Abstract
Deficiencies in the activity of cytochrome c oxidase (COX), the terminal enzyme in the respiratory chain, are a frequent cause of autosomal recessive mitochondrial disease in infants. These patients are clinically and genetically heterogeneous, and all defects so far identified in this group have been found in genes coding for accessory proteins that play important roles in the assembly of the COX holoenzyme complex. Many patients, however, remain without a molecular diagnosis. We have used a panel of retroviral vectors expressing human COX assembly factors in these patients to identify the molecular basis for the COX deficiency by functional complementation. Here we show that overexpression of COX15, a protein involved in the synthesis of heme A, the heme prosthetic group for COX, can functionally complement the isolated COX deficiency in fibroblasts from a patient with fatal, infantile hypertrophic cardiomyopathy. Mutation analysis of COX15 in the patient identified a missense mutation (C700T) on one allele, changing a conserved arginine to tryptophan (R217W), and a splice-site mutation in intron 3 on the other allele (C447-3G), resulting in a deletion of exon 4. This splicing error introduces a frameshift and a premature stop codon, resulting in an unstable mRNA and, likely, a null allele. Mitochondrial heme A content was reduced in the patient's heart and fibroblast mitochondria, and levels of heme O were increased in the patient's heart. COX activity and the total amount of fully assembled enzyme were reduced by 50%–70% in patient fibroblasts. Expression of COX15 increased heme A content and rescued COX activity. These results suggest that reduced availability of heme A stalls the assembly of COX. This study establishes COX15 as an additional cause, along with SCO2, of fatal infantile, hypertrophic cardiomyopathy associated with isolated COX deficiency.
Introduction
Cytochrome c oxidase (COX) deficiency is one of the most frequent causes of respiratory-chain defects in humans. Patients with COX deficiencies can present with a number of different clinical phenotypes, including Leigh syndrome, a French Canadian form of Leigh syndrome, hypertrophic cardiomyopathy and myopathy, fatal infantile COX deficiency, and reversible COX deficiency in skeletal muscle (Robinson 2000, Shoubridge 2001a, 2001b; Barrientos et al. 2002).
COX is the terminal enzyme of the respiratory chain and catalyzes the reduction of molecular oxygen. In mammals, it is composed of 13 subunits, 10 of which are encoded by nuclear genes. The three mtDNA encoded subunits form the catalytic core of the enzyme, but the exact function of the nuclear-encoded structural genes is largely unknown. The complex is embedded in the inner mitochondrial membrane and functions as a dimer. The enzyme contains two copper-binding sites (CuA and CuB), two hemes (a and a3), and a magnesium and a zinc ion. The biogenesis of the COX complex requires a large number of accessory or assembly proteins that are not themselves part of the complex but are necessary for various stages of the process, including transcription, translation and processing, and membrane insertion of the structural subunits, as well as synthesis, chaperoning, and addition of the prosthetic groups (Barrientos et al. 2002). Although the total number of proteins necessary for the biogenesis of the intact complex is not precisely known, >30 different genetic complementation groups for COX assembly have been identified in the yeast Saccharomyces cerevisiae (McEwen et al. 1986; Tzagoloff and Dieckmann 1990), of which >20 have been assigned to accessory proteins (Barrientos et al. 2002). More than half of these have known human homologues.
Mutations in the three mtDNA-encoded COX subunits have been reported in several patients in association with a number of different, mostly encephalomyopathic, clinical phenotypes (Shoubridge 2001a); however, mutations have not yet been found in any of the 10 nuclear-encoded COX structural subunits (Jaksch et al. 1998). In contrast, gene defects have been reported in four different factors important for the biogenesis of the COX complex, and unexpectedly, all are primarily associated with different clinical phenotypes: SURF1 (Leigh syndrome) (Tiranti et al. 1998; Zhu et al. 1998), SCO2 (hypertrophic cardiomyopathy) (Papadopoulou et al. 1999; Jaksch et al. 2000), SCO1 (ketoacidotic coma and hepatopathy) (Valnot et al. 2000a), and COX10 (tubulopathy and leukodystrophy) (Valnot et al. 2000b).
COX15, one of the COX assembly factors identified in yeast, plays a key role in the mitochondrial heme biosynthetic pathway in which protoheme (heme B) is converted to heme A, the prosthetic heme group in COX. Two splice variants of the gene (COX15.1 and COX15.2 [GenBank accession numbers NM_078470 and NM_004376]) that differ in the predicted C-terminal amino acid sequence and 3′ UTR have been described in humans, but the functional significance of the different isoforms is unknown (Petruzzella et al. 1998). The gene is essential for the biogenesis of COX in yeast (Glerum et al. 1997), the model organism in which the details of the pathway have been most intensively investigated. Heme A biosynthesis begins with farnesylation of the vinyl group at carbon C-2 of heme B, forming heme O, a reaction catalyzed by a heme A:farnesyltransferase encoded by COX10. The heme O methyl group on carbon C-8 is oxidized to a carboxymethyl group—a reaction hypothesized to be carried out by a three-component heme mono-oxygenase, consisting of Cox15p, ferredoxin, and ferredoxin reductase (Barros et al. 2001, 2002; Barros and Tzagoloff 2002)—and to a formyl group by an as-yet-uncharacterized enzyme. COX15 deletion mutants are blocked in heme A but not in heme O biosynthesis (Barros et al. 2001).
Here, we report pathogenic mutations in COX15 in a patient with isolated COX deficiency and an early onset, fatal hypertrophic cardiomyopathy. The molecular defect was identified by functional complementation in patient fibroblasts using a retroviral expression system. We show that overexpression of COX15 increases the synthesis of heme A, which, in turn, influences the assembly of the COX complex and rescues COX activity.
Material and Methods
Cell Lines
Primary cell lines were established from patient skin fibroblasts. The patient and control cell lines were transduced with a retroviral vector expressing the E6E7 genes of type-16 human papilloma virus and a retroviral vector expressing the protein component of human telomerase to immortalize the cells (Yao and Shoubridge 1999). The fibroblasts were grown at 37°C in an atmosphere of 5% CO2 in high glucose Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum.
