Abstract
Obesity is a multigenic trait that has a substantial genetic component. Animal models confirm a role for gene-gene interactions, and human studies suggest that as much as one-third of the heritable variance may be due to nonadditive gene effects. To evaluate potential epistatic interactions among five regions, on chromosomes 7, 10, and 20, that have previously been linked to obesity phenotypes, we conducted pairwise correlation analyses based on alleles shared identical by descent (IBD) for independent obese affected sibling pairs (ASPs), and we determined family-specific nonparametric linkage (NPL) scores in 244 families. The correlation analyses were also conducted separately, by race, through use of race-specific allele frequencies. Conditional analyses for a qualitative trait (body mass index [BMI] ⩾27) and hierarchical models for quantitative traits were used to further refine evidence of gene interaction. Both the ASP-specific IBD-sharing probability and the family-specific NPL score revealed that there were strong positive correlations between 10q (88–97 cM) and 20q (65–83 cM), through single-point and multipoint analyses with three obesity thresholds (BMI ⩾27, ⩾30, and ⩾35) across African American and European American samples. Conditional analyses for BMI ⩾27 found that the LOD score at 20q rises from 1.53 in the baseline analysis to 2.80 (empirical P=.012) when families were weighted by evidence for linkage at 10q (D10S1646) through use of zero-one weights (weight0-1) and to 3.32 (empirical P<.001) when proportional weights (weightprop) were used. For percentage fat mass, variance-component analysis based on a two-locus epistatic model yielded significant evidence for interaction between 20q (75 cM) and the chromosome 10 centromere (LOD = 1.74; P=.024), compared with a two-locus additive model (LOD = 0.90). The results from multiple methods and correlated phenotypes are consistent in suggesting that epistatic interactions between loci in these regions play a role in extreme human obesity.
Introduction
Obesity (MIM 601665) is an increasingly prevalent condition associated with adverse consequences for health and quality of life. Comorbid disorders include type 2 diabetes mellitus, hypertension, cardiovascular disease, and some cancers (Carroll 1998; Kopelman 2000; Price 2002; Shmulewitz et al. 2001; Wolk et al. 2001). Family studies demonstrate that obesity and thinness follow family lines (Price 1987; Maes et al. 1997; Price et al. 2000), and twin and adoption studies indicate that most family variance is genetic in origin (Stunkard et al. 1986, 1990; Price 1987; Sorensen et al. 1989; Grilo and Pogue-Geile 1991; Price and Gottesman 1991; Maes et al. 1997).
Gene-gene interactions may be common. Family studies estimate the genetic heritability of obesity at ∼40%, and twin studies place the figure higher, at ∼65% (Price 2002). The consistent differences in these estimates suggest that as much as one-third of the heritable variance may be due to nonadditive genetic variance, including allelic (dominance and recessivity) and nonallelic gene interactions.
There are some specific examples of known gene interactions in obesity. For example, the extent of obesity and diabetes resulting from a single gene mutant depends on genomic background (Coleman and Hummel 1975). Other animal models provide further support for a role for gene-gene interactions (Ollmann et al. 1997; Niswender et al. 2001). Gene-gene interactions have been reported for several human disorders as well. Linkage studies suggest a possible interaction between the gene calpain-10 and an unknown gene on chromosome 15, in type 2 diabetes and obesity (Cox et al. 1999; Horikawa et al. 2000). Molecular studies demonstrate that some genes are needed to mediate the phenotypic effects of others (e.g., neuropeptide Y mediates the effects of leptin) (Spiegelman and Flier 2001).
The goal of the present article is to evaluate evidence for interaction among five regions, on three chromosomes, for which evidence of linkage to obesity was found in our previous studies (Reed et al. 1996; Lee et al. 1999; Li et al. 1999; Price et al. 2001).
Subjects and Methods
Since the details of family recruitment have been described elsewhere (Price et al. 1998; Lee et al. 1999), we briefly report the methodological approach used in the present study.
Subjects
The families included in these analyses consist of 200 European American families having 542 siblings and 44 African American families having 125 siblings. Of these 244 families, there were 114 families (98 European American and 16 African American) having both parent's DNA, 124 families (100 European American and 24 African American) having one parent's DNA, and 6 families (2 European American and 4 African American) having neither parent's DNA. Sibship size ranged from two to nine in European Americans and from two to eight in African Americans. Most families (220) have two to five siblings and a median sibship size of three. BMI was calculated on the basis of measured height and weight—that is, as weight (in kg)/height (in m2). In the correlation analyses, the families of affected sibling pairs (ASPs) were included only if genotyping data were available for chromosomes 7, 10, and 20. Three overlapping obesity thresholds—BMI ⩾27, ⩾30, and ⩾35—were used for computation of identity by descent (IBD). For European American families, these three thresholds yield 456 (282 independent), 329 (223 independent), and 194 (152 independent) ASPs, respectively; for the African American families, there were 131 (78 independent), 96 (63 independent), and 64 (45 independent) ASPs. Definition of “independent ASP” follows the Genehunter program manual.
Markers
Thirty-seven markers were selected from five regions, on three chromosomes (7, 10, and 20), where evidence for linkage to obesity-related phenotypes had been found in our previous linkage studies (Reed et al. 1996; Lee et al. 1999; Li et al. 1999; Price et al. 2001). The five regions included 8 markers that span ∼8 cM flanking the leptin gene on 7q31 (D7S685, D7S2501, D7S504, D7S1875, −2548, Shintani [Shintani et al. 1996], D7S530, and D7S2452); 12 markers from three regions on chromosome 10, including 13.5 cM on 10p (D10S582, D10S197, D10S193, and D10S208), 8.4 cM on proximal 10q (D10S1646, D10S1647, D10S1685, D10S537, and D10S535), and 6.7 cM on distal 10q (D10S1679, D10S587, and D10S1656); and 17 markers from a 23.1-cM segment on 20q13 (D20S178, D20S887, D20S176, D20S196, D20S869, D20S857, D20S839, D20S606, D20S902, D20S840, D20S211, D20S876, D20S913, D20S120, D20S100, D20S102, and D20S149). The average heterozygosity was ∼0.75, and the average information content (multipoint) was 0.94 for chromosome 7 markers, 0.90 for chromosome 10 markers, and 0.94 for chromosome 20 markers.
