Abstract
RNAi, a gene-silencing pathway triggered by double-stranded RNA, is conserved in diverse eukaryotic species but has been lost in the model budding yeast, Saccharomyces cerevisiae. Here, we show that RNAi is present in other budding-yeast species, including Saccharomyces castellii and Candida albicans. These species use noncanonical Dicer proteins to generate siRNAs, which mostly correspond to transposable elements and Y’ subtelomeric repeats. In S. castellii, RNAi mutants are viable but have excess Y’ mRNA levels. In S. cerevisiae, introducing Dicer and Argonaute of S. castellii restores RNAi, and the reconstituted pathway silences endogenous retrotransposons. These results identify a novel class of Dicer proteins, bring the tool of RNAi to the study of budding yeasts, and bring the tools of budding yeast to the study of RNAi.
RNA-silencing pathways contribute to transposon silencing, viral defense, DNA elimination, heterochromatin formation, and posttranscriptional repression of cellular genes (1, 2). In the simplest form of silencing, known as RNA interference (RNAi), the RNaseIII endonuclease Dicer successively cleaves double-stranded RNA (dsRNA) into small interfering RNAs (siRNAs), which are loaded into the effector protein Argonaute to guide the cleavage of target transcripts (1, 3). RNAi arose in an early eukaryotic ancestor and appears to have been conserved throughout most of the fungal kingdom (4, 5) (Fig. 1A). A prominent exception is Saccharomyces cerevisiae, a budding yeast that lacks recognizable homologs of Argonaute, Dicer, and RNA-dependent RNA polymerase (RdRP), which in some RNAi pathways produces dsRNA. Indeed, RNAi has been presumed lost in all budding yeasts. Despite this perceived loss, Argonaute genes are present in some budding yeasts (6, 7), including Saccharomyces castellii and Kluyveromyces polysporus (both close relatives of S. cerevisiae) and Candida albicans (the most common yeast pathogen of humans (8)) (Fig. 1A). The presence of these genes in budding yeast has been enigmatic because other RNAi genes, especially Dicer, have not been found in these species. A similar conundrum appears in prokaryotes, in which certain bacteria have Argonaute homologs yet lack the other genes associated with RNAi or related RNA-silencing pathways (9).
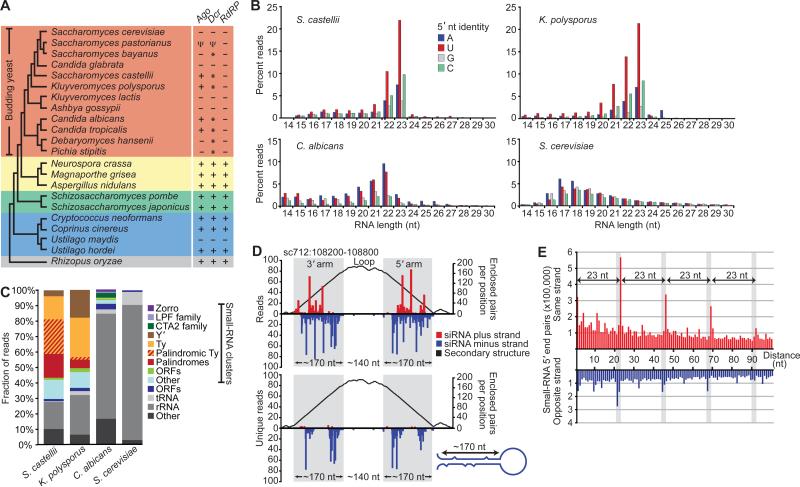
Fig. 1.
Endogenous siRNAs in budding yeasts. (A) Cladogram showing Basidiomycota (blue), Zygomycota (grey) and Ascomycota, subdivided into Saccharomycotina (budding yeasts, orange), Pezizomycotina (yellow) and Taphrinomycotina (green) (35, 36). The presence of canonical RNAi genes is indicated (+) (4, 5) and references therein). All genomes had a RNT1 ortholog, and several others had a second RNaseIII domain-containing gene (*), which has Dicer activity in S. castellii. Pseudogenes are indicated (Ψ). S. bayanus, which had a Dicer but not an Argonaute gene, appeared to lack siRNAs (fig. S1). (B) Length distribution of genome-matching sequencing reads representing small RNAs with the indicated 5’ nucleotide. Reads matching rRNA and tRNA are excluded. (C) Classification of loci to which 21–23-mers map, considering those that map to clusters in a pattern suggestive of siRNAs separately from those that do not. (D) A palindromic region generating siRNAs in S. castellii. 5’ termini of 22–23-mers were mapped to the genome, and counts (normalized to the number of genomic matches) are plotted for the plus and minus genomic strands. The top graph considers all reads; the bottom considers those matching the genome at only one locus. The predicted structure of the (–)-strand transcript is represented as a mountain plot (37). (E) Distribution of the genomic intervals separating the 5’ termini of sequenced 23-mers from S. castellii. Plotted is the frequency of each interval, when considering all pairs of reads less than 100 nt apart (excluding reads matching rRNA and tRNA).
siRNAs in budding yeasts
To search for RNA silencing in budding yeast, we looked for short guide RNAs, isolating 18–30-nt RNAs from S. castellii, K. polysporus, and C. albicans and preparing sequencing libraries representing the subset of small RNAs with 5’-monophosphates and 3’-hydroxyls (10), which are the chemical features of Dicer products. The small RNAs of S. castellii and K. polysporus were most enriched in 23-mers beginning with U, and those of C. albicans were most enriched in 22-mers beginning with A or U (Fig. 1B). These biases were reminiscent of those observed for Argonaute-bound guide RNAs of animals, plants, and other fungi (11-13). Analogous RNAs were not found in S. cerevisiae, as expected for a species lacking RNAi (Fig. 1B).
