Abstract
The Huntington’s disease (HD) mutation leads to a complex process of Huntingtin (Htt) aggregation into multimeric species that eventually form visible inclusions in cytoplasm, nuclei and neuronal processes. One hypothesis is that smaller, soluble forms of amyloid proteins confer toxic effects and contribute to early cell dysfunction. However, analysis of mutant Htt aggregation intermediates to identify conformers that may represent toxic forms of the protein and represent potential drug targets remains difficult. We performed a detailed analysis of aggregation conformers in multiple in vitro, cell and ex vivo models of HD. Conformation-specific antibodies were used to identify and characterize aggregation species, allowing assessment of multiple conformers present during the aggregation process. Using a series of assays together with these antibodies, several forms could be identified. Fibrillar oligomers, defined as having a β-sheet rich conformation, are observed in vitro using recombinant protein and in protein extracts from cells in culture or mouse brain and shown to be globular, soluble and non-sedimentable structures. Compounds previously described to modulate visible inclusion body formation and reduce toxicity in HD models were also tested and consistently found to alter the formation of fibrillar oligomers. Interestingly, these compounds did not alter the rate of visible inclusion formation, indicating that fibrillar oligomers are not necessarily the rate limiting step of inclusion body formation. Taken together, we provide insights into the structure and formation of mutant Htt fibrillar oligomers that are modulated by small molecules with protective potential in HD models.
Keywords: Huntington’s disease, Huntingtin protein, amyloid fibrils, Time-Lapsed Imaging, epigallocatechin-3-gallate, CEP-1347
INTRODUCTION
Huntington’s disease (HD), characterized by progressive movement abnormalities, psychiatric symptoms and cognitive deficits, is invariably fatal with no disease modifying treatment available. Aberrant accumulation and aggregation of mutant Huntingtin (Htt) are hallmarks of disease, representing an early therapeutic target for chemical compound screens. The tight threshold of above 40 glutamine repeats required for disease manifestation [1] can be functionally demonstrated by the propensity to form fibrils in vitro and in vivo [2, 3], implicating aggregation as a surrogate for the disease process. Aggregation was first demonstrated to be associated with HD by the discovery of intranuclear inclusions in both diseased mouse brain and human patient brain tissue [4, 5]. Further examination has suggested that inclusion bodies may initially reduce the levels of toxic soluble mutant Htt species and confer a lower risk of cell death in primary neurons [6]; therefore, specific SDS-soluble aggregation conformations may be linked to toxicity. Mechanistic studies of protein aggregation have led to a proposed common aggregation pathway in amyloid diseases [7-9] and current efforts seek to determine which species along this pathway are toxic to cells. Aggregation intermediates may be “on pathway” to form fibrils (e.g. protofibrils and soluble fibrils) or “off pathway” to form species such as prefibrillar and fibrillar oligomers (Figure 1) [9-12]. Prefibrillar oligomers have been previously defined as soluble, globular structures which do not stain with thioflavin-S [8]. As these oligomers preceded fibril formation and disappear upon the formation of mature fibrils, they therefore were termed ‘prefibrillar’ [13, 14], however later studies indicate that the formation of prefibrillar oligomers occurs through an independent pathway from the fibrillization pathway [15]. Annular protofibrils are a discrete class of pore-like structures which are thought to disrupt membrane permeability and are present in human AD brain samples. The pathway for annular protofibril formation is also independent of the pathway for fibril formation [16]. Fibrillar oligomers are morphologically similar yet immunogically distinct from prefibrillar oligomers and have a β-sheet rich conformation similar to that of fibrils [10], thus were named ‘fibrillar’ oligomers in spite of being spherical in nature. A previous study in AD described the increased presence of these fibrillar oligomers in patient brain tissue with levels of fibrillar oligomers correlating with cognitive decline. Prefibrillar oligomers and annular protofibrils were detected in brain tissue of both non-affected and affected individuals [17]. This supports the hypothesis that fibrillar oligomers are involved in the pathogenesis of amyloid diseases.
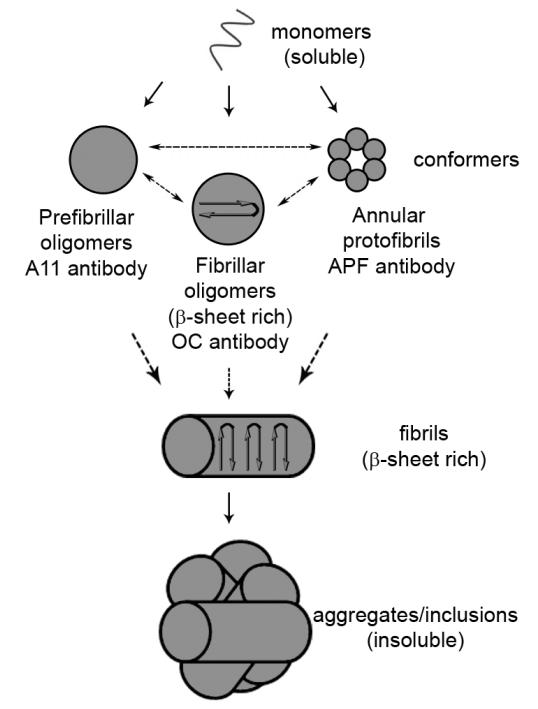
Figure 1. Schematic illustrating the possible conformers of aggregated mutant Htt and the conformation-specific antibodies that recognize the structures.
Misfolded monomers can assemble into multiple conformers of aggregated protein including prefibrillar oligomers, fibrillar oligomers and annular protofibrils. These structures are recognized by the A11 prefibrillar oligomer antibody, the OC fibrillar antibody and the APF annular protofibrils antibody, respectively. It is unknown if these conformers can convert into the other types of conformer, which is indicated by a dashed arrow between the various structures. Additionally, it is currently unknown which of these conformers is the precursor to fibril formation, also indicated by dashed arrows.
Antibodies that recognize specific conformations, such as prefibrillar oligomeric forms (A11) [8], annular protofibrillar forms (APF) [16] or fibrillar forms (OC) [10] (Figure 1) of amyloid proteins are valuable tools to define the presence of protein conformers based upon recognition of various amyloid structures, independent of disease protein sequence [8, 18]. This suggests that amyloid oligomers and soluble fibrils have a common structure that is distinct from other forms of the protein (e.g. monomers and visible aggregates). Additionally, a recent study showed that fibrillar oligomers seed the formation of more fibrillar oligomers, but not fibrils [12]. Therefore, fibrillar oligomers may have the same self-replicating capacity characteristic of prion proteins, supporting the concept that amyloid proteinopathies may involve prion-like mechanisms [19, 20] and that fibrillar oligomers may be toxic. Thus, it is important to develop a means of characterizing and detecting the modulation of fibrillar oligomers of amyloid forming proteins.