Genotyping
DNA was extracted from blood or lymphoblastoid cell lines. PCR amplification and gel analysis of radiolabeled or fluorescently labeled microsatellite markers were performed as described elsewhere (Lee et al. 1999; Price et al. 2001). Band patterns were independently scored by two individuals blinded to the phenotypes. All genotypes were checked for Mendelian inheritance by using the program Genehunter (Kruglyak et al. 1996), and errors were resolved by retyping or recoding as “unknown.” Any genetically unrelated parents and siblings were excluded, as were all half-siblings.
Map Locations
Genetic maps for markers were taken from the Whitehead Institute/MIT Center for Genome Research. Markers not found in the Whitehead Institute database were placed using the Genetic Location Database and the Genome Database.
ASP-Specific IBD-Sharing Probability, Family-Specific Nonparametric Linkage (NPL) Score, and Permutation Tests
ASP-specific IBD-sharing probabilities and family-specific NPL scores were obtained by using the computer program Genehunter (Kruglyak et al. 1996). IBD-sharing probability for each independent ASP and NPL score for each family were used for correlation analyses implemented in PROC CORR, of SAS, by use of the Pearson method. Because there were a large number of comparisons across regions (8×12×17), because single-point and multipoint analyses were conducted, and because there were three correlated phenotypes, Bonferroni correction may be too conservative. For 9,792 comparisons (8×12×17×2×3), the nominal P values required are 5.2 × 10−6 for P = .05 and 1.0 × 10−6 for P=.01. Therefore, a permutation test was used to determine statistical significance. Using the permuted data, we calculated the correlations on the basis of both the observed IBD-sharing probabilities and the NPL scores on chromosomes 7, 10, and 20. To eliminate the dependence in IBD-sharing probability among the ASPs within the pedigree, we permuted the IBD-sharing probability of one chromosome among all independent ASPs, for each pair of chromosomes examined. For NPL-based correlation, to control the effect of family size, we permuted the NPL score of one chromosome among the families with the same number of ASPs, for each pair of chromosomes examined. Each permutation generates a new data set in which the null hypothesis that there is no correlation between the markers on the two chromosomes is true. The correlation between markers on different chromosomes is then calculated for the permuted data. This procedure was repeated 1,000 times, which gives a distribution of the correlation under the null hypothesis of no interaction between the two chromosomes. Based on a two-sided test, the empirical significance level for each correlation was estimated by the proportion of permutation samples exhibiting a correlation as large or larger in absolute value than the corresponding absolute value of the observed correlation from the original data. This procedure generates the null distribution for each pair of markers.
The statistical-significance estimate based on the empirically derived null distribution does not reflect our having examined many marker pairs in the present study. To make the needed adjustment for multiple comparisons, we focused on the observed maximal correlations from the IBD analyses, to evaluate overall statistical significance. We focused on the IBD analyses, rather than the NPL scores, because the assumptions required are less stringent and are more likely to be met by the data. Specifically, we used the following procedure to obtain a corrected P value for the maximal correlation: we selected the maximal correlation score among all possible pairs of regions on chromosomes 7, 10, and 20; then, we permuted the IBD-sharing probability for markers in each of the regions and picked the highest correlation, from the permuted data, among all possible correlations for chromosomes 7, 10, and 20. The significance of the maximal correlations was based on the proportion of times that the observed maximum was equaled or exceeded by the maximum from the permuted data.
Conditional Analysis of a Qualitative Trait (BMI ⩾27)
For markers on chromosome 20, conditional LOD scores were computed using the program Genehunter-Plus (Kong and Cox 1997) on the basis of two weighting approaches, weight0-1 (family weight was 0 if the NPL score at D10S1646/D10S537 was ⩽0; otherwise, it was 1) and weightprop (family weight was the observed NPL score if the NPL score at D10S1646/D10S537 was >0; otherwise, it was 0). The conservative χ2 test—χ2 = 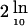 (LODconditional − LODbaseline), for 1 df—and the simulation approach (i.e., the random assignment to families of values for weight0-1 or weightprop) were used to assess the significance of the increase in LOD score (Cox et al. 1999).
(LODconditional − LODbaseline), for 1 df—and the simulation approach (i.e., the random assignment to families of values for weight0-1 or weightprop) were used to assess the significance of the increase in LOD score (Cox et al. 1999).
Linkage Analysis of Quantitative Traits
Multilocus quantitative-trait (BMI, percentage fat mass, waist circumference, and waist:hip circumference ratio) linkage analyses were conducted by Haseman-Elston regression, as implemented in the computer program MapMaker/Sibs (Kruglyak and Lander 1995), and by variance-component analysis using the computer program Solar (Almasy and Blangero 1998). After controlling for the linear effects of age within sex and race categories, we used standardized residuals of obesity phenotypes for Haseman-Elston regression analyses using MapMaker/Sibs. In variance-component analyses, age, sex, and race were adjusted as covariates for the obesity phenotypes, and the ascertainment scheme was accounted for by the identification of primary probands through use of Solar. The following serial hierarchical models were used to examine interaction: (1) two separate one-locus models with marker effects due to each marker locus individually, (2) a two-locus model with marker effects for two loci simultaneously but with no interaction term, and (3) a two-locus epistatic model wherein both marker effects have interaction terms.