Cardiac Tissue and Preparation of Mitochondria
Postmortem cardiac-tissue specimens from the patients and control individuals were removed and frozen <30 min after death. Heart and fibroblast homogenates (5%) prepared in 250 mM sucrose/10 mM Tris-HCl/1 mM EDTA (pH 7.4) were centrifuged twice for 10 min at 600 g to obtain postnuclear supernatant. Mitochondria were pelleted by centrifugation for 20 min at 10,000 g.
COX Assembly cDNA Constructs
Ten retroviral vectors were created with the Gateway Cloning system (Invitrogen). cDNAs from each of the human COX assembly genes SURF1, SCO1, SCO2, COX17, COX11, COX15.1 (variant 1), COX18, COX10, OXA1, and PET191 were amplified by OneStep RT-PCR (Qiagen) using specific primers modified for cloning into Gateway vectors. The PCR constructs were cloned into a Gateway-modified retroviral expression vector, pLXSH (Miller and Buttimore 1986). The fidelity of cDNA clones was confirmed by automated DNA sequencing.
Virus Production and Infection
Stable virus-producing cell lines were generated using procedures described elsewhere (Miller et al. 1993). In brief, the retroviral constructs were used to transfect a GP + E86 ecotropic packaging cell line (Markowitz et al. 1988), and the virus produced was used to infect the amphotropic packaging cell line PA317 (Miller and Buttimore 1986). Fibroblasts were infected by exposure to virus-containing medium in the presence of 4 μg/ml of polybrene, as described by Lochmuller et al. (1999).
Electrophoretic Methods
Blue-Native PAGE (BN-PAGE) (Schagger 1995) was used for separation of samples in the first dimension on 6%–15% polyacrylamide gradient gels, as described elsewhere (Klement et al. 1995). Mitoplasts were prepared from mitochondria by treatment with 0.8 μg of digitonin/mg of protein, as described elsewhere (Klement et al. 1995). Mitoplasts or mitochondria were solubilized with 1% lauryl maltoside, and 10–20 μg of the solubilized proteins were used for electrophoresis. In-gel qualitative assays for complex I and COX activity were performed as described by Zerbetto et al. (1997). COX and complexes I–III were detected by immunoblot analysis using monoclonal antibodies (Molecular Probes).
Enzyme Activity
COX and citrate synthase activities were measured in fibroblast cell extracts, as described elsewhere (Srere 1969; Capaldi et al. 1995). Protein concentration was measured by the Bradford method (Bradford 1976).
Extraction and Separation of Mitochondrial Hemes and HPLC
Mitochondrial hemes were prepared and analyzed by high pressure liquid chromatography (HPLC), as described elsewhere (Barros et al. 2001). Briefly, total heme was extracted from 100 (heart) to 600 (fibroblast) μg of mitochondrial protein, with 0.5 ml of acetone containing 2.5% HCl. The mixture was vortexed, centrifuged for 5 min at 15,000 g, and mixed with 600 μl of 50% acetonitrile. Insoluble material was removed by a second centrifugation. The extract was adjusted to ∼pH 3.5 with 1.65 M ammonium hydroxide, was clarified by centrifugation, and was applied to a 15 × 0.46 cm CSC-Kromacil 5 micron C18 column (Chromatography Sciences). Hemes were eluted at a flow rate of 1 ml/min using a 30%–50% acetonitrile gradient over the first 5 ml, followed by a 50%–75% linear acetonitrile gradient over the subsequent 35 ml. All gradient solutions contained 0.05% trifluoroacetic acid. The elution of heme compounds was monitored at 400 nm. The identity of the heme A and heme B peaks was confirmed by ESI tandem mass spectrometry.
PCR/Gene Sequencing
Primer pairs shown in table 1 were used for the PCR amplification and sequencing of the COX15 exons and for the RT-PCR amplification of fibroblast COX15 mRNA (see table 1). RT-PCR was performed on total RNA from fibroblasts with the OneStep RT-PCR kit (Qiagen). PCR amplifications were performed with the Expand High Fidelity PCR system (Roche Diagnostics) as follows: 1×Expand buffer with MgCl2, 125 μM dNTP, 2.5 μM primers, and 0.75 μM Expand enzyme mix. Reaction conditions were as follows: 94°C for 4 min, 35 cycles with denaturing phase of 94°C for 20 s, annealing phase of 55°C for 30 s, and extension phase of 72°C for 45 s, with a final extension phase of 72°C for 7 min.
Table 1.
COX15 Primers
|
Sequence |
|||
| Region | Upstream(5′→3′) | Downstream(5′→3′) | Product Size(bp) |
| cDNA 1124bp (EX1→EX9)a | tgaaggggaggcagtatctg | cagtgagcaaagccaaggag | 1,153 |
| cDNA 457bp (EX3→EX5) | cctctcgatggtagattggcattt | cggagggagtagcagtgacagtg | 457 |
| Exon 1 | gcctgcagtttccacttg | gggctctgatctgaacctat | 505 |
| Exon 2 | aaggctgatgcagtaatccagata | tcctcagtcaacctgtgcttctac | 402 |
| Exon 3/4 | atggcagctgtttctgacaa | aacaatgccaactaggaagctc | 837 |
| Exon 5 | caagatcccgccactg | gtccccatttaacgaacaat | 598 |
| Exon 6 | atggggtagaagggaaaaca | tgaagatgggggaatgaga | 429 |
| Exon 7 | ttggggtgggagcaggta | gtagggggacaggggtgaat | 564 |
| Exon 8 | gaagaggatggtggaaga | ttttgtagagatggggtttt | 474 |
| Exon 9 variant 1b | ggggaattttaggtttatca | tggcagacatttctttctc | 587 |
| Exon 9 variant 2c | gcccagctagttcctcttt | aggggttctggactcatcat | 704 |
| Intron 3/Exon4 | cttctaagcccccaaatccaaagtc | ttctgtcagtgtcatatcatgattcaaCctd |
192 |
Downstream primer is specific to exon 9 of the cox15 variant 1 mRNA transcript. This cDNA does not amplify 80 bp of 5′ and 64 bp of 3′ cDNA-coding sequence.
These primers are specific to COX15.1.
These primers are specific to COX15.2.
This primer has a mismatch at the underlined C (reference sequence=g).
The presence of the C700T mutation was verified by StyI digestion of an exon 5 PCR product. To confirm the intron 3 splice-acceptor site transversion mutation (C447-3G), a mismatch primer was used to amplify an intron 3/exon 4 region (table 1), and the PCR product was digested with ScrfI.
