analysis of each end point separately. Student t testing or Wilcoxon rank-sum analysis identified 4 signaling molecules whose phosphorylation levels were significantly different between the groups: CHK2 and β-catenin had higher phosphorylation levels in the tumor epithelium of patients with good response, whereas PDK1 and glycogen synthase kinase (GSK)-3α/β had lower phosphorylation levels in this cohort (Figures 2 and 3). The remaining 76 proteins were not statistically significantly different between these 2 groups.
Figure 2.
Comparisons of Phosphorylation Levels of Protein Kinases that were Statistically Different Between Poor and Good Responders
Figure 3.
Unsupervised 2-Way Hierarchical Clustering Analysis Using Only the Protein Kinases that were Statistically Different Between Poor and Good Responders
Discussion
Neoadjuvant RCT is the standard therapy for the treatment of locally advanced rectal cancer, the goal of which is to improve local control of disease, reduce the risk of local recurrence, and increase the rate of sphincter-saving rectal surgery.3 However the response rate is mild to weak because 50% of patients do not respond to neoadjuvant chemotherapy, and more than 80% of patients experience toxicity from chemotherapy or radiotherapy.4–6 Despite the initial description of new biomarkers predictive of response to neoadjuvant treatment in the past few years, none of them have translated to routine clinical practice.9 Alterations in protein signaling and signal transduction network pathways are now considered to underpin most human malignancies16,17 and are the targets of many of the new molecularly targeted inhibitors. Given their central role in tumorigenesis, we sought to determine if protein signaling alterations could be found between patients with rectal cancer who respond to RCT and those who do not. We used a well-controlled clinical study set of tumor specimens that were obtained before any therapy and immediately snap frozen within 15 minutes of surgery. We used the RPMA, a novel protein microarray technology that we developed for quantitative multiplexed analysis of protein signaling activation in microscopic tumor biopsy specimens to measure the activation state of 80 key signaling proteins at once. We postulated that aberrant cell signaling activation may provide a survival advantage to rectal cancer epithelium and that tumors with these pathway alterations could resist the apoptotic effects of RCT. Our results presented herein indicate that response to chemoradiotherapy in patients with rectal cancer could be predicted based on statistically significant pathway activation levels of GSK-3α/β, β-catenin, and PDK1 (P = .043, P = .050, and P = .043, respectively); all are members of the β-catenin pathway.
β-catenin is a transcription regulator that can activate a large number of oncogenic target genes once it translocates into the nucleus, as it associates with members of the Tcf family of DNA binding,18–21 such as c-myc, cyclin D1, vascular endothelial growth factor (VEGF), COX-2, matrix metallopeptidase-9 (MMP-9), and epidermal growth factor (EGF), and shuts down other cell cycle regulator genes such as p21. In the normal cell, β-catenin joins with α-catenin to anchor the cytoplasmic domain of E-cadherin. The unattached β-catenin is phosphorylated by the complex glycogen synthase kinase 3β (GSK-3β)-adenomatosis polyposis coli (APC)-axin; once phosphorylated, the β-catenin is then ubiquitinylated by the β-TrCP-E3-ligase complex and delivered to the proteasome for destruction.22 However, when the Wnt pathway is activated, it induces the GBP/Frat-1 protein to pull the GSK-3β from axin and sequester axin23; the cytosolic β-catenin can then move to the nucleus and promote gene synthesis. Activation of the β-catenin pathway is known to play a significant role in the development of colorectal cancer.18,20 Moreover, the PI3K/AKT pathway can positively regulate β-catenin–dependent transcription,24,25 and pharmacologic or genetic inhibition of PI3K and AKT activity allows suppression of β-catenin activation.25 Experimental studies demonstrated that the regulation of GSK-3β activity by a small-molecule inhibitor induces a 10-fold increase in apoptosis mediated by 5-FU.26