In this study we use biochemical approaches and conformation specific antibodies to visualize oligomeric species in multiple models of HD. As was previously shown for Aβ fibrillar oligomers [10, 12], we find that Htt fibrillar oligomers are globular in nature, soluble in SDS and that they are preferentially modified by aggregation modulators that have neuroprotective properties. Of note, altering levels of fibrillar oligomers did not alter the rate of visible aggregate formation, indicating that these species are not the rate limiting factor for inclusion body formation. This provides a unique and novel cell-based system for use in future studies to correlate the presence of fibrillar oligomers to toxicity in cells by modulating their levels using small molecules, thus providing alternative assays to inform our knowledge of the process of mutant Htt aggregation and in turn address drug-Htt interactions together with better pharmacodynamic readouts.
MATERIALS AND METHODS
Protein Purification
Glutathione S-transferase (GST)-HD fusion proteins were purified as described [21-23]. Cleavage of the GST moiety by a PreScission Protease (GE Healthcare) initiates aggregation. Fresh, unfrozen GST-HD fusion protein was used for each experiment. Solutions with fusion proteins were centrifuged at 20,000 X g for 30 min at 4°C to remove any preexisting aggregates before the addition of the PreScission protease.
Dot blot assay
The dot blot assay was performed as previously described [8, 15]. Briefly, 2 μL aliquots of each oligomerization reaction were applied onto a nitrocellulose membrane (Pierce). The membrane was blocked for 1 hour at room temperature with 10% nonfat milk in Tris-buffered saline containing 0.01% Tween 20 (TBS-T) and probed with A11 anti-prefibrillar oligomer antibody (kind gift from Dr. Glabe, 1:1500), OC anti-fibrillar oligomer antibody (kind gift from Dr. Glabe, 1:1500) or anti-Htt antibody (CAG53b, kind gift from Dr. Wanker, 1:1000). Peroxidase-conjugated AffiniPure goat anti-mouse secondary (Jackson ImmunoResearch Laboratories) was used at 1:5,000 for 1 h at room temperature. Blots were detected using ECL chemiluminescence detection reagent (Amersham).
SDS-AGE of PC12 Cell Homogenates
The analysis of mutant Htt fragments by SDS-AGE with Western analysis was performed as described [24-26]. The Htt14A2.6 PC12 line was generated and propagated as described [27]. This cell line expresses a truncated Htt peptide containing the first 17 amino acids and 103Qs fused in-frame to EGFP with a complete protein sequence of MATLEKLMKAFESLKSF(103Q)-EGFP. The cells were maintained in complete medium with continued selection: Dulbecco’s modified Eagle’s medium (5% glucose) with 10% horse serum, 5% fetal bovine serum and 200 mg/ml G418. Htt expression was induced with Ponasterone A (5 μM) (PA) for the indicated times. Three independent experiments were performed. At 24 and 48 h post-induction, cells were lysed in radioimmune precipitation assay buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA (from a concentrated stock at pH 8.0), 1% Nonidet P-40, 0.5% SDS) containing Complete Protease Inhibitor (Roche Diagnostics). A DC protein assay (Bio-Rad) was performed to determine protein concentration. Lysate (30 μg) was added in a 1:1 ratio to loading buffer (150 mM Tris, pH 6.8, 33% glycerol, 1.2% SDS) and loaded onto a 1% agarose gel containing 0.1% SDS and run until the dye front had migrated at least 12 cm to allow for maximum resolution of aggregates from the dye front. The proteins were then semidry-blotted (Owl HEP-1) onto a polyvinylidene difluoride membrane in transfer buffer (192 mM glycine, 25 mM Tris-base, 0.1% SDS, 15% MeOH). This blot was blocked for 1 hour in 5% bovine serum albumin in Tris-buffered saline containing 0.1% Tween 20 at room temperature. The blot was then probed with either OC anti-fibrillar oligomer antibody (Glabe, 1:1500), EM48 (Millipore, 1:1000) or EGFP (Clontech, 1:1000). Peroxidase-conjugated AffiniPure goat anti-mouse or goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories) was used at 1:50,000 for 1 h at room temperature. Blots were detected using PICO detection reagent (Pierce). The molecular mass standard used in SDS-AGE was ferritin (400 kDa).
SDS-AGE of R6/2 Mouse Brain Homogenates
At 8 weeks of age R6/2 and nontransgenic cortex were homogenized with 10 volumes of ice-cold sample buffer (100 mM Tris-HCl, pH 7.4, 150mM NaCl, and a protease inhibitor mixture) by a rotor/stator type tissue homogenizer and sonication with 10 pulses/min at 15% power. Samples were analyzed without centrifugation (total homogenates) and diluted 1:1 into non-reducing Laemmli sample buffer (150 mM Tris-HCl, pH 6.8, 33% glycerol, 1.2% SDS, and bromophenol blue). Bradford assays were used to determine protein concentration in the homogenates before the addition of Laemmli sample buffer. Total protein (50 μg) was loaded per SDS-AGE well. Mutant Htt oligomers were imaged after immunoreaction with OC and EM48 antibodies as described above or MAB2166 (Millipore, 1:1000).
Size-Exclusion Chromatography
6 μM recombinant Httex1Q53 was allowed to aggregate for 4 hours at 37°C. Samples were then clarified by centrifugation for 30 min at 20,000 x g and supernatant was injected and fractionated by Superdex200 10/300 size-exclusion column. The separation was performed at 4 °C with a flow rate of 0.5 mL/min in PBS. The elution was done with one column volume and fractions (500 μl per fraction) were applied to nitrocellulose membrane (0.1 μm) via slot blot manifold apparatus (Hoefer PR 648) without any washing steps or detergent treatment. Membranes were probed with OC anti-fibrillar oligomer antibody and EM48 anti-Htt antibody as described above. The amount of Htt conformers (OC or EM48) per fraction was quantified by densitometry of developed blots using NIH Image J. Protein standards were used to estimate size of Htt peaks. Void volume of elution was obtained with Blue dextran (Dead volume = V0).