Results
Clinical Presentation
The clinical history of patient N has been described elsewhere (Kennaway et al. 1990). In brief, patient N was the sixth child of a healthy 29-year-old woman and the first child of a nonconsanguinous white couple; all half-siblings were normal. Shortly after birth, patient N presented with midfacial hypoplasia, lactic acidosis, seizures, and hypotonia. Echocardiogram on day 6 showed no ventricular enlargement, but she died at age 24 d from a massive biventricular hypertrophic cardiomyopathy.
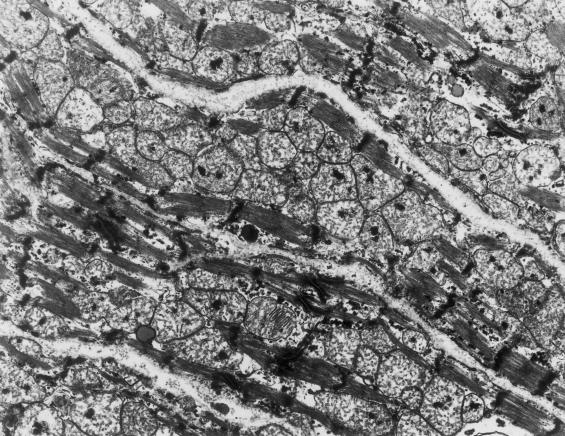
Pathologic examination of liver and muscle tissue (day 23) was normal. Autopsy findings included cardiac abnormalities, pneumonia, eosiniphilic infiltrates in multiple tissues, and abnormally small height, weight, head circumference, and internal organs (brain, lung, and kidney). Electron microscopy of skeletal muscle tissue was normal. Electron microscopy of the heart showed severe loss of myofibrillar material and greatly increased numbers of mitochondria with abnormal shapes, abnormally arranged cristae, and numerous matrix densities (fig. 1) compared with normal heart as shown by Matthews and Martin (1971, p. 119). COX activity was severely decreased in heart (7% of control; table 2) and in kidney and liver (<25% of control) and was in a low–normal range in muscle. Other respiratory chain activities were normal. Cytochrome spectra showed barely detectable levels of cytochrome aa3 in heart mitochondria, whereas it was 37% of normal in muscle mitochondria. Immunoblot analysis showed markedly low COX subunits II, III, VIa, VIb, VIc, and VIIa in the patient’s heart mitochondria, whereas subunits IV and Va were less affected (Kennaway et al. 1990). Analysis of mtDNA showed no mtDNA deletions and no pathogenic tRNA or COX I–III mutations.
Figure 1.
Abnormal ultrastructure of cardiac muscle in the patient. Electron micrograph of left ventricular cardiac tissue in the patient demonstrating replacement of myofibils with large numbers of morphologically abnormal mitochondria (×14,000).
Table 2.
Cytochrome C Oxidase Activity in Patient Heart and Fibroblasts
| Sample | COX/CS Activitya | % Control |
| Heart: | ||
| Control | 5.6 ± 3.2 (n = 6) | 100 |
| Patient N | 0.4 | 7 |
| Fibroblasts: | ||
| Control | .82 ± .21 (n = 16) | 100 |
| Control | .89 ± .18 (n = 5) | 100 |
| Patient N | .24 ± .11 (n = 6) | 28 |
| Patient N + COX15 | .51 ± .14 (n = 5) | 61 |
COX and citrate synthase (CS) were measured as described in the “Material and Methods” section. The number in parentheses indicates the number of independent measurements.
Assembly of COX
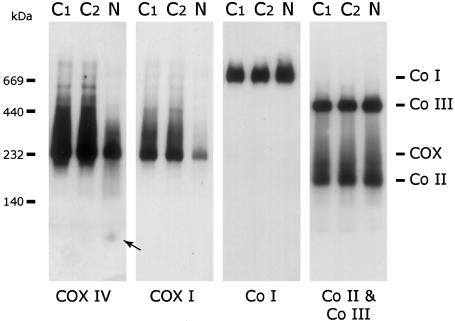
Blue-Native PAGE of patient heart tissue showed a decreased amount of fully assembled COX (fig. 2), in agreement with the low COX activity. The subcomplex, previously described in patients with SURF1 (Coenen et al. 1999; Tiranti et al. 1999), was not detected using either anti-COX IV or anti-COX I antibodies. Nonassembled subunit IV was present in heart tissue from the patient, consistent with the milder reduction of COX IV on immunoblot analysis. Other respiratory chain complexes were present at the same level as in control individuals. A similar pattern was found in patient fibroblasts, although the COX activity (28% of control; table 2) and COX protein levels (fig. 3A) were less affected than in heart tissue. In-gel assays showed no detectable COX activity in patient fibroblasts (fig. 3B) or heart (data not shown), whereas complex I activity was normal.
Figure 2.
Blue-Native PAGE analysis of COX levels in patient heart mitochondria. Heart mitochondria (10 μg protein) from two control subjects (C1 and C2) and the patient (N) were separated by BN-PAGE, and the content of COX, Complex I (Co I), Complex II (Co II), and Complex III (Co III) were determined by immunoblot analysis using antibodies against COX subunits I and IV (COX I and COX IV), the 39-kDa subunit of complex I, the 70-kDa subunit of complex II, and the Core1 protein of complex III. Patient heart mitochondria show a specific reduction in the total amount of fully assembled COX but no evidence of any subcomplexes. The arrow indicates the small amount of unassembled COX IV in the patient. The migration of molecular mass standards is indicated on the left.
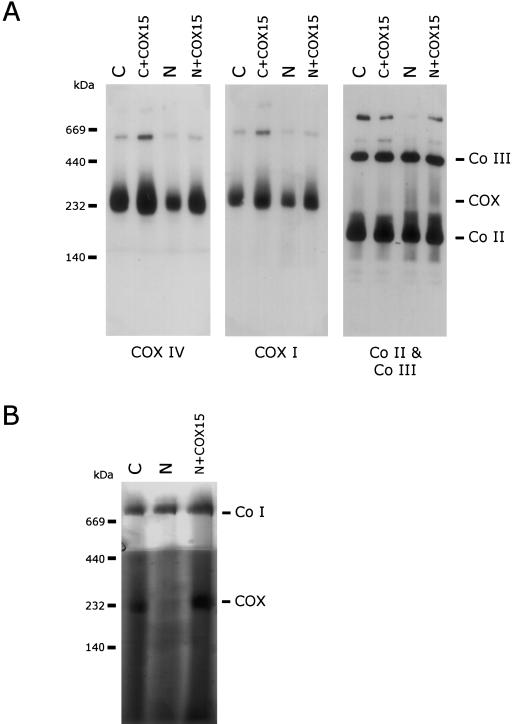
Figure 3.