Although some reads from the Argonaute-containing yeasts mapped to ribosomal RNA (rRNA) and tRNA and presumably represented degradation intermediates of abundant RNAs, many reads clustered at other types of genomic loci. The loci generating the most reads had sequence homology to repetitive elements, including LTR retrotransposons (Ty elements), LINE-like retrotransposons (Zorro elements), and subtelomeric repeats (Y’ elements) (Fig. 1C, table S1). Loci of S. castellii were also particularly enriched in long inverted repeats; these palindromic loci generated most of the reads with homology to Ty elements (Fig. 1C and D). In S. cerevisiae, essentially all the reads appeared to represent degradation fragments of rRNA, tRNA, and mRNA.
The reads matching inverted repeats suggested origins from paired regions of transcripts that folded back on themselves to form hairpins (Fig. 1D). These inferred hairpins had 100- to 400-bp stems with loops ranging from 19 to >1600 nt. In regions of imperfect duplex, where reads could be mapped unambiguously, the small RNAs tended to match only one genomic strand, further supporting the idea that they originated from hairpin transcripts (Fig. 1D, bottom). Other reads did not map to inverted repeats and instead mapped uniquely to both genomic strands in a pattern suggesting origins from long bimolecular duplexes involving transcripts from both strands.
Most siRNAs of the fission yeast Schizosaccharomyces pombe correspond to the outer repeats of the centromeres and direct heterochromatin formation and maintenance (14). We therefore examined whether any of our sequenced small RNAs matched centromeres. Of the three Argonaute-containing species from which we sequenced (Fig. 1B), only C. albicans had annotated centromeres, and almost none (<0.001%) of our C. albicans reads matched these genomic loci. Also arguing against a function analogous to that in S. pombe, budding yeasts lack recognizable orthologs of the H3K9 methyltransferase Clr4 and recognizable homologs of RdRP, Tas3, Chp1, and the HP1-like chromodomain protein Swi6—proteins all necessary for RNAi-dependent heterochromatin in S. pombe (14).
When mapped to the genome, the end of one 23-mer RNA was often next to the beginning of another 23-mer, suggesting that endonuclease cleavage simultaneously generated the 3’-terminus of one small RNA and the 5’-terminus of the next. Consistent with this hypothesis, systematic analysis of the intervals spanning the mapped ends of all 23-mer pairs revealed a clear phasing interval of 23 nt (Fig. 1E). Such phasing implied successive cleavage beginning at preferred starting points. Moreover, pairs from opposite strands had the same phasing interval but in a register 2 nt offset from that of the same-strand pairs. Together, the phasing and offset implied successive cleavage of dsRNA with a 2-nt 3’ overhang—the classic biogenesis of endogenous siRNAs by Dicer (3). Therefore, the small RNAs that appeared to derive from regions of dsRNA, i.e., those mapping in clusters to the arms of predicted hairpins and those mapping in clusters to both genomic strands, were classified as siRNAs.
Dicer in budding yeasts
The presence of siRNAs in Argonaute-containing budding yeasts implied that each of these species also had a Dicer-like activity. To assay for this activity, we monitored processing of a long dsRNA added to whole-cell extracts (15). Extracts from S. castellii, K. polysporus, and C. albicans—but not from S. cerevisiae—contained an activity that produced 22–23-nt RNAs, each preferentially from dsRNA rather than from single-stranded RNA (Fig. 2A). Moreover, for each extract the small-RNA length matched that of the most abundant length observed in vivo (Figs. 1B and 2A).
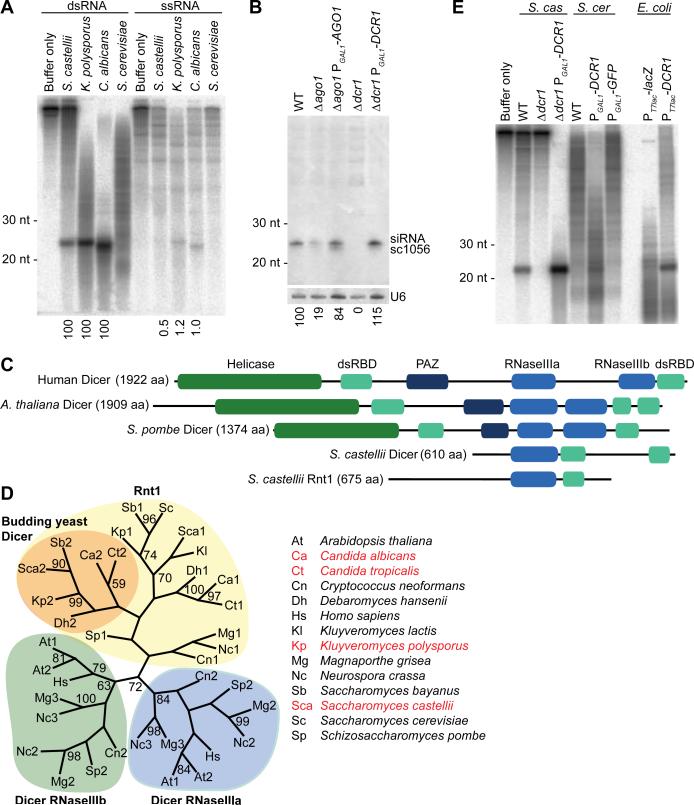
Fig. 2.