Atomic Force Microscopy
6 μM recombinant Httex1Q53 was allowed to aggregate for 3 hours at 37°C and injected onto the Superdex200 size-exclusion column as above. Fractions 31, 47 and 81 were collected and deposited on freshly cleaved mica (Ted Pella Inc., Redding, CA) and allowed to sit for 30 s. The substrate was then washed with 200 μL of ultrapure water and dried under a gentle steam of air. Images were taken with silicon cantilevers (Veeco Instruments, Santa Barbara, CA) with a nominal spring constant of 40 N/m and resonance frequency of ~300 kHz. Typical imaging parameters were drive amplitude 150–500 kHz with set points of 0.7–0.8 V, scan frequencies of 2–4 Hz, image resolution 512 by 512 points, and a scan size of 2–10 μm.
Filter-Retardation Assay
The same lysates used in the SDS-AGE assays were analyzed by cellulose acetate filter-retardation assays. Lysate (30 μg) was diluted to 200 μL with 2% SDS and filtered through cellulose acetate membrane (Schleicher & Schuell, 0.2-μm pore size) with a Bio-Rad dot blot filtration unit. The assay was performed as described [2]. The blot was then probed with primary and secondary antibodies and developed as described above.
Chemical Compounds
Epigallocatechin gallate (EGCG) was purchased from Sigma-Aldrich (E4143). CEP-1347 was received as a gift from Cephalon, Inc.
Immunocytochemistry and Quantitation of Aggregates
Cells were grown on UV-treated cover slips to approximately 50% confluency and induced with 5 μM PA for 48h, fixed in 2% formaldehyde, and incubated for 2 min with 0.1% Triton X-100 in PBS. Fixed cells were 4′,6-diamidino-2-phenylindole stained. Fluorescent microscopy was performed on a Zeiss AxioObserver.Z1 microscope and Axiovision software. At least 500 cells were counted from five to six fields in three independent experiments for each data point at 10X magnification. Aggregation is expressed as the percentage of cells with aggregates versus total number of EGFP-positive cells.
Quantification of Western blots
Quantification was done by densitometry with ImageJ. Relative levels of mutant Htt were normalized to control samples treated with vehicle in all of the assays.
Time-lapse microscopy
14A2.6 cells were grown as described above and 0.5×106 cells were plated onto collagen-coated glass bottom microwell dishes (MatTek, P35G-1.5-14-C). 24 hours after plating, cells were induced with PA and allowed to equilibrate in the imaging system. The amount of equilibration time varied between experiments and was factored in to the time elapsed for analysis. Cells were then allowed to grow for 48 hours in a VivaView live cell imaging system (Olympus). Cells were imaged at 20X magnification every 10 minutes for 48 hours in 9 locations per well (to monitor sufficient cell numbers) in three independent experiments. Results are shown as the number of hours before the appearance of the first aggregate in that location, indicative of the ‘lag phase’ of Htt aggregation.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5.04 software. All data are expressed as mean ± SEM. P < 0.05 was considered to be statistically significant. Statistical comparisons of densitometry results were performed by one-way ANOVA followed by Bonferroni’s multiple comparison or Dunnett’s multiple comparison tests.
RESULTS
Fibrillar oligomers are detected in HD Models
Fibrillar oligomers have been identified in human Alzheimer’s disease (AD) brain tissue and the presence of these species as detected by a conformation specific antibody (OC) correlates with symptoms of cognitive dysfunction [10, 17], suggesting disease relevance. Htt forms soluble oligomers and fibrillar structures both in vitro and in vivo in a polyglutamine dependent manner [9, 21, 22, 25], however the presence of fibrillar oligomers has not been systematically evaluated. Therefore, we investigated whether these structures are also present in models of HD and characterized their structural properties.
To begin to evaluate the presence of oligomeric species, aggregation of purified GST-Httex1Q20 and GST-Httex1Q53 (2.5μM) was initiated by incubation with a site-specific protease then allowed to proceed at 37C for 144 h. Aliquots were removed beginning at 48 hours post-initiation and at subsequent 24 hour time points up to 144 hours. Samples were spotted onto a nitrocellulose membrane and then probed as a Western blot with conformation specific and anti-Htt antibodies (Fig 2A-C). The prefibrillar oligomeric antibody (A11) was raised against a molecular mimic of soluble oligomeric Aβ40 and was found to be specific for prefibrillar oligomers of a number of amyloid proteins including Aβ, α-synuclein and polyglutamine [8]. The fibrillar oligomeric antibody (OC) was generated by immunization with homogeneous Aβ42 fibrils and specifically recognized fibrils, but not random coil monomer or prefibrillar (A11 positive) oligomers of Aβ. Additionally, OC recognizes 100,000 × g soluble fibrillar oligomers which are immunologically distinct from prefibrillar oligomers recognized by A11 [10]. At 144 hours, prefibrillar oligomers were detected with the A11 antibody in both the 20Q and 53Q samples (Fig. 2A). At this same time point, fibrillar oligomers are only detected in the 53Q samples using the OC fibrillar oligomer antibody (Fig. 2B), reflecting polyglutamine repeat-length dependence. All of the samples show Htt reactivity with the N-terminal Htt antibody CAG53b (Fig. 2C) [4]. These results indicate that recombinant Httex1p can form prefibrillar and fibrillar oligomers as defined by reactivity to antibodies specific for these conformations in vitro.
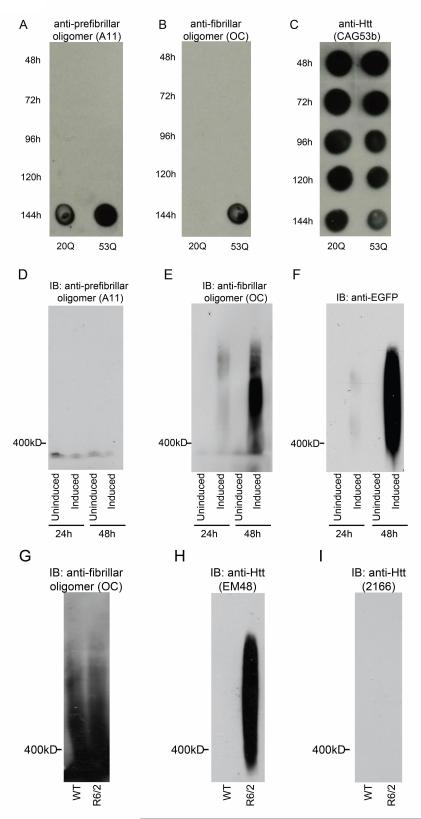
Figure 2. Fibrillar oligomers are present in multiple models of HD.