Rescue of COX content and activity in patient fibroblasts. Mitoplasts (20 μg protein) isolated from patient (N) and control (C) fibroblasts and patient and control fibroblasts overexpressing COX15 (N+COX15 and C+COX15, respectively) were analyzed by BN-PAGE. A, Western blot analysis using antibodies against COX subunits I and IV and against the 70-kDa subunit of Complex II (Co II) and the Core1 protein of Complex III (Co III). Overexpression of COX15 rescues COX levels in patient mitochondria. B, In-gel activity staining of COX and Complex I demonstrating restoration of COX activity in patient fibroblasts overexpressing COX15. The migration of molecular mass standards is indicated on the left.
Overexpression of COX Assembly Factors
To determine whether mutations in any of the known human COX assembly factors were responsible for the low COX activity in patient N, the cDNAs coding for 10 different factors (SURF1, SCO2, SCO1, COX10, COX11, COX15, COX17, COX18, OXA1, and PET191) were individually overexpressed in patient fibroblasts using a retroviral expression system. Overexpression of COX15 restored COX activity to ∼60% of the control levels when normalized to citrate synthase (table 2). COX activity was not restored by any other COX assembly factor (data not shown). Overexpression of COX15 in control cells or in other COX-deficient patient fibroblast lines (Leigh syndrome SURF1 and non-SURF1 patients) did not significantly affect COX activity.
To establish whether overexpression of COX15 in patient fibroblasts rescued the assembly of COX, BN-PAGE analysis was performed. Immunoblot analysis showed near-normal levels of fully assembled COX when compared with control fibroblasts, and ∼60% when compared with control fibroblasts overexpressing COX15 (fig. 3A), indicating that COX15 plays a role in the biosynthesis of a fully assembled COX. No subcomplexes of COX were present in samples overexpressing COX15. The in-gel activity assay showed restoration of COX activity in rescued fibroblasts (fig. 3B).
Mutation Detection in the COX15 Gene
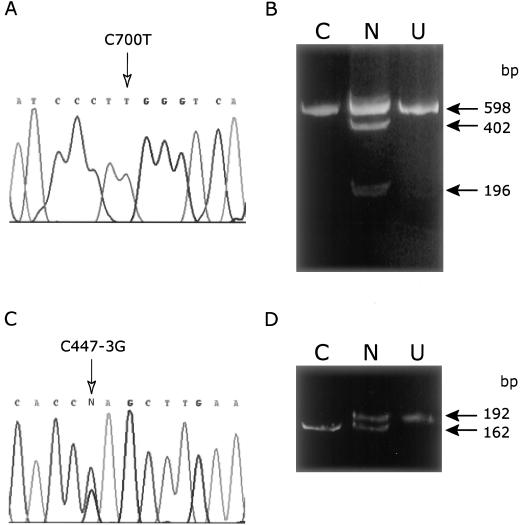
Direct sequencing of a 1,155-bp RT-PCR cDNA fragment of COX15 (variant 1) from patient N fibroblasts showed a single transition mutation (C700T) compared with the published sequence. This mutation, which appeared homozygous in the cDNA, occurs in exon 5 of the gene (fig. 4A) and corresponds to an Arg217Trp (R217W) amino acid substitution. The presence of the mutation was confirmed by restriction endonuclease digest (StyI) of both the 1,155-nucleotide cDNA product and a PCR-amplified exon 5 product from genomic DNA. In contrast to the results obtained by cDNA sequencing, analysis of genomic DNA showed that the C700T change was heterozygous (fig. 4B), suggesting that the other disease allele might be a nonsense mutation. The C700T substitution was not identified in 50 control individuals tested using the restriction endonuclease digest.
Figure 4.
Sequence analysis of COX15. DNA sequence analysis of COX15 cDNA (A) and genomic DNA (C) from patient fibroblasts identifies the two pathogenic mutations: a C700T transition mutation in exon 5 and a C447-3G mutation in the splice acceptor of intron 3. The mutations shown are on the sense strand. The C700T mutation was confirmed by RFLP analysis with StyI (B). When the mutation is present, the enzyme digests a 598-bp PCR product once, producing two fragments of 402 and 196 bp. The wild-type PCR product remains uncut. A PCR utilizing a mismatch reverse primer to create a ScrfI site in wild-type DNA was used to confirm the C447-3G mutation (D). ScrfI digests wild-type DNA to yield 162-bp and 30-bp fragments (latter not shown). The intron C447-3G mutation abolishes the ScrfI site. Panels B and D, C = control, N = patient N, U = undigested PCR product.
To identify the other disease allele, we amplified and sequenced all eight common exons from COX15 and both variants of exon 9, including the intron/exon boundaries, in patient N. This analysis identified a heterozygous intron 3 splice-acceptor site transversion mutation (C447-3G) at the conserved –3 position (fig. 4C). This heterozygous C→G mutation preceding the conserved AG acceptor site in intron 3 was confirmed using a ScrfI restriction endonuclease digest (fig. 4D). Patient N was also found to be homozygous for an SNP, T1171C (F374L), in variant 2 of exon 9 (dbSNP [rs2231687]).
RT-PCR amplification of a segment of COX15 cDNA encompassing exons 3–5 was performed on fibroblast total RNA from patient N to test whether the C447-3G mutation produced an abnormally spliced cDNA product. This analysis identified two bands corresponding to the expected product sizes, with and without the inclusion of exon 4 (data not shown). A single PCR product, whose size corresponded to the correctly spliced cDNA, was observed in control individuals. Sequencing of the smaller band from the patient showed the expected sequence with a clean deletion of exon 4. This deletion removes a 187-bp fragment of mRNA and, thus, predicts a frameshift mutation, resulting in a protein product truncated because of a premature stop codon (fig. 5). The quantity of the PCR product corresponding to the smaller cDNA was greatly reduced compared with the correctly spliced cDNA, presumably the result of the nonsense-mediated mRNA decay pathway.