Results
Baseline Multipoint Linkage Analyses
In the combined sample, multipoint linkage analyses indicated that there were five regions, on chromosomes 7, 10, and 20, that had at least marginal evidence for linkage to obesity phenotypes. Table 1 presents the four regions, on chromosomes 10 and 20, that have been linked to obesity-related phenotypes. Markers on 10p (51 cM) and 20q (65 cM) had evidence for linkage with BMI ⩾27, with an NPL score of Z>2.2. Markers on the chromosome 10 centromere were linked with quantitative traits, namely BMI and waist circumference. Markers on 10q (157 cM) and 20q (75 cM) were linked with waist:hip circumference ratio and percentage fat mass, respectively. These findings, based on a larger sample than that used in the original studies, are similar to results from our previous studies (Reed et al. 1996; Lee et al. 1999; Li et al. 1999; Price et al. 2001).
Table 1.
Four Regions on Chromosomes 10 and 20 with Linkage to Obesity Phenotypes in the Combined Sample
|
Map Distances(cM) |
Loci |
|||||||||
| Chromosome | No. of Markers | Start | End | Start | End | Program | Analysis (Statistic)a | Score | Phenotype(s)b | Position(cM) |
| 10p | 4 | 46.70 | 60.30 | D10S582 | D10S208 | Genehunter | NPL (Z) | 2.68 | BMI ⩾27 | 50.50 |
| 10 centromere | 5 | 88.70 | 97.00 | D10S1646 | D10S535 | Solar | VC (LOD) | 2.50 | Waist, QTL | 88.70 |
| Solar | VC (LOD) | 1.46 | BMI, QTL | 88.70 | ||||||
| MapMaker/Sibs | EMHE (t) | 2.24 | BMI, QTL | 88.70 | ||||||
| 10q | 3 | 151.60 | 158.30 | D10S1679 | D10S1656 | MapMaker/Sibs | NPL (Z) | 2.22 | WHR, QTL | 156.70 |
| 20q | 17 | 64.00 | 87.06 | D20S178 | D20S149 | Genehunter | NPL (Z) | 2.25 | BMI⩾27 | 64.50 |
| MapMaker/Sibs | NPL (Z) | 2.57 | %fat, QTL | 75.00 | ||||||
VC = variance component; EMHE = expectation-maximization Haseman-Elston.
Waist = waist circumference; WHR = waist:hip circumference ratio; %fat = percentage fat mass.
Correlation Analyses Based on ASP-Specific IBD-Sharing Probability
All of the pairwise correlations between markers on chromosome 7 and markers on chromosomes 10 and 20 approached 0.0. Table 2 includes the five highest correlation scores between chromosomes 10 and 20 on the basis of IBD-sharing probability of independent ASPs by race, obesity threshold, and analytic approach. For the African American sample, most of the observed highest correlations occurred within 5 cM (89–94 cM) of D10S1646 and 7 cM (69–83 cM) of D20S211. The rest were found between D10S582 and 20q (65–87 cM). The highest correlation based on multipoint analysis was found at D10S1646 (89 cM) and 20q (83–87 cM) through three thresholds. For the European American sample, however, most observed highest correlations occurred within 5 cM (46–51 cM) of D10S197 and 20q (72–83 cM). The rest were found between 10q (89–97 cM) and 20q (64–75 cM). The highest correlation by multipoint analysis was found between D10S197 and D20S100 through three thresholds. For the combined sample, the locations of the observed highest correlations were shifted slightly on chromosomes 10 and 20. One occurred between D10S582 and 20q (82–87 cM), and another occurred between D10S1646 and 20q (64–68 cM).
Table 2.
Correlation between Loci on Chromosomes 10 and 20 on the Basis of ASP-Specific IBD-Sharing Probability
|
African American Results |
European American Results |
Combined Results |
||||||||||
| Analysis and BMI | N | Markers (Distance [in cM]) | r | P | N | Markers (Distance [in cM]) | r | P | N | Markers (Distance [in cM]) | r | P |
| Single point: | ||||||||||||
| ⩾27 | 78 | D10S582 (46.70)/D20S102 (83.26) | .329 | .003 | 282 | D10S582 (46.70)/D20S902 (74.36) | .198 | .001 | 360 | D10S582 (46.70)/D20S102 (83.26) | .194 | <.001 |
| D10S537 (94.10)/D20S149 (87.06) | .304 | .004 | D10S582 (46.70)/D20S839 (72.40) | .190 | <.001 | D10S582 (46.70)/D20S149 (87.06) | .188 | <.001 | ||||
| D10S1647 (91.50)/D20S196 (69.40) | .299 | .006 | D10S537 (94.10)/D20S211 (75.36) | .184 | .001 | D10S1646 (88.70)/D20S876 (71.40) | .187 | <.001 | ||||
| D10S1647 (91.50)/D20S211 (75.36) | .299 | .010 | D10S582 (46.70)/D20S100 (82.26) | .166 | .004 | D10S537 (94.10)/D20S211 (75.36) | .187 | <.001 | ||||
| D10S582 (46.70)/D20S887 (64.50) | .298 | .009 | D10S582 (46.70)/D20S857 (71.40) | .162 | .007 | D10S582 (46.70)/D20S196 (69.40) | .183 | <.001 | ||||