A-C, Recombinant Httex1Q20 and Httex1Q53 were allowed to aggregate at 37C for 144h. Aliquots were spotted onto nitrocellulose at times indicated. Blots were probed with A, A11 anti-prefibrillar oligomer antibody B, OC anti-fibrillar oligomer antibody and C, CAG53b anti-Htt antibody. D-F, Htt expression was induced in PC12 cells stably expressing truncated Httex1p. At 24 and 48h post-induction cells were lysed and the lysates were analyzed by SDS-AGE. Cells without induction of Htt expression were used as negative controls. Blots were probed with D, A11 anti-prefibrillar oligomer antibody, E, OC anti-fibrillar oligomer antibody and F, anti-GFP antibody. G-I, Striatal brain tissue homogenates from 8 week old non-transgenic and R6/2 transgenic mice were analyzed by SDS-AGE. Blots were probed with G, OC anti-fibrillar oligomer antibody, H, EM48 anti-Htt antibody and I, MAB2166 anti-C-terminal Htt antibody.
We next examined cellular models of HD for the presence of aggregation conformers using SDS-AGE gels to visualize high molecular weight aggregated Htt [24-26]. SDS-AGE uses low amounts of SDS and no reducing agent, which allows the visualization of large (>400kD), SDS-soluble, oligomeric species. Lysates from PC12 cells stably expressing inducible, truncated Httex1p with 103Q fused to EGFP were first analyzed. The A11 prefibrillar oligomer antibody did not show any reactivity with SDS-AGE analysis (Fig. 2D). High molecular weight species were detected upon mutant Httex1p induced expression, showing reactivity with both the OC fibrillar oligomer antibody (Fig. 2E) and EGFP antibody for detection of tagged Htt (Fig. 2F). The presence of the OC-reactive species increases over time from 24 to 48 hours post-induction, which reflects optimal expression [27], indicating that fibrillar oligomers are present in this cell model of HD and accumulate in a dose-dependent manner. Additionally, we have previously shown that oligomers resolved by SDS-AGE do not react with the 3B5H10 monoclonal antibody which appears to recognize monomeric or very small (<400kD) oligomeric form of Htt based upon the inability to resolve these reactive species from the dye front [26]. Taken together, these data suggest that the β-sheet rich, OC-positive fibrillar oligomeric species are resolved by SDS-AGE analysis.
Finally, we investigated whether these same species are present in brain tissue dissected from R6/2 mice to determine if fibrillar oligomers can be detected ex vivo. Striatal tissue from 8 week old R6/2 transgenic mice expressing a human Htt exon 1 transgene [28] containing ~200 Qs (Fig. 2G-I), a stage when visible aggregation can be observed, was analyzed and showed that oligomeric Htt of a similar size to the fibrillar oligomers from cell lysates could only be detected in the R6/2 mice and not in non-transgenic control lysates (Fig. 2H) using Htt antibody (EM48). OC oligomer antibody reactivity to mutant Htt was not detected preferentially in R6/2 mouse tissue, potentially masked by the presence of endogenous protein recognized by the antibody that is independent of mutant Htt (Fig. 2G). The OC antibody is not sequence specific, therefore this may reflect the presence of a large immunoreactive protein in the striatal samples which adopts a β-sheet rich conformation recognized by this antibody. This protein is not endogenous Htt, as there is no reactivity with the C-terminal Htt antibody MAB2166 (Fig. 2I). Given our previous findings that oligomers of protein from recombinant systems, cell lysates and transgenic mice show similar resolution patterns on SDS-AGE gels [25] and that the only conformer we see resolved by SDS-AGE is the OC-positive fibrillar oligomer; the EM48-positive oligomers resolved by SDS-AGE from the R6/2 mouse samples likely represent fibrillar oligomeric species.
Huntingtin fibrillar oligomers are SDS-soluble globular structures
Oligomers are resolved by SDS-AGE; however these species could represent SDS insoluble or soluble conformers. To determine whether Htt fibrillar oligomers are SDS-soluble, filter-retardation assays were performed using lysates from stably expressing PC12 cells (Fig. 3A,B). Samples were boiled in 2% SDS to ensure that all SDS soluble species were dissolved and then applied to a cellulose acetate membrane and probed with OC fibrillar oligomer (Fig. 3A) or EGFP antibodies (Fig. 3B). This assay detects all SDS-insoluble material greater or equal to the pore size (0.2μm) of the membrane [2]. Since no reactivity is detected using the oligomer antibody in lysates, we conclude that the OC-positive fibrillar oligomers using SDS-AGE analysis are SDS-soluble. Further, although the OC antibody can recognize both oligomers and fibrils, our previous data showed that the oligomers resolved by SDS-AGE are non-sedimentable at 20,000 x g, indicating that these SDS-soluble species are fibrillar oligomers but are not fibrils [25], as defined by sedimentation analysis.
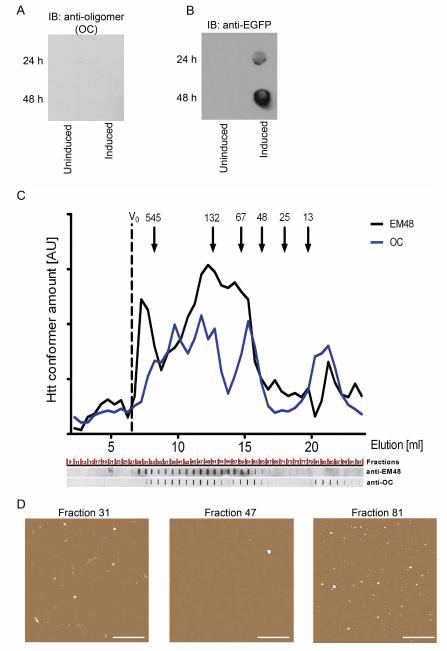
Figure 3. Fibrillar oligomers are SDS-soluble globular structures.