Figure 5.
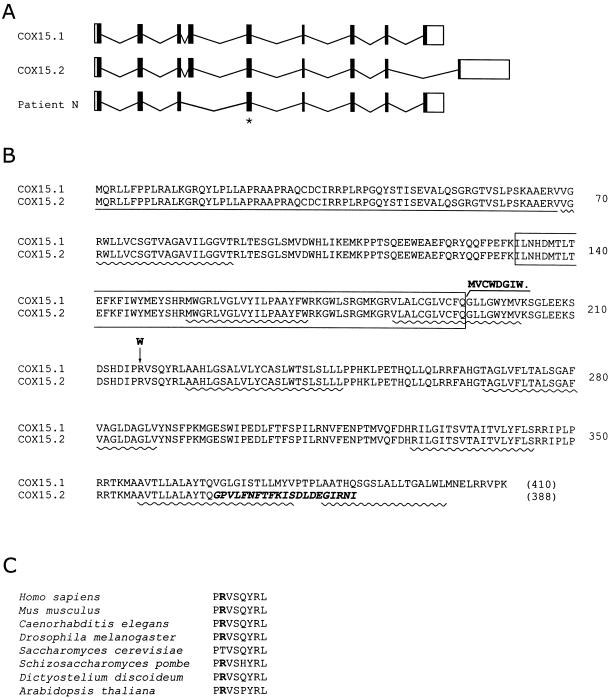
Genomic structure of COX15, predicted protein sequences of human COX15 variants, and location of patient mutations. A, Genomic structure of COX15. The solid vertical bars represent the 9 exons, and the open boxes represent the 5′ and 3′ untranslated regions of the gene. Horizontal lines represent introns. Two-splice variants of the human gene have been described (COX15.1 and COX15.2) that differ in the 3′ end of the mRNA. The splice-site mutation in patient N results in skipping of exon 4 in the mature transcript. The missense mutation occurs in exon 5 and is indicated with an asterisk. B, The predicted protein sequence of the human COX15.1 and COX15.2 variants are shown in single amino acid code. The size of predicted proteins is shown in parentheses. The C-terminal region in which the COX15.2 species diverges from COX15.1 is indicated in bold italics. The missense mutation (R217W) and the predicted C-terminal end of the truncated protein resulting from the splice-site allele in patient N are shown in bold. Exon 4, which is deleted by the splice-site mutation, is boxed. The predicted (MITOPROT II) canonical mitochondrial targeting sequence is underlined, and the predicted transmembrane domains (ENSEMBL) are indicated with wavy lines. C, The short evolutionarily conserved sequence containing the missense mutation is shown.
Sequencing of COX15 in five additional patients with hypertrophic cardiomyopathy and COX deficiency did not reveal any further mutations in these cases.
Role of COX15 in the Biosynthesis of Heme A
Yeast Cox15p is involved in the biosynthesis of heme A, the modified prosthetic heme group present in the COX holoenzyme complex (Barros et al. 2001, 2002). The initial step in this pathway, the hydroxylation of heme O, is thought to be catalyzed by a three-component mono-oxygenase consisting of Cox15p, ferredoxin, and ferredoxin reductase. Yeast cox15 mutants have no detectable heme A and have low levels of heme O (Barros and Tzagoloff 2002). To investigate the effect of the COX15 mutations in patient N on heme A levels, total heme was isolated from patient and control heart and fibroblast mitochondria and was analyzed by HPLC. Patient N heart mitochondria contained markedly reduced levels of heme A and increased levels of heme O (fig. 6A). To quantify the extent of these changes, the levels of heme A were normalized to heme B, the amount of which was a relatively constant fraction of mitochondrial protein in the control and patient samples (table 3). The heme A content was reduced to 6% of control, which corresponds well with the magnitude of the decrease in COX activity and the cytochrome aa3 levels in heart tissue from the patient. Heme O was undetectable in control samples but accumulated to levels similar to that of heme A in patient heart, consistent with a defect in the heme A biosynthetic pathway.
Figure 6.
Analysis of mitochondrial hemes in heart tissue and fibroblasts. Total hemes were extracted from isolated mitochondria with acidic acetone and separated by reverse phase HPLC. The heme B, heme A, and heme O peaks were identified from the elution times of known standards. The areas under the heme B, heme A, and heme O peaks were quantified and are reported in arbitrary units (in parentheses). Chromatograms are shown from heart mitochondria (A) and from fibroblast mitochondria (B) isolated from cells with and without overexpression of COX15. The insert shows that no heme O was detectable in the extract from patient fibroblasts.
Table 3.
Relative Levels of Heme A and Heme O in Patient Heart and Fibroblasts[Note]
|
Relative Level of |
|||
| Sample | Heme A/B | Heme O/B | Heme A/O |
| Heart: | |||
| Control | .221 ± .084 | 0 | … |
| Patient N | .014 | .014 | 1.066 |
| Fibroblasts: | |||
| Control | .064 | 0 | … |
| Control + COX15 | .139 | 0 | … |
| Patient N | .030 | 0 | … |
| Patient N + COX15 | .090 | 0 | … |
Note.— Mitochondrial hemes were extracted and analyzed, as described in the “Material and Methods” section. The values reported in the table are calculated from integrals of hemes A, B, and O. The value in control heart is an average of eight independent measurements, the values in patient heart are result of two independent measurements.
Low heme A was also detected in patient fibroblasts; however, no heme O peak could be detected (table 3; fig. 6B). Overexpression of COX15 in patient fibroblasts increased the heme A levels threefold, raising the normalized heme A levels to 65% of that in control fibroblast lines also overexpressing COX15. The magnitude of this increase agrees well with the increase in COX activity in the complemented patient fibroblast line. Overexpression of COX15 in control fibroblasts increased the heme A content twofold (table 3). The COX content (fig. 3) and COX activity (table 2) in these control lines were also slightly increased, although the increase in activity was not statistically significant (P>.9). Thus, the increase in heme A synthesis in patient fibroblasts overexpressing COX15 rescues the activity and assembly of the COX complex.