| ⩾30 | 63 | D10S1647 (91.50)/D20S196 (69.40) | .377 | .001 | 223 | D10S582 (46.70)/D20S902 (74.36) | .224 | .001 | 286 | D10S582 (46.70)/D20S149 (87.06) | .239 | <.001 |
| D10S582 (46.70)/D20S102 (83.26) | .377 | .001 | D10S582 (46.70)/D20S839 (72.40) | .216 | <.001 | D10S1646 (88.70)/D20S196 (69.40) | .221 | <.001 | ||||
| D10S1646 (88.70)/D20S196 (69.40) | .374 | .002 | D10S582 (46.70)/D20S606 (70.40) | .196 | .005 | D10S582 (46.70)/D20S902 (74.36) | .221 | <.001 | ||||
| D10S1647 (91.50)/D20S887 (64.50) | .363 | .001 | D10S582 (46.70)/D20S100 (82.26) | .196 | .003 | D10S537 (94.10)/D20S606 (70.40) | .219 | <.001 | ||||
| D10S582 (46.70)/D20S149 (87.06) | .363 | .002 | D10S537 (94.10)/D20S606 (70.40) | .194 | .002 | D10S1646 (88.70)/D20S887 (64.50) | .216 | .001 | ||||
| ⩾35 | 45 | D10S1647 (91.50)/D20S211 (75.36) | .430 | .001 | 152 | D10S582 (46.70)/D20S839 (72.40) | .266 | .004 | 197 | D10S582 (46.70)/D20S149 (87.06) | .269 | .001 |
| D10S1646 (88.70)/D20S196 (69.40) | .409 | .006 | D10S582 (46.70)/D20S857 (71.40) | .232 | .001 | D10S582 (46.70)/D20S102 (83.26) | .241 | <.001 | ||||
| D10S582 (46.70)/D20S149 (87.06) | .408 | .003 | D10S582 (46.70)/D20S149 (87.06) | .216 | .007 | D10S1646 (88.70)/D20S196 (69.40) | .230 | <.001 | ||||
| D10S1646 (88.70)/D20S211 (75.36) | .394 | .005 | D10S535 (97.00)/D20S211 (75.36) | .214 | .004 | D10S582 (46.70)/D20S196 (69.40) | .226 | .002 | ||||
| D10S1647 (91.50)/D20S902 (74.36) | .376 | .011 | D10S537 (94.10)/D20S606 (70.40) | .212 | .006 | D10S537 (94.10)/D20S606 (70.40) | .213 | <.001 | ||||
| Multipoint: | ||||||||||||
| ⩾27 | 78 | D10S1646 (88.70)/D20S149 (87.06) | .371 | <.001 | 282 | D10S197 (50.60)/D20S100 (82.26) | .205 | <.001 | 360 | D10S582 (46.70)/D20S149 (87.06) | .200 | <.001 |
| D10S1646 (88.70)/D20S876 (71.40) | .354 | <.001 | D10S197 (50.60)/D20S102 (83.26) | .194 | <.001 | D10S582 (46.70)/D20S102 (83.26) | .196 | <.001 | ||||
| D10S1647 (91.50)/D20S149 (87.06) | .351 | <.001 | D10S582 (46.70)/D20S102 (83.26) | .191 | <.001 | D10S582 (46.70)/D20S100 (82.26) | .195 | <.001 | ||||
| D10S1646 (88.70)/D20S102 (83.26) | .343 | .001 | D10S582 (46.70)/D20S100 (82.26) | .189 | <.001 | D10S1646 (88.70)/D20S913 (78.36) | .182 | .002 | ||||
| D10S1646 (88.70)/D20S913 (78.36) | .337 | .001 | D10S193 (59.10)/D20S100 (82.26) | .178 | .001 | D10S1646 (88.70)/D20S100 (82.26) | .177 | .001 | ||||
| ⩾30 | 63 | D10S1646 (88.70)/D20S149 (87.06) | .418 | .001 | 223 | D10S197 (50.60)/D20S100 (82.26) | .222 | <.001 | 286 | D10S582 (46.70)/D20S102 (83.26) | .236 | <.001 |
| D10S1646 (88.70)/D20S102 (83.26) | .403 | .001 | D10S582 (46.70)/D20S102 (83.26) | .218 | <.001 | D10S582 (46.70)/D20S100 (82.26) | .233 | <.001 | ||||
| D10S1646 (88.70)/D20S887 (64.50) | .399 | .002 | D10S582 (46.70)/D20S100 (82.26) | .216 | <.001 | D10S582 (46.70)/D20S149 (87.06) | .230 | <.001 | ||||
| D10S1647 (91.50)/D20S149 (87.06) | .397 | .001 | D10S197 (50.60)/D20S102 (83.26) | .206 | .001 | D10S1646 (88.70)/D20S887 (64.50) | .218 | <.001 | ||||
| D10S1646 (88.70)/D20S100 (82.26) | .393 | .002 | D10S193 (59.10)/D20S100 (82.26) | .195 | .001 | D10S1646 (88.70)/D20S178 (64.00) | .214 | <.001 | ||||
| ⩾35 | 45 | D10S1646 (88.70)/D20S102 (83.26) | .481 | <.001 | 152 | D10S197 (50.60)/D20S100 (82.26) | .227 | .004 | 197 | D10S1646 (88.70)/D20S178 (64.00) | .268 | <.001 |
| D10S1646 (88.70)/D20S100 (82.26) | .473 | <.001 | D10S1646 (88.70)/D20S178 (64.00) | .225 | .003 | D10S1646 (88.70)/D20S887 (64.50) | .250 | .002 | ||||
| D10S1646 (88.70)/D20S887 (64.50) | .435 | .001 | D10S582 (46.70)/D20S102 (83.26) | .220 | .008 | D10S582 (46.70)/D20S149 (87.06) | .239 | .001 | ||||
| D10S1646 (88.70)/D20S120 (79.36) | .435 | .002 | D10S197 (50.60)/D20S102 (83.26) | .219 | .003 | D10S1646 (88.70)/D20S869 (68.40) | .239 | .002 | ||||
| D10S1646 (88.70)/D20S606 (70.40) | .429 | .001 | D10S582 (46.70)/D20S100 (82.26) | .215 | .010 | D10S582 (46.70)/D20S102 (83.26) | .231 | .002 | ||||
When we used the permutation procedure to account for multiple comparisons in the multipoint analyses, the corrected P values for maximal correlation score were .019 (correlation 0.200), .007 (correlation 0.236), and .018 (correlation 0.268), for BMI ⩾27, ⩾30, and ⩾35, respectively. As expected, correction for multiple testing reduced the level of significance by as much as an order of magnitude, but the correlations remained significant, with P<.02.