A-B, Htt expression was induced in PC12 cells stably expressing truncated Httex1p. At 24 and 48h post-induction cells were lysed and the lysates were analyzed by filter-retardation assay. Cells without induction of Htt expression were used as negative controls. Blots were probed with A, OC anti-fibrillar oligomer antibody and B, anti-GFP antibody. C, Recombinant Httex1Q53 was allowed to aggregate at 37C for 4h and then separated on a size-exclusion column. Fractions were spotted onto nitrocellulose and probed with either OC anti-fibrillar oligomer antibody or EM48 anti-Htt antibody. D, Fractions corresponding to EM48 reactivity only (Fraction 31), both EM48 and OC reactivity (Fraction 47) and OC reactivity only (Fraction 81) were analyzed by AFM. Scale bar = 1μm.
In order to define the SDS soluble structures recognized by the conformation-specific antibody OC, size-exclusion chromatography was performed on aggregated recombinant Httex1Q53. 6μM Httex1Q53 was allowed to aggregate for 3 h at 37°C, which allows the formation of oligomers but not larger aggregates [22]. The sample was injected onto a Superdex200 size-exclusion column and fractions collected and spotted onto a nitrocellulose membrane, and then probed with OC anti-fibrillar oligomer antibody and EM48 anti-Htt antibody (Fig 3C). Similar to findings in cell lysates on SDS-AGE gels, overlapping reactivity was observed depending on the antibody used. EM48, which recognizes aggregated Htt, reacted with species in fraction 31, corresponding to the largest structure, both antibodies recognized fraction 47 and the species in fraction 81 demonstrated greater reactivity with the OC anti-fibrillar oligomer antibody. The medium-sized structures eluting in fraction 47 are recognized by both the anti-Htt and the OC antibody and are likely to be the structures resolved on the SDS-AGE gels given that the species on the gels are also recognized by both antibodies. The final OC reactive peak beginning at fraction 80 on the chromatogram eluted at a much lower apparent molecular weight than expected. We hypothesize that the fibrillar oligomers interacted non-specifically with the column matrix which influenced the retention time of the oligomers. The low level of EM48 reactivity to this later fraction indicates that it is likely Htt, but that the EM48 epitope might not be as accessible as in the other fractions. To evaluate the morphologies of the eluted species, fractions 31, 47 and 81 were imaged on mica using atomic force microscopy (AFM) (Fig. 3D). AFM images reveal mostly large fibril-like structures estimated to be 100-200nm in length in fraction 31. Fractions 47 and 81 have only globular structures present which are similar to those seen with fibrillar oligomers of Aβ [12]. These findings indicate that the structures recognized by the OC anti-fibrillar oligomer antibody are globular in nature, despite having the β-sheet rich structure recognized by the OC antibody which is typically associated with fibrillar structures.
Green tea (-)-epigallocatechin-gallate decreases fibrillar oligomers and SDS-insoluble Htt in a stably-expressing truncated Httex1p model of HD
Given that fibrillar oligomeric species are present in several HD models, we next asked whether formation of this species is a dynamic process that can be altered by small molecules that change visible inclusion formation. Small molecules have been identified in various types of aggregation screens that select for modulation of visible aggregate formation and reduced neurotoxicity sin HD models [27, 29-31]. However, using visible aggregates as a measure may be insufficient to identify alterations in the aggregation process and may represent a less informative outcome measure for therapeutic intervention.
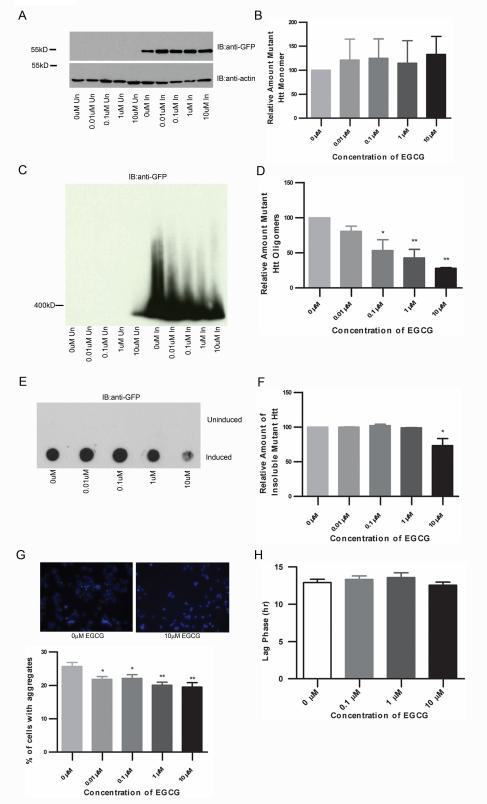
Epigallocatechin gallate (EGCG) is an antioxidant in green tea that inhibits the formation of insoluble aggregates of recombinant Htt in vitro and reduces toxicity and visible aggregate load in a yeast model of HD [32]. EGCG also reduces the formation of small Htt oligomers by promoting the formation of larger ones visualized by AFM analysis [32]. ECGC was therefore tested for modulation of fibrillar oligomers in inducible PC12 cells. While the relative amount of monomeric Htt does not appear to change with EGCG treatment (one-way ANOVA: F= 0.1567, P=0.9545) (Fig. 4A-B), there is a dose-dependent decrease in the level of mutant Htt oligomers as determined by SDS-AGE analysis (one-way ANOVA: F= 9.551, P=0.0019) (Fig. 4C-D). EGCG significantly decreases the amount of SDS-insoluble mutant Htt (one-way ANOVA: F= 6.623, P=0.03) (Fig. 4E-F) and as previously shown, also decreases the number of visible inclusions in the 14A2.6 cells (one-way ANOVA: F= 4.579, P=0.0019) (Fig. 4G, supplemental figure 1). Relative levels of monomeric, oligomeric and insoluble Htt were determined by densitometry of blots probed with anti-EGFP antibody.
Figure 4. EGCG decreases oligomers and SDS-insoluble Htt in a stably expressing truncated Httex1p model of HD.