The Dicer of budding yeast. (A) In vitro processing of radiolabeled dsRNA or single-stranded RNA (ssRNA) in extracts from the indicated budding-yeast species. Products were resolved on a denaturing gel. The fraction of product normalized to that observed with dsRNA is indicated below as a percentage. (B) RNA blot probing for an endogenous siRNA (sc1056) in the indicated deletion and rescue strains. The blot was reprobed for U6 small nuclear RNA, and the siRNA percent signal normalized to that of U6 is indicated below. (C) Domain architectures of representative Dicer proteins and the two S. castellii proteins containing an RNaseIII domain. (D) Maximum-likelihood tree based on amino acid alignment of RNaseIII domains from Dicer proteins and Rnt1 homologs. Orange shading highlights budding-yeast Dicer candidates indicated by asterisks in Figure 1A. Budding-yeast species encoding Argonaute are listed in red. Bootstrap values higher than 50% are shown. (E) In vitro dicing in extracts from recombinant S. castellii (S. cas), S. cerevisiae (S. cer), or E. coli strains with the indicated deletions and additions, analyzed as in (A).
Despite the observed Dicer-like activity, a gene with the domain architecture of known Dicers was not found in any budding-yeast genome (Fig. 1A) (10). Because we had evidence for cleavage of dsRNA with 2-nt 3’ overhangs, a hallmark of RNaseIII activity, we relaxed the search criteria to consider any gene with an RNaseIII domain. S. cerevisiae had only one gene, RNT1, with a recognizable RNaseIII domain. RNT1 helps process rRNA and other noncoding RNAs (16), and presumed orthologs were found throughout the fungal kingdom. S. castellii had a second RNaseIII-domain-containing gene, and a potential ortholog of this gene was found in each of the other Argonaute-containing budding yeasts (Fig. 1A). Anticipating that this second gene encoded the Dicer of budding yeasts, we named it DCR1.
To test whether the Dicer candidate is required for siRNA accumulation, we deleted DCR1 in S. castellii—the closest relative to S. cerevisiae among the sequenced Argonaute-containing species. This procedure required establishing strains and protocols to better enable molecular genetic analysis in this species (15, 17). In the Δdcr1 mutant, siRNAs failed to accumulate (Fig. 2B, fig. S2, and table S1). Deletion of the Argonaute homolog, which we named AGO1, also reduced siRNA accumulation, as expected if loading into Argonaute protected siRNAs from degradation (Fig. 2B, fig. S2, and table S1). For both mutants, ectopically expressing the deleted gene rescued siRNA accumulation (Fig. 2B). These results indicate that the core components of endogenous RNAi pathways—Dicer, Argonaute, and siRNAs—are present in some species of the budding-yeast clade.
In S. pombe and other fungi, known Dicer genes resemble those in plants and animals, complete with tandem RNaseIII domains, 2–3 dsRBDs, a PAZ domain, and an N-terminal helicase domain (15, 18, 19) (Fig. 2C). In budding yeasts, DCR1 has two dsRBDs but only a single RNaseIII domain and no helicase or PAZ domains. Because RNaseIII domains work in pairs to nick both strands of an RNA duplex (19, 20), we suspect that S. castellii Dcr1 acts as a homodimer. Dicers of insects, plants, and mammals, which already have two RNaseIII domains, do not homodimerize but do form heterodimeric complexes with cofactors that provide additional dsRBDs (21-23). A homodimeric S. castellii Dcr1 complex would already possess four dsRBDs, which might obviate the need for such a cofactor.
Except for its second dsRBD, the domain architecture of the budding-yeast Dicer resembled that of RNT1 rather than that of canonical Dicer genes (Fig. 2C). Furthermore, the amino acid sequence of its RNaseIII domain was more similar to that of the RNT1 RNaseIII domain than to that of any previously identified Dicer RNaseIII domain (Fig. 2D). These observations suggest that budding-yeast Dicer might have emerged from a duplication of RNT1 early in the budding-yeast lineage, perhaps coincident with the loss of canonical Dicer. The unusual ancestry and domain structure of DCR1 might explain why its activity, and thus RNAi more generally, went undetected for so long in budding yeast.
Biochemical analyses of Dcr1 and Ago1
Dicing activity of S. castellii extracts was lost in the Δdcr1 mutant and restored by Dcr1 overexpression (Fig. 2E). To determine if Dcr1 is active in the absence of S. castellii cofactors, we expressed the protein in S. cerevisiae and E. coli (Fig. 2E). Expression in E. coli conferred robust activity, indicating that S. castellii Dcr1 is sufficient to dice dsRNA at precise intervals. In other Dicers, the PAZ domain is an essential component of a molecular ruler that imparts cleavage precision (19). The budding-yeast Dicers, which lack this domain, must achieve this measuring function differently.
To establish a biochemical link between Ago1 and the siRNAs of S. castellii, we sequenced the small RNAs that co-purified with tagged Ago1 expressed from its native promoter. Compared to the input RNA, the population of Ago1-associated RNAs was even more enriched for 22–23-nt RNAs and was depleted in matches to both rRNA and tRNA, with concomitant enrichment for matches to palindromes, Ty elements, and Y’ elements (fig. S3 and table S2). These biochemical results supported the genetic link between AGO1 and the siRNAs (Fig. 2B) and provided a set of small RNAs suitable for annotating the siRNA-producing loci of S. castellii (table S3).