Discussion
This study establishes mutations in COX15 as an additional cause of isolated COX deficiency associated with fatal infantile hypertrophic cardiomyopathy. This is the fifth COX assembly factor that has been associated with autosomal recessive COX deficiency. Our results also provide strong evidence that COX15 plays an essential role in mitochondrial heme modification in human beings, as it does in yeast.
To identify the underlying molecular defect in autosomal recessive COX deficiencies in this and similar cases, we have taken a functional complementation approach in which we have used a panel of retroviral expression vectors to overexpress human COX assembly genes, identified by homology to their yeast counterparts. COX activity in fibroblasts from patient N was specifically and reproducibly restored to ∼60% of control levels by transduction with a COX15.1 cDNA construct. No significant changes in COX activity were observed in the patient cells with any of the other COX assembly gene constructs. Overexpression of COX15 did not significantly alter the COX activity in 12 fibroblast lines from patients with isolated COX deficiency resulting from SURF1, COX10, or SCO2 mutations or other, unidentified gene defects. These data indicated that there is no functional overlap among the COX assembly factors and strongly suggested that mutations in COX15 were responsible for the COX deficiency in patient N.
Sequence analysis of COX15 revealed two heterozygous mutations at evolutionarily conserved positions common to the two-splice variants of COX15. The C700T missense mutation results in the substitution of tryptophan for a highly conserved arginine. The R217W substitution occurs in the region of a 21–amino acid segment between transmembrane domains three and four (fig. 5) in a string of eight highly conserved residues, PRVSQYRL. The arginine residue is present in all sequenced mammalian and some yeast species. The splice acceptor site mutation, which occurs in the −3 position of intron 3, is predicted to abolish this site and produce a misspliced cDNA with a premature stop codon (fig. 5). Aberrant splicing of the mRNA produced by this allele was demonstrated by the identification of a short transcript with a clean deletion of exon 4. The relative rarity of this transcript (compared with that transcribed from the other allele) and the highly truncated protein predicted by the premature stop codon suggest that the allele is functionally null.
The COX15 gene was originally identified by functional complementation studies of respiratory deficient yeast with an isolated COX assembly defect (Glerum et al. 1997). A human COX15 homologue was subsequently identified by homology to the yeast gene and mapped to chromosome 10 (Petruzzella et al. 1998). In both yeast and human beings, the COX15 protein product localizes to the inner mitochondrial membrane and has several predicted transmembrane domains (fig. 5) (Glerum et al. 1997; Petruzzella et al. 1998). In yeast, Cox15p functions in the biosynthesis of heme A, the prosthetic group of cytochrome a and a3. The synthesis of heme A begins with farnesylation of the vinyl group at carbon C-2 of protoheme (heme B) to form heme O, a reaction catalyzed by Cox10p (heme A:farnesyltransferase) (Tzagoloff et al. 1993; Glerum and Tzagoloff 1994). Cox15p mediates hydroxylation of the methyl group at the C-8 position of the heme O molecule, a reaction thought to occur in concert with two other inner mitochondrial membrane proteins, ferredoxin (Yah1p) and ferredoxin reductase (Arh1p), which together function as a heme mono-oxygenase (Barros et al. 2001, 2002; Barros and Tzagoloff 2002). The enzyme responsible for converting the C-8 hydroxymethyl to a formyl group has not yet been identified. The heme profiles of different COX assembly mutants in yeast suggest positive regulation of heme B farnesylation by the hydroxylated intermediate formed at the subsequent step or by Cox15p itself (Barros and Tzagoloff 2002). A deletion of COX15 in yeast results in undetectable levels of heme A but detectable levels of heme O (Barros et al. 2001). This pattern contrasts with that seen in a COX10 deletion, where heme O and heme A are undetectable, consistent with a role of Cox10p in the synthesis of heme O from protoheme (Tzagoloff et al. 1993).
Patient N had severely decreased levels of mitochondrial heme A and increased heme O levels in her heart as compared with control levels. Fibroblasts from the patient also showed a marked reduction in heme A but no evidence for an accumulation of heme O. Cytochrome aa3 was absent in patient heart mitochondria when analyzed by difference spectroscopy. These findings are similar to the pattern observed in yeast COX15 deletion mutants, suggesting a similar functional role for COX15 in mammalian mitochondria and a similar pathogenesis for the COX deficiency. The function of the two-splice variants of COX15 remains unknown. The pattern of expression of the mRNAs coding for the two isoforms in different tissues is similar to that seen with other COX assembly genes, apparently reflecting tissue-specific mitochondrial content, although the steady-state concentration of COX15.2 is much less than that of COX15.1 (Petruzzella et al. 1998). COX15.2 differs from COX15.1 in both the region coding for the C-terminal end of the protein and the 3′-UTR (fig. 5). In our complementation experiments, we used only COX15.1 and never obtained complete rescue of COX activity in patient fibroblasts. Similar experiments in fibroblast cell lines from patients with SURF1, SCO2, SCO1, or COX10 mutations, using the appropriate expression vectors, have resulted in virtually complete rescue of COX activity in most cases, so it is unlikely that incomplete rescue is a trivial artifact of the expression system we used. One possibility is that both isoforms of COX15 are necessary for optimal function of the enzyme, and we are currently testing this hypothesis.
Patient N manifested with a severe COX deficiency in cardiac muscle and a milder COX deficiency in skeletal muscle, liver, kidney, and fibroblasts. The decrease of COX activity in patient fibroblasts (∼70%) was accompanied by a similar decrease (∼50%) in the amount of fully assembled COX complex, suggesting that the residual holoenzyme complex in fibroblasts is fully active and that the COX deficiency in this case results from an inability to assemble sufficient enzyme because of reduced availability of heme A. Studies on a human leukemia cell line have identified three intermediates (subcomplexes) in the assembly of the holoenzyme complex that have been called S1–S3. (Nijtmans et al. 1998). The first steps in the assembly process are thought to involve the addition of heme a, heme a3, copper at the CuB site, and COX IV to S1 (the COX I subunit) to form the S2 subcomplex (Nijtmans et al. 1998). The formation of S3 involves addition of copper to the CuA site and most of the other nuclear-encoded subunits, except VIa and VIIa or -b, which are added in the final stage of assembly to form S4, the fully assembled enzyme. The fact that no subcomplexes could be detected in mitochondria from patient heart tissue by BN-PAGE analysis suggests the decrease in the COX complex content is caused by a block in assembly at an early stage, most likely at S1. This conclusion is consistent with the above model in which heme A is required for the formation of the S2 subcomplex, and is further supported by the observation that the S2 subcomplex accumulates in patients with SURF1 mutations (Coenen et al. 1999; Tiranti et al. 1999), showing that this species is not an inherently unstable intermediate in the assembly process.