A-F, 14A2.6 cells were treated with EGCG at the concentrations listed for 48 hours. Cells were lysed and analyzed for different conformations of mutant Htt. A-B, SDS-PAGE analysis of lysates shows no significant difference in monomeric Htt (one-way ANOVA: F= 0.1567, P=0.9545). C-D, There is a dose-dependent decrease in the level of mutant Htt oligomers (one-way ANOVA: F= 9.551, P=0.0019; Dunnett’s multiple comparison test: 0μM vs. 0.01μM P=n.s., 0μM vs. 0.1μM P<0.05, 0μM vs. 1μM P<0.01, 0μM vs. 10μM P<0.01). E-F, EGCG treatment decreases the amount of SDS-insoluble mutant Htt (one-way ANOVA: F= 6.623, P=0.03; Dunnett’s multiple comparison test: 0μM vs. 0.01μM P=n.s., 0μM vs. 0.1μM P=n.s., 0μM vs. 1μM P=n.s., 0μM vs. 10μM P<0.05). G, 14A2.6 cells were treated with EGCG at the concentrations listed for 48 hours. Cells were then fixed and the number of cells containing aggregates was counted by fluorescence microscopy. EGCG treatment significantly decreases the percentage of cells containing at least one visible aggregate (one-way ANOVA: F= 4.579, P=0.0019; Dunnett’s multiple comparison test: 0μM vs. 0.01μM P<0.05, 0μM vs. 0.1μM P<0.05, 0μM vs. 1μM P<0.01, 0μM vs. 10μM P<0.01). H, EGCG treatment does not appear to affect aggregation kinetics (one-way ANOVA: F= 0.8071, P=0.4927).
It is possible that the fibrillar oligomers are the ‘seeds’ of inclusion body formation and therefore the rate limiting step of inclusion body formation. To test this hypothesis, we utilized live-cell imaging to monitor inclusion body formation in real time and treated the cells with EGCG to decrease the levels of fibrillar oligomers (supplementary video file 2). We find that EGCG treatment does not alter the kinetics of visible aggregate formation (one-way ANOVA: F= 0.8071, P=0.4927) (Fig. 4H), suggesting that reduced oligomeric species does not affect the kinetics of inclusion body formation.
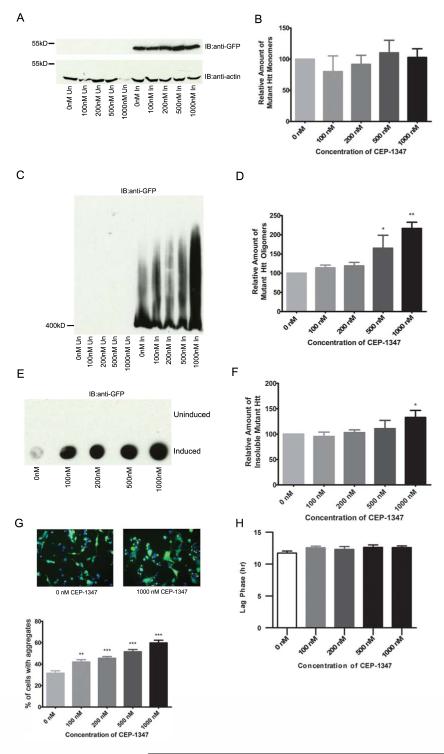
CEP-1347 increases fibrillar oligomers and SDS-insoluble Htt in a stably-expressing truncated Httex1p model of HD
To determine if fibrillar oligomers are also modulated by a compound that increases visible inclusion formation, CEP-1347 was tested. This small molecule is an inhibitor of Mixed Lineage Kinases (MLKs), including MLK1, MLK2, MLK3, and dual leucine zipper kinase (DLK) [33, 34], which activate JNK and induce apoptosis [35, 36]. CEP-1347 is protective against cell based expanded repeat Htt phenotypes as well as in Drosophila and R6/2 mouse models of the disease [37]. The mechanism of protection from CEP-1347 appears to involve coordinated regulation of multiple signaling pathways and altered aggregation. Relevant to the studies presented here, CEP-1347 increases the number of cells containing visible aggregates in inducible PC12 cells [37], therefore the effect on fibrillar oligomers was determined. The amount of monomeric Htt is not altered with CEP-1347 treatment (one-way ANOVA: F= 0.4630, P=0.7622) (Fig. 5A-B), however the levels of fibrillar oligomers significantly increased in a dose-dependent manner (one-way ANOVA: F= 7.096, P=0.0056) (Fig. 5C-D). The amount of SDS-insoluble Htt also increases with CEP-1347 treatment (one-way ANOVA: F= 3.195, P=0.0415) (Fig. 5E-F) and as previously described, the percentage of cells containing visible aggregates is significantly increased (one-way ANOVA: F= 24.75, P=0.0019) (Fig. 5G, supplemental figure 3). Relative levels of monomeric, oligomeric and insoluble Htt were determined by densitometry. Similar to EGCG, treatment with CEP-1347 does not appear to affect aggregation kinetics (one-way ANOVA: F= 1.289, P=0.2775) (Fig. 5H quantitation, video file, supplemental video file 4). This indicates that fibrillar oligomers are not the rate limiting step of inclusion body formation, as altering the levels of fibrillar oligomers does not alter the rate of inclusion body formation.
Figure 5. CEP-1347 increases oligomers and SDS-insoluble Htt in a stably expressing truncated Httex1p model of HD.
A-F, 14A2.6 cells were treated with CEP-1347 at the concentrations listed for 48 hours. Cells were lysed and analyzed for different conformations of mutant Htt. A-B, SDS-PAGE analysis of lysates shows no significant difference in monomeric Htt (one-way ANOVA: F= 0.4630, P=0.7622) C-D, There is a dose-dependent increase in the level of mutant Htt oligomers (one-way ANOVA: F= 7.096, P=0.0056; Dunnett’s multiple comparison test: 0nM vs. 100nM P=n.s., 0nM vs. 200nM P=n.s., 0nM vs. 500nM P<0.05, 0nM vs. 1000nM P<0.01). E-F, CEP-1347 treatment appears to increase the amount of insoluble Htt by filter-retardation assay (one-way ANOVA: F=3.195, P=0.0415). G, 14A2.6 cells were treated with CEP-1347 at the concentrations listed for 48 hours. Cells were then fixed and the number of cells containing aggregates was counted by fluorescence microscopy. CEP-1347 treatment significantly increases the percentage of cells containing at least one visible aggregate (one-way ANOVA: F= 24.75, P=0.0019; Dunnett’s multiple comparison test: 0nM vs. 100nM P<0.01, 0nM vs. 200nM P<0.001, 0nM vs. 500nM P<0.001, 0nM vs. 1000nM P<0.001) H, Treatment with CEP-1347 does not appear to affect aggregation kinetics (one-way ANOVA: F= 1.289, P=0.2775).