The impact of RNAi on the S. castellii transcriptome
To investigate the molecular consequences of RNAi, we performed high-throughput sequencing of polyadenylated RNA (mRNA-Seq (24)) from wild-type, Δago1, and Δdcr1 strains (table S4). The two annotated open reading frames (ORFs) that changed most in RNAi deletion strains were also the two with the highest density of antisense siRNA reads (Fig. 3A, red points). One was the consensus Y’ ORF (fig. S5), which increased >7 fold in both deletion mutants. The other was an ORF within a palindromic Ty fragment, which increased >4 fold in the Δdcr1 mutant but less in the Δago1 mutant. For other ORFs, transcript-abundance changes were modest and not correlated with siRNA density (fig. S6), although changes in Δago1 and Δdcr1 mutants did correlate with each other (R2 = 0.39, Fig. 3A). This correlation might reflect a general response to the loss of RNAi (although we cannot exclude contributions of a common response to the hygromycin- and kanamycin-resistance genes used to delete AGO1 and DCR1, respectively).
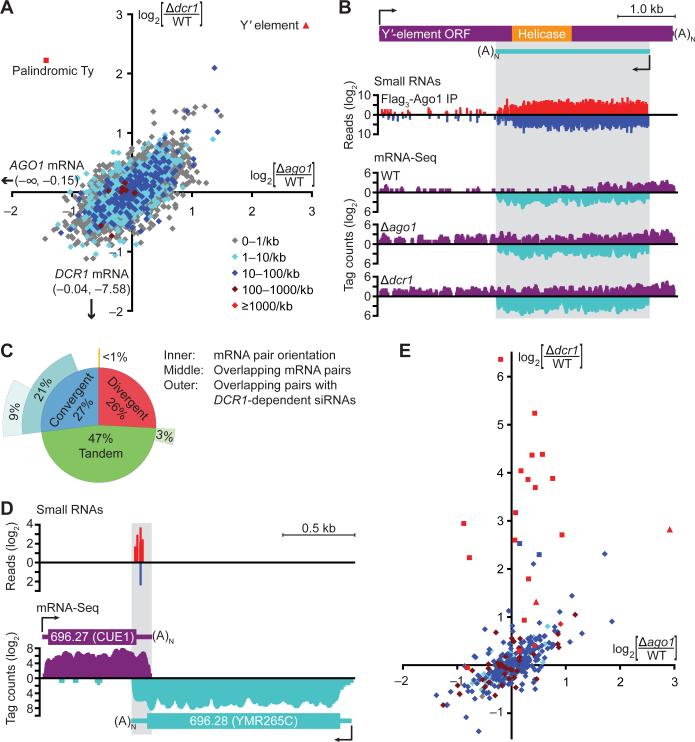
Fig. 3.
The impact of RNAi on the S. castellii transcriptome. (A) Strand-specific mRNA-Seq analysis of annotated ORF transcripts in wild-type (WT) and RNAi-mutant strains. Plotted is the log2 ratio of transcript abundance in Δago1 versus wild-type (x-axis) and Δdcr1 versus wild-type (y-axis). Colors indicate the density (reads/kb) of antisense small (22–23-nt) RNAs that co-purified with Ago1. A Ty ORF fragment (annotated as Scas_712.50) embedded within a palindromic siRNA-producing locus is indicated (square). Annotated Y’-element ORFs were replaced by one consensus Y’ ORF (triangle, fig. S5). Because the mRNA-Seq protocol included poly(A) selection, which retains the 3’ but not 5’ fragments of cleaved mRNAs, full-length transcript abundance was calculated using tags mapping to the 5’ half of each ORF. Similar trends were observed when using tags mapping across the ORF (fig. S4). (B) Analysis of the S. castellii Y’ element. The numbers of siRNA 5’ ends (small RNAs) and mRNA tags (mRNA-Seq) mapping to the consensus Y’ element are plotted for each position (sense, above axis; antisense, below axis). (C) Gene-pair organization and overlap in S. castellii. Inner ring, fraction of neighboring annotated ORFs with the indicated orientation; middle ring, fraction of transcript pairs with overlapping 3’ ends (convergent), overlapping 5’ ends (divergent), or continuous transcription in between (tandem); outer ring, fraction of convergent transcript pairs generating siRNAs in the overlapping region. (D) A pair of convergent transcripts that generate siRNAs in the region of overlap. Plots are as in (B). (E) mRNA-Seq analysis of inferred siRNA-generating transcripts. The plot is as in (A), using the same colors to indicate siRNA-read density and shapes to indicate transcripts mapping to Y’ elements (triangle), palindromes (square), and others (diamonds).
Because many siRNAs mapped antisense to or outside of ORFs, the mRNA-Seq data revealing the S. castellii polyadenylated transcriptome enabled the systematic identification of siRNA precursor transcripts. We focused on three types of siRNA precursors: sense-antisense transcript pairs from ORF loci, partially overlapping mRNAs, and transcripts producing the most siRNA-like reads, regardless of annotation.