The molecular basis for the tissue-specific patterns of COX deficiency, and the resultant tissue-specific clinical phenotypes, in patients with mutations in COX assembly genes remains elusive. SCO2 is the only other known gene defect associated with hypertrophic cardiomyopathy and isolated COX deficiency (Papadopoulou et al. 1999; Jaksch et al. 2000, 2001). These patients have a severe reduction in COX activity in heart that is similar to that observed in the patient with COX15 we describe here. However, the deficiency in patients with SCO2 also occurs in skeletal muscle, whereas the COX defect in the muscle of patient N was moderate. Most strikingly, patients with mutations in COX10, which prevents the synthesis of heme A by blocking heme O synthesis, rather than heme A synthesis itself, have a completely different phenotype—tubulopathy and leukodystrophy (Valnot et al. 2000b). However, mutations in COX10 and COX15 have so far only been identified in single affected families, and it is possible that a range of clinical phenotypes will eventually be associated with mutations in these genes.
As far as is known, the COX assembly factors are housekeeping genes that are transcribed in proportion to other mitochondrial genes. There is no evidence yet for tissue-specific expression of any of the known accessory proteins. As reagents are developed to analyze the proteins themselves, it will be possible to look at post-transcriptional or post-translational effects that may turn up tissue-specific patterns of expression. Another possibility is that the tissue specificity reflects differences in the regulation of the pathways of COX assembly in different tissues. Consistent with this notion is the observation that, although heme O levels increased in heart mitochondria of patient N, this species was undetectable in fibroblast mitochondria, raising the possibility of tissue-specific regulation of the heme-biosynthetic pathway.
In conclusion, this study establishes mutations in COX15 as an additional cause, along with SCO2, of fatal infantile, hypertrophic cardiomyopathy associated with COX deficiency. Sequencing of both of these genes in five other cases with a similar clinical and biochemical phenotype failed to identify pathogenic mutations, indicating that there is additional genetic heterogeneity in this group of disorders.
Acknowledgments
We acknowledge the contributions of N. R. M. Buist and B. Powell to the care of this patient. We thank T. Johns, G.-H. Guercin, B. Lauzon, K. Manning, and D. Dennie for excellent technical assistance. This research was supported by a grant from the Canadian Institutes of Health Research (CIHR) to E.A.S and D.M.G. C.G.C. is supported by a Natural Sciences and Engineering Research Council Postgraduate Scholarship “A” Studentship. H.A. holds a postdoctoral fellowship from the Muscular Dystrophy Association of Canada–CIHR partnership. SCL holds postdoctoral fellowships from the Heart and Stroke Foundation of Canada and the Tomlinson Foundation of McGill University. D.M.G. is a CIHR New Investigator and Alberta Heritage Foundation for Medical Research Scholar. E.A.S. is an International Scholar of the Howard Hughes Medical Institute and a Senior Investigator of the CIHR.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=SNP (for the T1171C polymorphism [rs2231687])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the COX15.1 variant mRNA sequence [NM_078470] and COX15.2 variant mRNA sequence [NM_004376]) [Google Scholar]
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for cytochrome c oxidase deficiency [MIM 220110], SCO2 mutations [MIM 602472 and MIM 604377], and COX15 [MIM 603646])
References
- Barrientos A, Barros MH, Valnot I, Rotig A, Rustin P, Tzagoloff A (2002) Cytochrome oxidase in health and disease. Gene 286:53–63 [DOI] [PubMed] [Google Scholar]
- Barros MH, Carlson CG, Glerum DM, Tzagoloff A (2001) Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Lett 492:133–138 [DOI] [PubMed] [Google Scholar]
- Barros MH, Nobrega FG, Tzagoloff A (2002) Mitochondrial ferredoxin is required for heme A synthesis in Saccharomyces cerevisiae. J Biol Chem 277:9997–10002 [DOI] [PubMed] [Google Scholar]
- Barros MH, Tzagoloff A (2002) Regulation of the heme A biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett 516:119–123 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 [DOI] [PubMed] [Google Scholar]
- Capaldi RA, Marusich MF, Taanman JW (1995) Mammalian cytochrome-c oxidase: characterization of enzyme and immunological detection of subunits in tissue extracts and whole cells. Methods Enzymol 260:117–132 [DOI] [PubMed] [Google Scholar]
- Coenen MJ, van den Heuvel LP, Nijtmans LG, Morava E, Marquardt I, Girschick HJ, Trijbels FJ, Grivell LA, Smeitink JA (1999) SURFEIT-1 gene analysis and two-dimensional blue native gel electrophoresis in cytochrome c oxidase deficiency. Biochem Biophys Res Commun 265:339–344 [DOI] [PubMed] [Google Scholar]
- Glerum DM, Muroff I, Jin C, Tzagoloff A (1997) COX15 codes for a mitochondrial protein essential for the assembly of yeast cytochrome oxidase. J Biol Chem 272:19088–19094 [DOI] [PubMed] [Google Scholar]
- Glerum DM, Tzagoloff A (1994) Isolation of a human cDNA for heme A:farnesyltransferase by functional complementation of a yeast cox10 mutant. Proc Natl Acad Sci USA 91:8452–8456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaksch M, Hofmann S, Kleinle S, Liechti-Gallati S, Pongratz DE, Muller-Hocker J, Jedele KB, Meitinger T, Gerbitz KD (1998) A systematic mutation screen of 10 nuclear and 25 mitochondrial candidate genes in 21 patients with cytochrome c oxidase (COX) deficiency shows tRNA(Ser)(UCN) mutations in a subgroup with syndromal encephalopathy. J Med Genet 35:895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaksch M, Ogilvie I, Yao J, Kortenhaus G, Bresser HG, Gerbitz KD, Shoubridge EA (2000) Mutations in SCO2 are associated with a distinct form of hypertrophic cardiomyopathy and cytochrome c oxidase deficiency. Hum Mol Genet 9:795–801 [DOI] [PubMed] [Google Scholar]