This study has 2 main limitations. First, the small size may distort the statistical analysis, so it must be confirmed with a larger sample size. The second limitation is that the chemotherapy schedule in the preoperative treatment was heterogeneous—in particular, 6 patients received a combinations of 2 drugs (5-FU/capecitabine and oxaliplatin), which theoretically may improve tumor downstaging; however, the data available did not find a difference between patients receiving capecitabine with or without oxaliplatin and patients receiving 5-FU with or without oxaliplatin (phase III studies STAR [Studio Terapia Adiuvante Retto]-01 and ACCORD [Action Clinique Coordonnées en cancérologie Digestive] 12/405).27,28
Conclusion
Our study provides evidence about deregulation of the β-catenin pathway in patients who do not respond to neoadjuvant chemoradiotherapy. If these results are validated in larger studies, a number of intriguing clinical applications could arise. In the future, patients with rectal cancer could be prescreened for RCT before treatment using protein pathway analysis to ascertain the activation level of the β-catenin pathway in a tumor biopsy specimen. Those patients with relatively low pathway activation could be given RCT with an enriched chance of therapeutic and clinical success. Patients whose tumors have high β-catenin pathway activation could be selected for other therapeutic strategies (ie, surgery alone) and save them from RCT therapy that would not work. Furthermore, clinical trials could be initiated to evaluate the intriguing possibility of selecting and stratifying patients with rectal cancer whose tumors harbor high β-catenin pathway activation and treating them first with targeted inhibitors that decrease activation of the β-catenin pathway and then applying standard-of-care RCT; this would possibly convert a patient who would not otherwise respond to RCT to one who responds to such treatment. Such paradigms have been proposed before for other tumors in which pathway activation was found to underpin a lack of response to standard chemotherapy.29
Clinical Practice Points.
In patients with locally advanced rectal cancer, the use of neoadjuvant RCT may cause important toxicity, and at present there are no markers to predict benefit.
RPMA maps the state of key signal transduction pathways from human specimens, detecting activation and inhibition of protein kinases by phosphorylation.
Analyzing specimens from endoscopic biopsies obtained before neoadjuvant treatment, we found that the activating state of the PI3K-AKT pathway can stratify patients who could benefit most from neoadjuvant treatment. If confirmed in larger studies, these data will help to select patients for neoadjuvant treatment.
References
- 1.Phillips RK, Hittinger R, Blesovsky L, et al. Local recurrence following ’curative’ surgery for large bowel cancer: I. The overall picture. Br J Surg. 1984;71:12–16. doi: 10.1002/bjs.1800710104. [DOI] [PubMed] [Google Scholar]
- 2.Minsky BD, Mies C, Recht A, et al. Resectable adenocarcinoma of the rectosigmoid and rectum. I. Patterns of failure and survival. Cancer. 1988;61:1408–1416. doi: 10.1002/1097-0142(19880401)61:7<1408::aid-cncr2820610722>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 3.Sauer R, Becker H, Hohenberger W, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med. 2004;351:1731–1740. doi: 10.1056/NEJMoa040694. [DOI] [PubMed] [Google Scholar]
- 4.O’Neill BD, Brown G, Heald RJ, et al. Non-operative treatment after neoadjuvant chemoradiotherapy for rectal cancer. Lancet Oncol. 2007;8:625–633. doi: 10.1016/S1470-2045(07)70202-4. [DOI] [PubMed] [Google Scholar]
- 5.Gérard JP, Conroy T, Bonnetain F, et al. Preoperative radiotherapy with or without concurrent fluorouracil and leucovorin in T3-4 rectal cancers: results of FFCD 9203. J Clin Oncol. 2006;24:4620–4625. doi: 10.1200/JCO.2006.06.7629. [DOI] [PubMed] [Google Scholar]
- 6.Bosset JF, Collette L, Calais G, et al. Chemotherapy with preoperative radiotherapy in rectal cancer. N Engl J Med. 2006;355:1114–1123. doi: 10.1056/NEJMoa060829. [DOI] [PubMed] [Google Scholar]