The potential for dsRNA composed of sense-antisense transcripts from ORF loci was indicated by widespread low-level antisense transcription of ORFs, with antisense mRNA-Seq tags mapping to over half of all annotated ORFs. Moreover, small RNAs mapped antisense to nearly one-third of ORFs (Fig. 3A) and as a class were reduced in RNAi mutants and enriched by Ago1 immunoprecipitation (fig. S3 and table S2). Supporting a precursor-product relationship, the abundance of the sense-antisense duplexes (inferred from mRNA-Seq data) correlated with that of small RNAs deriving from these loci (fig. S7). The most striking example of siRNAs arising from sense-antisense transcript pairs was within the Y’ ORF, which was most affected by the loss of the RNAi machinery (Fig. 3A and B). Y’ elements are conserved protein-coding repeats. In S. cerevisiae they are located near both ends of most chromosomes (25) (fig. S7), and synteny suggests analogous locations in S. castellii (26). The S. castellii elements had a robustly expressed antisense transcript with many siRNAs mapping to the region of sense-antisense overlap (Fig. 3B).
We considered partially overlapping mRNAs as another potential source of siRNA-generating dsRNA, after using the mRNA-Seq data to extend the 5’ and 3’ boundaries of 5297 S. castellii protein-coding transcripts. Although only 1% of divergent transcript pairs and 7% of tandem transcript pairs overlapped, 78% of convergent transcript pairs overlapped (Fig. 3C, median overlap of 92 nt at their 3’ ends, fig. S7). At least 43% of these convergent and overlapping gene pairs (comprising 9% of all gene pairs) generated DCR1-dependent siRNAs in the region of overlap (Fig. 3C and fig. S3); one such pair is illustrated (Fig. 3D). A recent study reported pervasive overlapping transcripts in S. cerevisiae (27). Our results revealing analogous overlap in S. castellii show that, in contrast to previous speculation, this phenomenon is not restricted to RNAi-deficient organisms and is an ancestral feature of these Saccharomyces species (15).
We next inferred precursor transcripts without considering whether or not they overlapped ORFs (table S5). A hidden Markov model analyzing the Ago1-associated small RNAs identified the genomic loci producing abundant siRNAs, and analysis of the mRNA-Seq data from Δdcr1 strains revealed the corresponding transcripts. In addition to recovering the more prolific ORF-overlapping siRNA precursors, this analysis identified the transcript illustrated in Figure 1D and transcripts of 84 other non-protein-coding siRNA-generating genes of S. castellii (annotated as NCS1–NCS85, tables S3 and S5). Transcripts producing fewer siRNAs in RNAi-competent cells changed modestly but similarly in both deletion mutants (Fig. 3E), as observed when analyzing only ORF transcripts (Fig. 3A). Transcripts producing the most siRNAs—which were predominantly from palindromic loci—increased dramatically in the Δdcr1 mutant but were relatively unchanged in the Δago1 mutant (Fig. 3E and table S5), indicating that Dcr1 alone was sufficient to reduce these transcripts to wild-type levels. This mode of posttranscriptional down-regulation may be unique to palindromic transcripts, which can fold into hairpin structures that are ideal Dcr1 substrates but refractory to intermolecular pairing with Ago1-associated siRNAs.
Taken together, our results indicate that more than a thousand genomic loci in S. castellii generate siRNAs. The consequences of siRNAs derived from the widespread antisense and overlapping transcription in S. castellii are unknown. With the exception of the Y’ mRNA, the loss of the RNAi machinery did not substantially affect the levels of mRNAs corresponding to these siRNAs (Figs. 3A and S6). Perhaps in other growth conditions the regulatory impact of non-Y’ siRNAs might be more pronounced. The specificity for Y’-element regulation could arise from requiring both an abundance of antisense siRNAs and the ability to base pair with a target transcript. Although palindromic loci generate many siRNAs, the hairpin structure of these transcripts might block pairing with siRNAs, and although coding mRNAs are relatively unstructured, most generate only low levels of siRNAs. These two requirements would explain the observed impact of RNAi on the S. castellii transcriptome.
Engineering RNAi in S. castellii
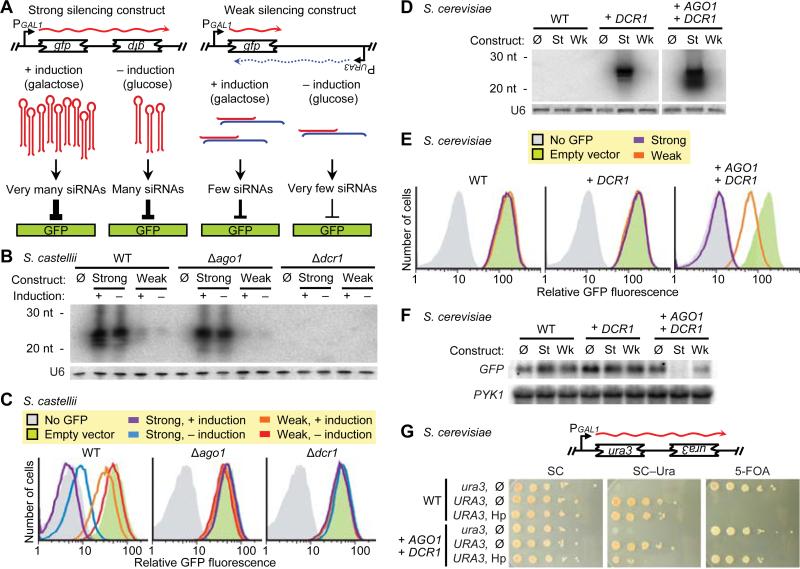
To confirm that siRNAs can silence a gene in S. castellii and to create tools for monitoring RNAi in budding yeast, we generated two constructs (strong and weak) designed to silence a green-fluorescent-protein (GFP) reporter gene (Fig. 4A). Both silencing constructs were under the control of an inducible promoter, and each was integrated into the chromosomes of wild-type, Δago1, and Δdcr1 strains expressing GFP. The two constructs and two induction conditions produced a gradient of GFP siRNAs (Fig. 4B). In cells containing both AGO1 and DCR1, the amount of GFP silencing, as measured by fluorescence-activated cell sorting (FACS), corresponded to the level of GFP siRNAs, with the highest level of siRNA production repressing fluorescence to background autofluorescence (Fig. 4C). As expected, silencing depended on DCR1 for siRNA production and on AGO1 for siRNA function (Fig. 4B and C). These results confirmed that siRNAs could function to silence a gene and demonstrated that the targeted transcript could originate from a locus distinct from that producing the siRNAs.