Correlation Analyses Based on Family-Specific NPL Score
Table 3 presents correlation scores based on family-specific NPL score, with empirical P<.01 in at least one of the three samples. All the correlations were localized within 10q (89–97 cM) and 20q (65–75 cM). Most observed correlations occurred across both subgroups. For the African American sample, the highest correlation was found between D10S1646 and 20q (69–70 cM) for single (0.542 [empirical P<.001]) and multipoint (0.429 [empirical P=.002]) analyses. For the European American sample, the highest correlation was 0.361 (empirical P=.002) for single-point analysis and 0.302 (empirical P<.001) for multipoint analysis. For the combined sample, the maximal correlation of 0.311 (empirical P = .002) was found between D10S1646 and D20S211 (210 families with BMI ⩾35). The P values (corrected for multiple testing) for the maximal correlation score from the NPL analyses were .013 (correlation 0.273), .065 (correlation 0.207), and .002 (correlation 0.311), for BMI ⩾27, ⩾30, and ⩾35, respectively.
Table 3.
Correlation between Loci on Chromosomes 10 and 20 on the Basis of Family-Specific NPL Score
|
Correlation Score (Empirical P) |
||||
| Analysis and BMI | Markers (Distance [in cM]) | African Americans(N=43) | European Americans(N=167) | Combined(N=210) |
| Single point: | ||||
| ⩾27 | D10S1647 (91.50)/D20S887 (64.50) | .342 (.012) | .339 (.003) | .344 (.002) |
| D10S1647 (91.50)/D20S196 (69.40) | .362 (.008) | .334 (.014) | .340 (.008) | |
| D10S1646 (88.70)/D20S606 (70.40) | .171 (.142) | .329 (<.001) | .299 (.009) | |
| D10S1685 (94.00)/D20S211 (75.36) | .241 (.061) | .313 (.002) | .297 (.002) | |
| D10S537 (94.10)/D20S211 (75.36) | .135 (.202) | .334 (<.001) | .296 (.004) | |
| D10S1685 (94.00)/D20S196 (69.40) | .434 (.001) | .242 (.040) | .285 (.021) | |
| D10S535 (97.00)/D20S211 (75.36) | .094 (.276) | .328 (.002) | .282 (.020) | |
| ⩾30 | D10S537 (94.10)/D20S606 (70.40) | .348 (.013) | .293 (<.001) | .302 (<.001) |
| D10S1646 (88.70)/D20S887 (64.50) | .421 (<.001) | .218 (.027) | .273 (<.001) | |
| D10S1646 (88.70)/D20S196 (69.40) | .535 (<.001) | .180 (.087) | .245 (.001) | |
| ⩾35 | D10S537 (94.10)/D20S606 (70.40) | .188 (.113) | .354 (.001) | .329 (<.001) |
| D10S537 (94.10)/D20S211 (75.36) | .253 (.051) | .335 (.006) | .323 (.003) | |
| D10S535 (97.00)/D20S211 (75.36) | .060 (.348) | .361 (.002) | .311 (.002) | |
| D10S1646 (88.70)/D20S211 (75.36) | .437 (.001) | .277 (.035) | .310 (.003) | |
| D10S1646 (88.70)/D20S196 (69.40) | .542 (<.001) | .197 (.016) | .266 (.005) | |
| Multipoint: | ||||
| ⩾27 | D10S535 (97.00)/D20S839 (72.40) | .281 (.033) | .270 (.001) | .273 (.005) |
| D10S1685 (94.00)/D20S176 (67.40) | .352 (.011) | .248 (.002) | .271 (.009) | |
| D10S535 (97.00)/D20S902 (74.36) | .283 (.035) | .264 (.001) | .267 (.009) | |
| D10S1685 (94.00)/D20S196 (69.40) | .336 (.014) | .248 (.001) | .265 (.010) | |
| D10S1685 (94.00)/D20S869 (68.40) | .341 (.014) | .245 (.002) | .263 (.013) | |
| D10S1646 (88.70)/D20S839 (72.40) | .195 (.106) | .282 (<.001) | .260 (.006) | |
| ⩾30 | D10S1646 (88.70)/D20S606 (70.40) | .334 (.017) | .168 (.015) | .207 (.007) |
| D10S1646 (88.70)/D20S211 (75.36) | .252 (.050) | .177 (.010) | .198 (.006) | |
| ⩾35 | D10S1646 (88.70)/D20S211 (75.36) | .352 (.011) | .302 (<.001) | .311 (.002) |
| D10S1646 (88.70)/D20S876 (77.36) | .402 (.004) | .281 (.001) | .304 (.002) | |
| D10S1646 (88.70)/D20S120 (79.36) | .408 (.004) | .281 (<.001) | .304 (.001) | |
| D10S1646 (88.70)/D20S913 (78.36) | .410 (.004) | .276 (.001) | .302 (.001) | |
| D10S1646 (88.70)/D20S840 (74.86) | .335 (.013) | .284 (<.001) | .293 (.002) | |
| D10S1646 (88.70)/D20S606 (70.40) | .429 (.002) | .259 (.001) | .287 (.002) | |
| D10S1646 (88.70)/D20S902 (74.36) | .391 (.005) | .241 (.002) | .269 (.005) | |
Conditional and Epistatic Interaction Analyses
Figures 1 and 2 show the LOD scores based on the combined sample for the baseline and conditional multipoint allele-sharing analyses (BMI ⩾27) of chromosome 20, weighted by the evidence for linkage at D10S537 and D10S1646, respectively. Compared with baseline analyses, conditional analyses had the largest increment in LOD score at 20q (75 cM) for both weighting approaches. The LOD score at the peak rose from 1.53 in the baseline analysis to 2.80 (χ2=5.85; nominal P=.016; empirical P=.012) when families were weighted by evidence for linkage at 10q (D10S1646) through use of weight0-1 and to 3.32 (χ2=8.24; nominal P=.004; empirical P<.001) when weightprop was used (fig. 1). Similarly, the LOD score at the peak rose to 2.21 (χ2 = 3.13; nominal P = .077; empirical P = .040) when families were weighted by evidence for linkage at 10q (D10S537) through use of weight0-1 and to 2.93 (χ2 = 6.45; nominal P = .011; empirical P = .007) when weightprop was used (fig. 2). For quantitative traits, oligogenic analyses were performed using both additive and epistatic two-locus models, to look for evidence of joint effects. Figure 3 shows that there was a significant epistatic effect on percentage fat mass, between 20q (75 cM) and proximal 10p (LOD=1.74; P=.024), compared with a two-locus additive model (LOD=0.90). The P value obtained from the likelihood-ratio test of an epistatic two-locus model was compared with the two-locus additive model. No significant epistatic effects on other quantitative phenotypes were found in these regions.