DISCUSSION
HD is an amyloid disease caused by an abnormal expansion of polyglutamine in the Htt protein. Similar to Alzheimer’s disease, predictions have been made that soluble forms of amyloid in HD may confer toxicity [6]. To expand our knowledge of this possibility, a detailed analysis of aggregation conformers in multiple models of HD was performed and the structure of Htt fibrillar oligomers characterized in order to determine levels and sizes of these species which accrue in HD.
Characterization of oligomeric species
In this study, we show that an expanded polyQ mutant Htt fragment that causes mutant Htt-mediated aggregation and cellular toxicity [27] forms SDS-soluble fibrillar oligomers of greater than 400kD in a cellular model of HD. The Htt fibrillar oligomers react to the OC anti-fibrillar oligomer antibody, are globular structures and are soluble in SDS.
Recombinant fragments of Htt (e.g.Httex1Q53) can not only form prefibrillar oligomers, as shown previously [21], but fibrillar oligomers as well. It is known that recombinant Httex1Q44 can form globular and protofibrillar aggregation conformers that are rich in β-structure [9]. Our results indicate that these β-rich conformers are likely fibrillar oligomers, which are implicated in pathogenesis of neurodegenerative diseases [17]. We have also developed an assay to quantify the relative levels of fibrillar oligomers utilizing the SDS-AGE technique in assays to monitor modulation of these species. This species was identified in lysates from an inducible stably-expressing truncated Httex1p-GFP cell model, which is relevant as this cell line has been used to screen for chemical inhibitors of aggregation [27, 38]. Therefore, SDS-AGE gel assays can be used to analyze lysates from this cell line which had been treated with aggregation modulators to identify desirable outcomes for compound screens.
We verified that oligomers of the same size as those present in cell lysates are detected in striatal tissue of a mouse model of HD. Unfortunately; the OC anti-fibrillar oligomer antibody recognized a species independent of the presence of mutant Htt even though oligomers of Htt could only be detected in the transgenic samples and not the wild-type samples. Because the OC antibody is not sequence specific, it is possible that a large protein present in the striatal samples adopts a β-sheet rich conformation that can be recognized by this antibody, masking any signal from the Htt oligomers. This protein is not endogenous Htt as there was no reactivity with MAB2166, an antibody that recognizes a C-terminal region of Htt and will recognize endogenous Htt. Given the similarity between oligomers originating from recombinant protein, cell models and mouse brain tissue [25]; and since other types of aggregation conformers (i.e. β-rich monomers, prefibrillar oligomers and annular protofibrils) were not detected by antibodies specific for those conformations, we conclude that the species resolved by SDS-AGE are likely fibrillar oligomers. As such, we can utilize antibodies against the Htt protein or epitope tags as a surrogate for OC antibody reactivity.
OC reactivity has been shown for fibrillar oligomers as well as mature fibrils [10]. Therefore, it was important to establish that the species resolved on the SDS-AGE assay were in fact oligomeric. The globular structures of Htt recognized by the OC anti-fibrillar oligomer antibody are similar to the structure of Aβ fibrillar oligomers [12]. The data presented indicates that the structures recognized by the OC anti-oligomer antibody are globular in nature despite having the β-sheet rich structure typically associated with fibrillar structures. We established that OC-positive species from cell lysates are soluble in SDS as they are not retained by filter-retardation assay. Additionally, the oligomers were not altered after sedimentation of the fibrillar material [25]. Together, these results indicate that the OC-positive species resolved by SDS-AGE are soluble fibrillar oligomers and not mature fibrils.
Pharmacologic modifiers of aggregation
To evaluate whether the fibrillar oligomers defined in the various assays are also modulated by small molecules previously established to alter aggregation, EGCG and CEP-1347 were tested and showed decreased and increased levels of fibrillar oligomers, respectively. EGCG has previously been shown to reduce expanded repeat Htt aggregation [32] and to reduce the formation of Htt prefibrillar (A11-positive) oligomers by promoting formation of larger ones [32]. The larger oligomers are likely “off-pathway” as EGCG has been shown to refold amyloidogenic polypeptides into unstructured, “off-pathway” oligomers [39]. These “off-pathway” oligomers are not likely to be fibrillar oligomers as fibrillar oligomers have a defined structure and decrease in quantity with EGCG treatment whereas the “off-pathway” oligomers increased in quantity with EGCG. The data here indicates that EGCG reduces fibrillar oligomers and inclusion body formation in a cell model of HD. Both of these outcomes could be beneficial in the treatment of HD and indicate that fibrillar oligomers may track with pathogenesis.
EGCG also appears to decrease the amount of visible aggregate formation without altering the ‘seeding’ which is thought to trigger the polymerization into inclusion bodies [40, 41]. This is intriguing as one might expect that a decrease in visible aggregate formation could result from a decrease in the formation of the aggregation ‘seeds’. These results suggest that increasing the formation of ‘off-pathway’ oligomers could shift the equilibrium away from inclusion body formation, but that this shift does not prevent the formation of the ‘seed’ that leads to aggregate formation. This result also suggests that fibrillar oligomers are not the aggregation ‘seeds’ as EGCG reduces the presence of fibrillar oligomers but did not alter the kinetics of aggregation, which correlates to previous work done with Aβ showing that fibrillar oligomers did not seed the nucleation of Aβ fibrils [12]. This is consistent with additional compound testing with other aggregation inhibitors (e.g. methylene blue and cystamine, unpublished data in review and supplementary figure 5, respectively), which also reduce the presence of fibrillar oligomers and visible aggregates but do not affect the rate of visible inclusion formation. Further characterization of the interaction between off-pathway species and aggregation conformers will be necessary to draw firm conclusions regarding this interaction.
CEP-1347 increased the number of cells containing visible aggregates in a clone of stably-expressing expanded repeat Htt polypeptide in PC12 cells [37] and increased levels of fibrillar oligomers in this line. This finding was surprising given that if fibrillar oligomers are a toxic aggregation intermediate, one might anticipate that an increase in this species would result in an increase in toxicity. However; it is possible that an increase in inclusion body formation is in some way reducing the toxicity of the fibrillar oligomers in this mode or that the effects of CEP-1347 on signal transduction pathways and apoptosis [35-37] could be a confound by preventing mutant Htt-mediated cell death in spite of an accumulation of these fibrillar oligomers. Additionally, CEP-1347 is increasing visible aggregate formation without increasing the rate of aggregation. This indicates that CEP-1347 might be able to stabilize the aggregate seed once it has formed, allowing for the formation of inclusion bodies in a higher percentage of cells. This could shift the equilibrium away from a toxic intermediate toward the formation of inclusion bodies resulting in a protective effect in cells. Ultimately, given the complexity of potential mechanisms of CEP-1347, it is difficult to ascertain how significant the increase of oligomers levels is to disease state. Future investigation into more specific compounds will lead to a more refined understanding of the role of fibrillar oligomers in disease progression.