Fig. 4.
Engineering RNAi in S. castellii and S. cerevisiae. (A) Schematic for silencing of a GFP reporter. The strong silencing construct included inverted repeats of a gfp fragment and was designed to produce a hairpin transcript (38). The weak silencing construct contained one copy of the fragment, which is transcribed convergently to produce dsRNA. (B) RNA blot probing for siRNAs antisense to GFP, using total RNA from the indicated S. castellii strains with integrated empty vector (Ø) or silencing construct (strong or weak), either induced with galactose (+) or uninduced (–). The blot was reprobed for U6 small nuclear RNA. (C) FACS histograms showing GFP fluorescence in the indicated S. castellii strains expressing the indicated silencing constructs. (D) RNA blot probing for siRNAs antisense to GFP in S. cerevisiae strains expressing either no S. castellii genes (WT) or the indicated integrated S. castellii genes, and either the strong (St), the weak (Wk), or no (Ø) silencing construct. The blot was reprobed for U6 small nuclear RNA. (E) FACS histograms showing GFP fluorescence in the indicated S. cerevisiae strains expressing the indicated silencing constructs. All strains were induced; silencing from uninduced constructs was similar for the strong construct and undetectable for the weak construct (fig. S9). (F) RNA blot probing for GFP mRNA in the indicated S. cerevisiae strains expressing the indicated silencing constructs. The blot was reprobed for PYK1 mRNA as a loading control. (G) Silencing an endogenous gene. S. cerevisiae strains containing non-functional and functional URA3 genes (ura3 and URA3, respectively) and expressing the indicated S. castellii genes and either the diagramed hairpin construct (Hp) or no silencing construct (Ø) were tested for Ura3p expression by plating serial dilutions on complete media (SC), media lacking uracil (SC–Ura), and media containing 5-FOA (to which cells producing Ura3p are sensitive).
Reconstitution of RNAi in S. cerevisiae
Our observation that some budding yeasts closely related to S. cerevisiae contain a functional RNAi pathway suggested that the S. cerevisiae lineage lost RNAi recently and that perhaps introducing the two RNAi proteins found in S. castellii—Ago1 and Dcr1—could restore the pathway. To test this possibility, we used a GFP-reporter system based on our S. castellii system. GFP-positive strains of S. cerevisiae were generated that expressed either the strong, the weak, or no silencing construct. Introducing Dcr1 was sufficient to generate some GFP siRNAs from the weak construct and abundant GFP siRNAs from the strong silencing construct (Fig. 4D). When Ago1 and Dcr1 were both present, we observed intermediate silencing with the weak construct and robust silencing with the strong construct (Fig. 4E), with a >100-fold decrease in mRNA accompanying the decrease in fluorescence (Fig. 4F and fig. S10). Moreover, a hairpin construct targeting URA3 reduced growth in the absence of uracil and enabled growth on 5-fluoroorotic acid (5-FOA), demonstrating that the RNAi pathway reconstituted in S. cerevisiae can silence an endogenous gene with phenotypic consequences (Fig. 4G).
The ability to reconstitute RNAi in S. cerevisiae using only Ago1 and Dcr1 raises the possibility that the S. castellii RNAi pathway requires only these two proteins. This simplicity would make budding-yeast RNAi distinct from all known RNAi pathways, which use additional proteins involved in, for example, Argonaute loading (e.g., R2D2 in Drosophila melanogaster (28)) or maturation of the silencing complex (e.g., QIP in Neurospora crassa (29)). The four dsRBDs that would be present in a Dcr1 homodimer might explain the absence of a separate loading factor. Alternatively, overexpression of Ago1, Dcr1, and a hairpin precursor might be sufficient to enact RNAi in S. cerevisiae, but they might require additional factors for efficient silencing when expressed at physiological levels in S. castellii. Another possibility is that the reconstituted pathway uses components that have been maintained in S. cerevisiae since its recent loss of RNAi.
RNAi and transposon silencing
The Δago1 and Δdcr1 mutants of S. castellii were viable, with no obvious growth disadvantage in minimal or rich media at a range of temperatures, no observed decrease in mating, sporulation, or chromosome stability, and no altered sensitivity to a replication inhibitor (hydroxyurea) or to microtubule destabilizing agents (thiobendazole and benomyl). However, both Δago1 and Δdcr1 mutants had difficulty retaining introduced plasmids, demonstrating that the loss of RNAi has detectable phenotypic consequences (fig. S11).