Figure 1.
Multipoint analyses of chromosome 20 for BMI ⩾27 in the combined sample. Conditional allele-sharing multipoint analyses were performed by weighting the families on the basis of the evidence for linkage at D10S1646.
Figure 2.
Multipoint analyses of chromosome 20 for BMI ⩾27 in the combined sample. Conditional allele-sharing multipoint analyses were performed by weighting the families on the basis of the evidence for linkage at D10S537.
Figure 3.
Epistatic-interaction analyses for percentage fat mass by variance-component approach in the combined sample. Oligogenic analyses were performed using both additive and epistatic two-locus models to look for evidence of joint effects.
Discussion
Obesity has a substantial heritable component and has been found to be associated with or linked to >250 genomic regions (Perusse et al. 2001; Rankinen et al. 2002), but the identification of genes responsible for common forms of obesity continues to be difficult. Heritability studies suggest that nonadditive gene effects could account for as much as one-third of all genetic variation in obesity. However, to date, few studies have attempted to incorporate gene interaction in obesity-linkage analyses.
Since the mode of inheritance is largely unknown and is likely to be complex, the present study evaluated the evidence for gene interaction primarily by using nonparametric methods. We calculated correlations, in ASP-specific IBD-sharing probabilities and family-specific NPL scores, between unlinked regions on chromosomes 7, 10, and 20. Since we did not find any positive correlation, with nominal P<.01, between these regions and the chromosome 7 loci that have been linked to obesity phenotypes, the present study focused detailed analyses on chromosomes 10 and 20. Based on single-point and multipoint analyses and thresholds, the observed correlation scores revealed that there is an interaction between genes on 10q and on 20q. This conclusion was also supported by results from conditional analyses and hierarchical models, by using a variance-component approach.
The major challenge for gene-gene–interaction detection is that a potentially large number of comparisons is possible. We have adopted strategies to control for overall type I error rates while increasing the likelihood that possible gene-gene interactions will be identified. Accordingly, the overall statistical significance was assessed through simulation studies. Our major findings for correlations in IBD-sharing probabilities were significant even after correction for multiple testing, through permutation analyses. The relative simplicity of the IBD correlational approach requires few assumptions, and they should be met by many data sets. For this reason, we believe it should be preferred over correlations in NPL scores.
To date, several independent linkage studies have suggested the existence of obesity-predisposition loci on 20q (Norman et al. 1998; Lee et al. 1999; Hunt et al. 2001; Perusse et al. 2001; Deng et al. 2002; Rankinen et al. 2002). The reported regions appear to be too broad for study differences to be due to poor gene localization. The breadth of the linked interval, as well as the appearance of multiple peaks, suggests that there are multiple susceptibility genes in this region. In fact, 20q is rich in genes involved in signaling and contains several putative candidate genes for obesity. ASIP (agouti-signaling protein) is a protein inhibitor of MC3R and MC4R (melanocortin receptors 3 and 4, respectively) (Fong et al. 1997); it has been reported that inactivation of MC3R results in increased body fat at the expense of lean body mass (Chen et al. 2000) and that mutations of ASIP lead to obesity in mice (Miller et al. 1993). CEBPB (CAAT/enhancer-binding protein β) is related to adipocyte differentiation (Yeh et al. 1995). Mutations of GNAS1 (guanine nucleotide-binding protein α-stimulating activity polypeptide 1) were associated with Albright hereditary osteodystrophy partly characterized by obesity (Gunay-Aygun et al. 1997). It is noteworthy that conditional analyses based on evidence of linkage at either D10S1646 or D10S537, spanning 10 cM on 20q, resulted in similar increased evidence for linkage. This finding may reflect poor localization of the linkage signal; however, it may also indicate a complex interaction among common polymorphisms at different loci involved in common expression pathways that differ in frequency among families.