- 7.Buie WD, MacLean AR, Attard JA, et al. Neoadjuvant chemoradiation increases the risk of pelvic sepsis after radical excision of rectal cancer. Dis Colon Rectum. 2005;48:1868–1874. doi: 10.1007/s10350-005-0154-1. [DOI] [PubMed] [Google Scholar]
- 8.Matthiessen P, Hallböök O, Andersson M, et al. Risk factors for anastomotic leakage after anterior resection of the rectum. Colorectal Dis. 2004;6:462–469. doi: 10.1111/j.1463-1318.2004.00657.x. [DOI] [PubMed] [Google Scholar]
- 9.Kuremsky JG, Tepper JE, McLeod HL. Biomarkers for response to neoadjuvant chemoradiation for rectal cancer. Int J Radiat Oncol Biol Phys. 2009;74:673–688. doi: 10.1016/j.ijrobp.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Paweletz CP, Charboneau L, Bichsel VE, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 11.Petricoin EF, Bichsel VE, Calvert VS, et al. Mapping molecular networks using proteomics: a vision for patient-tailored combination therapy. J Clin Oncol. 2005;23:3614–3621. doi: 10.1200/JCO.2005.02.509. [DOI] [PubMed] [Google Scholar]
- 12.Pierobon M, Calvert V, Belluco C, et al. Multiplexed cell signaling analysis of metastatic and nonmetastatic colorectal cancer reveals COX2-EGFR signaling activation as a potential prognostic pathway biomarker. Clin Colorectal Cancer. 2009;8:110–117. doi: 10.3816/CCC.2009.n.018. [DOI] [PubMed] [Google Scholar]
- 13.VanMeter AJ, Rodriguez AS, Bowman ED, et al. Laser capture microdissection and protein microarray analysis of human non-small cell lung cancer: differential epidermal growth factor receptor (EGPR) phosphorylation events associated with mutated EGFR compared with wild type. Proteomics. 2008;7:1902–1924. doi: 10.1074/mcp.M800204-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wulfkuhle JD, Speer R, Pierobon M, et al. Multiplexed cell signaling analysis of human breast cancer applications for personalized therapy. J Proteome Res. 2008;7:1508–1517. doi: 10.1021/pr7008127. [DOI] [PubMed] [Google Scholar]
- 15.Mandard AM, Dalibard F, Mandard JC, et al. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer. 1994;73:2680–2686. doi: 10.1002/1097-0142(19940601)73:11<2680::aid-cncr2820731105>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 16.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 19.Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 20.Korinek V, Barker N, Morin PJ, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 21.Morin PJ, Sparks AB, Korinek V, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 22.Pećina-Slaus N. Tumor suppressor gene E-cadherin and its role in normal and malignant cells. Cancer Cell Int. 2003:3–17. doi: 10.1186/1475-2867-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 24.Albanese C, Wu K, D’Amico M, et al. IKKalpha regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Biol Cell. 2003;14:585–599. doi: 10.1091/mbc.02-06-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agarwal A, Das K, Lerner N, et al. The AKT/I kappa B kinase pathway promotes angiogenic/metastatic gene expression in colorectal cancer by activating nuclear factor-kappa B and beta-catenin. Oncogene. 2005;24:1021–1031. doi: 10.1038/sj.onc.1208296. [DOI] [PubMed] [Google Scholar]
- 26.Tan J, Zhuang L, Leong HS, et al. Pharmacologic modulation of glycogen synthase kinase-3beta promotes p53-dependent apoptosis through a direct Bax-mediated mitochondrial pathway in colorectal cancer cells. Cancer Res. 2005;65:9012–9020. doi: 10.1158/0008-5472.CAN-05-1226. [DOI] [PubMed] [Google Scholar]
- 27.Aschele C, Cionini L, Lonardi S, et al. Primary tumor response to preoperative chemoradiation with or without oxaliplatin in locally advanced rectal cancer: pathologic results of the STAR-01 randomized phase III trial. J Clin Oncol. 2011;29:2773–2780. doi: 10.1200/JCO.2010.34.4911. [DOI] [PubMed] [Google Scholar]
- 28.Gérard JP, Azria D, Gourgou-Bourgade S, et al. Comparison of two neoadjuvant chemoradiotherapy regimens for locally advanced rectal cancer: results of the phase III trial ACCORD 12/0405-Prodige 2. J Clin Oncol. 2010;28:1638–1644. doi: 10.1200/JCO.2009.25.8376. [DOI] [PubMed] [Google Scholar]
- 29.Petricoin EF, Espina V, Araujo RP, et al. Phosphoprotein pathway mapping: Akt/ mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res. 2007;67:3431–3440. doi: 10.1158/0008-5472.CAN-06-1344. [DOI] [PubMed] [Google Scholar]