We suspected that budding-yeast RNAi might also silence transposable elements. RNAi and related processes silence and eliminate transposons in other eukaryotes (2), and a large fraction of our budding-yeast siRNAs corresponded to transposable elements. For example, most S. castellii siRNAs mapped to fragments of Ty retrotransposons (Fig. 1C). Despite the abundance of Ty fragments, indicative of former activity in the S. castellii lineage (fig. S12), we have not yet found an active retrotransposon in the current, albeit incomplete, S. castellii genome sequence. Therefore, to test the effect of RNAi on transposition, we turned to the RNAi-competent S. cerevisiae strain.
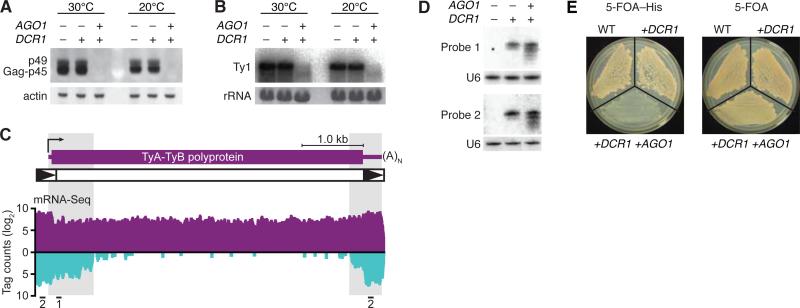
Compared to the strain with no RNAi genes or the one with only DCR1, the RNAi-competent strain had much less Ty1 Gag protein and mRNA (Fig. 5A and B). The dsRNA triggering this repression of protein and mRNA from native Ty1 elements could have come from elements expressing their own antisense transcripts (30) or from neighboring elements or fragments oriented with potential to produce convergent or hairpin transcripts (31). Analysis of published mRNA-Seq data from S. cerevisiae (32) revealed regions with many tags antisense to Ty1 elements (Fig. 5C), and these regions produced siRNAs in S. cerevisiae strains containing DCR1 (Fig. 5D). To examine whether RNAi can suppress retrotransposition, we ectopically expressed a Ty1 element marked with HIS3, which enabled transposition to be detected as plasmid-independent complementation of histidine auxotrophy (33). Consistent with our molecular findings for endogenous elements, the RNAi-competent strain permitted much less transposition (Fig. 5E). These results, combined with our sequencing data (Fig. 1C), indicate that a major role of budding-yeast RNAi is to silence transposons.
Fig. 5.
Silencing of Ty1 retrotransposons by RNAi in S. cerevisiae. (A) Immunoblot probing for Ty1 Gag protein (p45) and its precursor (p49) (39) in S. cerevisiae strains expressing the indicated S. castellii genes. Strains were grown under standard (30°C) or transposition-inducing (20°C) conditions. The blot was reprobed for actin. (B) RNA blot probing for Ty1 mRNA, analyzing the same cultures as in (A). Ethidium bromide-stained rRNA is shown. (C) mRNA-Seq analysis of S. cerevisiae Ty1 elements. The average numbers of mRNA tags to a consensus Ty1 element are plotted (sense, above axis; antisense, below axis). The schematic shows a Ty1 transcript (purple) and element, with long terminal repeats as black triangles. Locations of the two probes used in panel D are indicated. (D) RNA blot probing for siRNAs processed from endogenous Ty1 dsRNA. The blot was reprobed for U6 small nuclear RNA. (E) HIS3-marked Ty1 transposition assay. Galactose-induced S. cerevisiae strains expressing the indicated S. castellii genes were tested for transposition by growth on plates with 5-FOA and lacking histidine (5-FOA–His). Cells grow without histidine and are resistant to 5-FOA when the HIS3-marked Ty1 element has transposed into the genome and the URA3-marked plasmid carrying the original HIS3-marked element has been lost (33). Also shown is growth on media selective for plasmid loss but not transposition (5-FOA).
Adding the minimal RNAi components conferred transposon silencing to a species normally lacking the RNAi pathway. The recipient strain had no obvious abnormalities while endogenous transposon protein and mRNA were both drastically reduced, which illustrates the ability of RNAi to preferentially target transposon genes rather than other cellular genes. While specific for transposable elements, the pathway appears general for any element requiring an RNA transcript—including those it had not previously encountered—by exploiting internally initiated antisense transcripts as well as the intrinsic propensity of these elements to generate hairpin and convergent transcripts as their genomic load increases.
Concluding remarks
We have uncovered an RNAi pathway present in several different budding-yeast species that appears distinct from the well-characterized pathway of fission yeast. The two known components of the pathway have a patchy phylogenetic distribution among budding yeasts (Fig. 1A), indicating that the pathway can be lost easily. Indeed, if transposon silencing is the critical function of the RNAi pathway, then a species in which transposons have been completely silenced for a long evolutionary period is likely to lose all intact elements and thereby lose selection to retain the RNAi pathway, opening the door to re-invasion. Perhaps also contributing to RNAi loss is its potential inhibition of dsRNA viruses and their associated satellite dsRNAs. In S. cerevisiae, the M satellite element of the reovirus-like L-A virus encodes a secreted toxin that kills neighboring cells lacking element-encoded immunity (34). If cells that have lost RNAi are better able to retain this system, they might have a selective advantage despite having lost an efficient transposon-defense pathway.