Although French and German studies and our study of the present cohort found markers on 10p to be linked to obesity (Hager et al. 1998; Hinney et al. 2000; Price et al. 2001), candidate genes for obesity within the identified regions are not well characterized. A French study suggested that there is a major gene locus, on 10p (D10S197; maximum LOD score 4.85), that is implicated in the development of human obesity. It is very interesting that, in the correlation analyses based on ASP-specific IBD-sharing probability, D10S197 (peak-NPL-score locus in the French study and the present study cohort) and D10S582 (∼3 cM from D10S197) gave a strong correlation with markers on 20q in the European American sample only. However, the strongest correlation in family-specific NPL score was found between proximal 10q markers and 20q13 markers across both subgroups in the present study. The French study also found a secondary linkage peak at the 10q centromeric location. Both the conditional analyses for a qualitative trait (BMI ⩾27) and the hierarchical model for a quantitative trait (percentage fat mass) revealed that there was an epistatic interaction between the two regions. Proximal 10q appears to be somewhat richer in genes than 10p is, but the association between these genes and obesity is largely unknown. Because the observed interactions in the present study are at a statistical (rather than biological) level, independent study will be needed to confirm and further characterize these interactions.
Conclusion
The present study provides evidence that genes in proximal 10q and 20q may interact to increase susceptibility to human obesity. More attention should be paid to epistatic gene-interaction effects in the identification of genes for this complex disorder. Caution should also be used in the interpretation of linkage evidence in the presence of interaction. Independent replication in multiple studies remains critical for the identification of genes controlling complex traits.
Acknowledgments
This research was supported in part by National Institutes of Health grants R01DK44073, R01DK48095, and R01DK56210 (to R.A.P.) and R01GM59507 (to H.Z.). We gratefully acknowledge the generous cooperation of the participating families.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- Genetic Location Database, The, http://cedar.genetics.soton.ac.uk/public_html/ldb.html (for map location)
- Genome Database, The, http://gdbwww.gdb.org/ (for map location)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for obesity [MIM 601665])
- Whitehead Institute/MIT Center for Genome Research, http://www-genome.wi.mit.edu/ (for map location)
References
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KK (1998) Obesity as a risk factor for certain types of cancer. Lipids 33:1055–1059 [DOI] [PubMed] [Google Scholar]
- Chen AS, Marsh DJ, Trumbauer ME, Frazier EG, Guan XM, Yu H, Rosenblum CI, Vongs A, Feng Y, Cao L, Metzger JM, Strack AM, Camacho RE, Mellin TN, Nunes CN, Min W, Fisher J, Gopal-Truter S, MacIntyre DE, Chen HY, Van der Ploeg LH (2000) Inactivation of the mouse melanocortin-3 receptor results in increased fat mass and reduced lean body mass. Nat Genet 26:97–102 [DOI] [PubMed] [Google Scholar]
- Coleman DL, Hummel KP (1975) Symposium IV: diabetic syndrome in animals. Influence of genetic background on the expression of mutations at the diabetes locus in the mouse. II. Studies on background modifiers. Isr J Med Sci 11:708–713 [PubMed] [Google Scholar]
- Cox NJ, Frigge M, Nicolae DL, Concannon P, Hanis CL, Bell GI, Kong A (1999) Loci on chromosomes 2 (NIDDM1) and 15 interact to increase susceptibility to diabetes in Mexican Americans. Nat Genet 21:213–215 [DOI] [PubMed] [Google Scholar]
- Deng HW, Deng H, Liu YJ, Liu YZ, Xu FH, Shen H, Conway T, Li JL, Huang QY, Davies KM, Recker RR (2002) A genomewide linkage scan for quantitative-trait loci for obesity phenotypes. Am J Hum Genet 70:1138–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong TM, Mao C, MacNeil T, Kalyani R, Smith T, Weinberg D, Tota MR, Van der Ploeg LH (1997) ART (protein product of agouti-related transcript) as an antagonist of MC-3 and MC-4 receptors. Biochem Biophys Res Commun 237:629–631 [DOI] [PubMed] [Google Scholar]
- Grilo CM, Pogue-Geile MF (1991) The nature of environmental influences on weight and obesity: a behavior genetic analysis. Psychol Bull 110:520–537 [DOI] [PubMed] [Google Scholar]
- Gunay-Aygun M, Cassidy SB, Nicholls RD (1997) Prader-Willi and other syndromes associated with obesity and mental retardation. Behav Genet 27:307–324 [DOI] [PubMed] [Google Scholar]
- Hager J, Dina C, Francke S, Dubois S, Houari M, Vatin V, Vaillant E, Lorentz N, Basdevant A, Clement K, Guy-Grand B, Froguel P (1998) A genome-wide scan for human obesity genes reveals a major susceptibility locus on chromosome 10. Nat Genet 20:304–308 [DOI] [PubMed] [Google Scholar]
- Hinney A, Ziegler A, Oeffner F, Wedewardt C, Vogel M, Wulftange H, Geller F, Stubing K, Siegfried W, Goldschmidt HP, Remschmidt H, Hebebrand J (2000) Independent confirmation of a major locus for obesity on chromosome 10. J Clin Endocrinol Metab 85:2962–2965 [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, et al (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26:163–175 (erratum 26:502) [DOI] [PubMed] [Google Scholar]