- Jaksch M, Paret C, Stucka R, Horn N, Muller-Hocker J, Horvath R, Trepesch N, Stecker G, Freisinger P, Thirion C, Muller J, Lunkwitz R, Rodel G, Shoubridge EA, Lochmuller H (2001) Cytochrome c oxidase deficiency due to mutations in SCO2, encoding a mitochondrial copper-binding protein, is rescued by copper in human myoblasts. Hum Mol Genet 10:3025–3035 [DOI] [PubMed] [Google Scholar]
- Kennaway NG, Carrero-Valenzuela RD, Ewart G, Balan VK, Lightowlers R, Zhang YZ, Powell BR, Capaldi RA, Buist NR (1990) Isoforms of mammalian cytochrome c oxidase: correlation with human cytochrome c oxidase deficiency. Pediatr Res 28:529–535 [DOI] [PubMed] [Google Scholar]
- Klement P, Nijtmans LG, Van den Bogert C, Houstek J (1995) Analysis of oxidative phosphorylation complexes in cultured human fibroblasts and amniocytes by blue-native-electrophoresis using mitoplasts isolated with the help of digitonin. Anal Biochem 231:218–224 [DOI] [PubMed] [Google Scholar]
- Lochmuller H, Johns T, Shoubridge EA (1999) Expression of the E6 and E7 genes of human papillomavirus (HPV16) extends the life span of human myoblasts. Exp Cell Res 248:186–193 [DOI] [PubMed] [Google Scholar]
- Markowitz D, Goff S, Bank A (1988) Construction and use of a safe and efficient amphotropic packaging cell line. Virology 167:400–406 [PubMed] [Google Scholar]
- Matthews JL, Martin JH (1971) Atlas of human histology and ultrastructure. Lea and Febiger, Philadelphia [Google Scholar]
- McEwen JE, Ko C, Kloeckner-Gruissem B, Poyton RO (1986) Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae: characterization of mutants in 34 complementation groups. J Biol Chem 261:11872–11879 [PubMed] [Google Scholar]
- Miller AD, Buttimore C (1986) Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol 6:2895–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AD, Miller DG, Garcia JV, Lynch CM (1993) Use of retroviral vectors for gene transfer and expression. Methods Enzymol 217:581–599 [DOI] [PubMed] [Google Scholar]
- Nijtmans LG, Taanman JW, Muijsers AO, Speijer D, Van den Bogert C (1998) Assembly of cytochrome-c oxidase in cultured human cells. Eur J Biochem 254:389–394 [DOI] [PubMed] [Google Scholar]
- Papadopoulou LC, Sue CM, Davidson MM, Tanji K, Nishino I, Sadlock JE, Krishna S, Walker W, Selby J, Glerum DM, Coster RV, Lyon G, Scalais E, Lebel R, Kaplan P (1999) Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat Genet 23:333-337 [DOI] [PubMed] [Google Scholar]
- Petruzzella V, Tiranti V, Fernandez P, Ianna P, Carrozzo R, Zeviani M (1998) Identification and characterization of human cDNAs specific to BCS1, PET112, SCO1, COX15, and COX11, five genes involved in the formation and function of the mitochondrial respiratory chain. Genomics 54:494–504 [DOI] [PubMed] [Google Scholar]
- Robinson BH (2000) Human cytochrome oxidase deficiency. Pediatr Res 48:581–585 [DOI] [PubMed] [Google Scholar]
- Schagger H (1995) Native electrophoresis for isolation of mitochondrial oxidative phosphorylation protein complexes. Methods Enzymol 260:190–202 [DOI] [PubMed] [Google Scholar]
- Shoubridge EA (2001a) Cytochrome c oxidase deficiency. Am J Med Genet 106:46–52 [DOI] [PubMed] [Google Scholar]
- ——— (2001b) Nuclear genetic defects of oxidative phosphorylation. Hum Mol Genet 10:2277–2284 [DOI] [PubMed] [Google Scholar]
- Srere PA (1969) Citrate synthase. Methods Enzymol 13:3–11 [Google Scholar]
- Tiranti V, Galimberti C, Nijtmans L, Bovolenta S, Perini MP, Zeviani M (1999) Characterization of SURF-1 expression and Surf-1p function in normal and disease conditions. Hum Mol Genet 8:2533–2540 [DOI] [PubMed] [Google Scholar]
- Tiranti V, Hoertnagel K, Carrozzo R, Galimberti C, Munaro M, Granatiero M, Zelante L, Gasparini P, Marzella R, Rocchi M, Bayona-Bafaluy MP, Enriquez JA, Uziel G, Bertini E, Dionisi-Vici C (1998) Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am J Hum Genet 63:1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Dieckmann CL (1990) PET genes of Saccharomyces cerevisiae. Microbiol Rev 54:211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Nobrega M, Gorman N, Sinclair P (1993) On the functions of the yeast COX10 and COX11 gene products. Biochem Mol Biol Int 31:593–598 [PubMed] [Google Scholar]
- Valnot I, Osmond S, Gigarel N, Mehaye B, Amiel J, Cormier-Daire V, Munnich A, Bonnefont JP, Rustin P, Rotig A (2000a) Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal-onset hepatic failure and encephalopathy. Am J Hum Genet 67:1104–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valnot I, von Kleist-Retzow JC, Barrientos A, Gorbatyuk M, Taanman JW, Mehaye B, Rustin P, Tzagoloff A, Munnich A, Rotig A (2000b) A mutation in the human heme A:farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum Mol Genet 9:1245–1249 [DOI] [PubMed] [Google Scholar]
- Yao J, Shoubridge EA (1999) Expression and functional analysis of SURF1 in Leigh syndrome patients with cytochrome c oxidase deficiency. Hum Mol Genet 8:2541–2549 [DOI] [PubMed] [Google Scholar]
- Zerbetto E, Vergani L, Dabbeni-Sala F (1997) Quantification of muscle mitochondrial oxidative phosphorylation enzymes via histochemical staining of blue native polyacrylamide gels. Electrophoresis 18:2059–2064 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Yao J, Johns T, Fu K, De Bie I, Macmillan C, Cuthbert AP, Newbold RF, Wang J, Chevrette M, Brown GK, Brown RM, Shoubridge EA (1998) SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet 20:337–343 [DOI] [PubMed] [Google Scholar]