With the discovery and characterization of the budding-yeast pathway, RNAi can be used as a tool to silence genes in S. cerevisiae, S. castellii, and presumably other budding yeasts. RNAi might be particularly useful in C. albicans, an obligate diploid for which both gene deletions and genetic screens are not trivial (8). Even in S. cerevisiae, RNAi might have advantages for repressing repetitive gene families. RNAi also enables an inducible repression system that might provide an alternative to existing technologies, which involve either non-physiological expression of the gene of interest (e.g. the GAL/GLU system) or generation of temperature-sensitive mutations. Perhaps more importantly, the tools of budding yeast can now be applied to the study of RNAi, either by developing reagents to investigate the endogenous pathway in S. castellii or by applying existing technologies to examine the reconstituted pathway in S. cerevisiae. While anticipating a productive future for RNAi research in budding yeasts, we note that if in the past S. castellii rather than S. cerevisiae had been chosen as the model budding yeast, the history of RNAi research would have been dramatically different.
Supplementary Material
Acknowledgments
We thank A. Hochwagen, G. Ruby, and H. Guo for helpful discussions, V. Agarwal for protein homology modeling, D. Garfinkel for Ty1-VLP antiserum and other reagents, M. Cohn and A. Regev for yeast strains, W. Johnston and the Whitehead Genome Technology Core for high-throughput sequencing, and S. Pääbo for academic advising. Supported by Science Foundation Ireland and IRCSET (K.W.), NIH grants GM040266 (G.F.), GM0305010 (G.F.), and GM067031 (D.B.), an NSF graduate research fellowship (D.W.), and a Boehringer-Ingelheim Fonds predoctoral fellowship (I.D.). D.B. is an Investigator of the Howard Hughes Medical Institute.
References and Notes
- 1.Tomari Y, Zamore PD. Genes Dev. 2005;19 doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 2.Malone CD, Hannon GJ. Cell. 2009;136 doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farazi TA, Juranek SA, Tuschl T. Development. 2008;135 doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 4.Nakayashiki H, Kadotani N, Mayama S. J Mol Evol. 2006;63 doi: 10.1007/s00239-005-0257-2. [DOI] [PubMed] [Google Scholar]
- 5.Laurie JD, Linning R, Bakkeren G. Curr Genet. 2008;53 doi: 10.1007/s00294-007-0165-7. [DOI] [PubMed] [Google Scholar]
- 6.Scannell DR, et al. Proc Natl Acad Sci U S A. 2007;104 [Google Scholar]
- 7.Axelson-Fisk M, Sunnerhagen P. Comparative Genomics: Using Fungi as Models. 2006;15 [Google Scholar]
- 8.Berman J, Sudbery PE. Nat Rev Genet. 2002;3 doi: 10.1038/nrg948. [DOI] [PubMed] [Google Scholar]
- 9.Hall TM. Structure. 2005;13 doi: 10.1016/j.str.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Grimson A, et al. Nature. 2008;455 [Google Scholar]
- 11.Lau NC, Lim LP, Weinstein EG, Bartel DP. Science. 2001;294 doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 12.Montgomery TA, et al. Cell. 2008;133 doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 13.Buhler M, Spies N, Bartel DP, Moazed D. Nat Struct Mol Biol. 2008;15 doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grewal SI, Jia S. Nat Rev Genet. 2007;8 doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 15.Materials and methods are available as supporting material on Science Online.
- 16.Lamontagne B, Larose S, Boulanger J, Elela SA. Curr Issues Mol Biol. 2001;3 [PubMed] [Google Scholar]
- 17.Astromskas E, Cohn M. Yeast. 2007;24 doi: 10.1002/yea.1488. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Nature. 2001;409 doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 19.MacRae IJ, Doudna JA. Curr Opin Struct Biol. 2007;17 doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Cell. 2004;118 [Google Scholar]
- 21.Liu Q, et al. Science. 2003;301 [Google Scholar]
- 22.Vazquez F, Gasciolli V, Crete P, Vaucheret H. Curr Biol. 2004;14 doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 23.Chendrimada TP, et al. Nature. 2005;436 doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lister R, et al. Cell. 2008;133 doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louis EJ, Haber JE. Genetics. 1992;131 doi: 10.1093/genetics/131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byrne KP, Wolfe KH. Genome Res. 2005;15 doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagalakshmi U, et al. Science. 2008;320 [Google Scholar]
- 28.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. Science. 2004;306 doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 29.Maiti M, Lee HC, Liu Y. Genes Dev. 2007;21 doi: 10.1101/gad.1497607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berretta J, Pinskaya M, Morillon A. Genes Dev. 2008;22 doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Genome Res. 1998;8 doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 32.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Science. 2009;324 doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garfinkel DJ, Mastrangelo MF, Sanders NJ, Shafer BK, Strathern JN. Genetics. 1988;120 doi: 10.1093/genetics/120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickner RB. Microbiol Rev. 1996;60 doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hedtke SM, Townsend TM, Hillis DM. Syst Biol. 2006;55 doi: 10.1080/10635150600697358. [DOI] [PubMed] [Google Scholar]
- 36.Fitzpatrick DA, Logue ME, Stajich JE, Butler G. BMC Evol Biol. 2006;6 doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuker M, Stiegler P. Nucleic Acids Res. 1981;9 doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sigova A, Rhind N, Zamore PD. Genes Dev. 2004;18 doi: 10.1101/gad.1218004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawakami K, et al. Genetics. 1993;135 doi: 10.1093/genetics/135.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.