- Hunt SC, Abkevich V, Hensel CH, Gutin A, Neff CD, Russell DL, Tran T, Hong X, Jammulapati S, Riley R, Weaver-Feldhaus J, Macalma T, Richards MM, Gress R, Francis M, Thomas A, Frech GC, Adams TD, Shattuck D, Stone S (2001) Linkage of body mass index to chromosome 20 in Utah pedigrees. Hum Genet 109:279–285 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman PG (2000) Obesity as a medical problem. Nature 404:635–643 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439–454 [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Reed DR, Li WD, Xu W, Joo EJ, Kilker RL, Nanthakumar E, North M, Sakul H, Bell C, Price RA (1999) Genome scan for human obesity and linkage to markers in 20q13. Am J Hum Genet 64:196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WD, Reed DR, Lee JH, Xu W, Kilker RL, Sodam BR, Price RA (1999) Sequence variants in the 5′ flanking region of the leptin gene are associated with obesity in women. Ann Hum Genet 63:227–234 [DOI] [PubMed] [Google Scholar]
- Maes HH, Neale MC, Eaves LJ (1997) Genetic and environmental factors in relative body weight and human adiposity. Behav Genet 27:325–351 [DOI] [PubMed] [Google Scholar]
- Miller MW, Duhl DM, Vrieling H, Cordes SP, Ollmann MM, Winkes BM, Barsh GS (1993) Cloning of the mouse agouti gene predicts a secreted protein ubiquitously expressed in mice carrying the lethal yellow mutation. Genes Dev 7:454–467 [DOI] [PubMed] [Google Scholar]
- Niswender KD, Morton GJ, Stearns WH, Rhodes CJ, Myers MG Jr, Schwartz MW (2001) Intracellular signalling: key enzyme in leptin-induced anorexia. Nature 413:794–795 [DOI] [PubMed] [Google Scholar]
- Norman RA, Tataranni PA, Pratley R, Thompson DB, Hanson RL, Prochazka M, Baier L, Ehm MG, Sakul H, Foroud T, Garvey WT, Burns D, Knowler WC, Bennett PH, Bogardus C, Ravussin E (1998) Autosomal genomic scan for loci linked to obesity and energy metabolism in Pima Indians. Am J Hum Genet 62:659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollmann MM, Wilson BD, Yang YK, Kerns JA, Chen Y, Gantz I, Barsh GS (1997) Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science 278:135–138 [DOI] [PubMed] [Google Scholar]
- Perusse L, Chagnon YC, Weisnagel SJ, Rankinen T, Snyder E, Sands J, Bouchard C (2001) The human obesity gene map: the 2000 update. Obes Res 9:135–169 [DOI] [PubMed] [Google Scholar]
- Price RA (1987) Genetics of human obesity. Ann Behav Med 9:9–14 [Google Scholar]
- ——— (2002) Genetics and common obesities: background, current status, strategies and future prospects. In: Wadden T (ed) Obesity, theory and therapy. Guilford Publications, New York [Google Scholar]
- Price RA, Gottesman II (1991) Body fat in identical twins reared apart: roles for genes and environment. Behav Genet 21:1–7 [DOI] [PubMed] [Google Scholar]
- Price RA, Li WD, Bernstein A, Crystal A, Golding EM, Weisberg SJ, Zuckerman WA (2001) A locus affecting obesity in human chromosome region 10p12. Diabetologia 44:363–366 [DOI] [PubMed] [Google Scholar]
- Price RA, Reed DR, Guido NJ (2000) Resemblance for body mass index in families of obese African American and European American women. Obes Res 8:360–366 [DOI] [PubMed] [Google Scholar]
- Price RA, Reed DR, Lee JH (1998) Obesity related phenotypes in families selected for extreme obesity and leanness. Int J Obes Relat Metab Disord 22:406–413 [DOI] [PubMed] [Google Scholar]
- Rankinen T, Perusse L, Weisnagel SJ, Snyder EE, Chagnon YC, Bouchard C (2002) The human obesity gene map: the 2001 update. Obes Res 10:196–243 [DOI] [PubMed] [Google Scholar]
- Reed DR, Ding Y, Xu W, Cather C, Green ED, Price RA (1996) Extreme obesity may be linked to markers flanking the human OB gene. Diabetes 45:691–694 [DOI] [PubMed] [Google Scholar]
- Shintani M, Ikegami H, Yamato E, Kawaguchi Y, Fujisawa T, Nakagawa Y, Hamada Y, Ueda H, Miki T, Ogihara T (1996) A novel microsatellite polymorphism in the human OB gene: a highly polymorphic marker for linkage analysis. Diabetologia 39:1398–1401 [DOI] [PubMed] [Google Scholar]
- Shmulewitz D, Auerbach SB, Lehner T, Blundell ML, Winick JD, Youngman LD, Skilling V, Heath SC, Ott J, Stoffel M, Breslow JL, Friedman JM (2001) Epidemiology and factor analysis of obesity, type II diabetes, hypertension, and dyslipidemia (syndrome X) on the island of Kosrae, Federated States of Micronesia. Hum Hered 51:8–19 [DOI] [PubMed] [Google Scholar]
- Sorensen TI, Price RA, Stunkard AJ, Schulsinger F (1989) Genetics of obesity in adult adoptees and their biological siblings. BMJ 298:87–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman BM, Flier JS (2001) Obesity and the regulation of energy balance. Cell 104:531–543 [DOI] [PubMed] [Google Scholar]
- Stunkard AJ, Harris JR, Pedersen NL, McClearn GE (1990) The body-mass index of twins who have been reared apart. N Engl J Med 322:1483–1487 [DOI] [PubMed] [Google Scholar]
- Stunkard AJ, Sorensen TI, Hanis C, Teasdale TW, Chakraborty R, Schull WJ, Schulsinger F (1986) An adoption study of human obesity. N Engl J Med 314:193–198 [DOI] [PubMed] [Google Scholar]
- Wolk A, Gridley G, Svensson M, Nyren O, McLaughlin JK, Fraumeni JF, Adam HO (2001) A prospective study of obesity and cancer risk (Sweden). Cancer Causes Control 12:13–21 [DOI] [PubMed] [Google Scholar]
- Yeh WC, Cao Z, Classon M, McKnight SL (1995) Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev 9:168–181 [DOI] [PubMed] [Google Scholar]