In summary, we provide evidence that mutant Htt forms OC-positive fibrillar oligomers in various models of HD. We establish that the relative levels of fibrillar oligomers can be monitored using SDS-AGE analysis. The results from the pilot sampling of compounds that increase or decrease visible aggregation are consistent with the idea that modulation of fibrillar oligomers tracks with neuroprotective effects in various assays. Testing the effect of a greater number of small molecules on levels fibrillar oligomers is required to ascertain the exact correlation between fibrillar oligomers and toxicity. Of several aggregation inhibitors tested (EGCG as described and others including cystamine and methylene blue, not shown), each showed reduction of fibrillar oligomers. For the one compound that can increase inclusion formation (CEP-1347, as described), fibrillar oligomer formation was also increased, suggesting that these types of assays can be used to evaluate specific conformers to uncover optimal therapeutic targets.
Of particular note in this study is the development of a battery of assays that allow for the ability to query not only one readout, such as inclusion body formation, but to instead assess all intermediate forms (monomeric, oligomeric and insoluble forms), as well as the rate at which they form, in a variety of models. While we have presented data here showing oligomer and SDS-insoluble Htt levels moving in the same direction, levels of each can be altered independently [25], suggesting that there is not one single readout for identifying therapeutic potential. As such, utilizing a combinatorial approach that allows for the identification of modulation of all forms of Htt is critical for the advancement of treatments. Furthermore, while these data have focused on models of Htt aggregation, the same techniques can be employed in all forms of proteinopathies (e.g. Alzheimer’s and Parkinson’s diseases) to screen for modulation of aggregation conformers, thereby increasing our understanding of disease mechanisms for multiple disease states.
Supplementary Material
Supplemental figure 1. Representative data for EGCG treatment. A-E, Representative images used to count visible inclusions in 14A2.6 cells treated with EGCG.
Supplemental figure 3. Representative data for CEP-1347 treatment. A-E, Representative images used to count visible inclusions in 14A2.6 cells treated with CEP-1347.
Supplemental figure 4. Representative video illustrating the lag phase of inclusion body formation in 14A2.6 cells treated with CEP-1347. Treatment with CEP-1347 does not appear to affect the rate of inclusion body formation.
Supplemental figue 2. Representative video illustrating the lag phase of inclusion body formation in 14A2.6 cells treated with EGCG. Treatment with EGCG does not appear to affect the rate of inclusion body formation.
Supplemental figure 5. Cystamine decreases oligomers and SDS-insoluble Htt in a stably expressing truncated Httex1p model of HD. A, Cystamine appears to decrease the levels of soluble fibrillar oligomers in 14A2.6 cells by SDS-AGE analysis. B, Cystamine decreases the amount of SDS-insoluble Htt in 14A2.6 cells by filter-retardation assay.
ACKNOWLEDGMENTS
We acknowledge Jack Riedling for insightful discussions and editorial assistance in the preparation of this manuscript. We would also like to thank Gillian Bates for the kind gift of the R6/2 and control mouse tissue. Funding was provided by the Hereditary Disease Foundation (LMT), Huntington’s Disease Society of America (LMT), CHDI, Inc. (LMT), NIH R01NS054753 (PJM) and NS52789 (LMT), CIRM ETA II award TR2-01767 (BJC) and NIH5600672751 (subaward LMT).
REFERENCES
- [1].Gusella JF, MacDonald ME. Huntington’s disease. Semin Cell Biol. 1995;6(1):21–8. doi: 10.1016/1043-4682(95)90011-x. [DOI] [PubMed] [Google Scholar]
- [2].Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90(3):549–58. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- [3].Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99(16):10417–22. doi: 10.1073/pnas.152161099. Epub 2002/07/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90(3):537–48. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- [5].DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–3. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- [6].Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–10. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- [7].Glabe CG. Conformation-dependent antibodies target diseases of protein misfolding. Trends Biochem Sci. 2004;29(10):542–7. doi: 10.1016/j.tibs.2004.08.009. [DOI] [PubMed] [Google Scholar]
- [8].Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–9. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- [9].Poirier MA, Li H, Macosko J, Cai S, Amzel M, Ross CA. Huntingtin spheroids and protofibrils as precursors in polyglutamine fibrilization. J Biol Chem. 2002;277(43):41032–7. doi: 10.1074/jbc.M205809200. Epub 2002/08/13. [DOI] [PubMed] [Google Scholar]
- [10].Kayed R, Head E, Sarsoza F, Saing T, Cotman CW, Necula M, et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2(1):18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sathasivam K, Lane A, Legleiter J, Warley A, Woodman B, Finkbeiner S, et al. Identical oligomeric and fibrillar structures captured from the brains of R6/2 and knock-in mouse models of Huntington’s disease. Hum Mol Genet. 2010;19(1):65–78. doi: 10.1093/hmg/ddp467. Epub 2009/10/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wu JW, Breydo L, Isas JM, Lee J, Kuznetsov YG, Langen R, et al. Fibrillar oligomers nucleate the oligomerization of monomeric amyloid beta but do not seed fibril formation. J Biol Chem. 2010;285(9):6071–9. doi: 10.1074/jbc.M109.069542. Epub 2009/12/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Baglioni S, Casamenti F, Bucciantini M, Luheshi LM, Taddei N, Chiti F, et al. Prefibrillar amyloid aggregates could be generic toxins in higher organisms. J Neurosci. 2006;26(31):8160–7. doi: 10.1523/JNEUROSCI.4809-05.2006. Epub 2006/08/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Harper JD, Wong SS, Lieber CM, Lansbury PT. Observation of metastable Abeta amyloid protofibrils by atomic force microscopy. Chemistry & biology. 1997;4(2):119–25. doi: 10.1016/s1074-5521(97)90255-6. Epub 1997/02/01. [DOI] [PubMed] [Google Scholar]
- [15].Necula M, Kayed R, Milton S, Glabe CG. Small molecule inhibitors of aggregation indicate that amyloid beta oligomerization and fibrillization pathways are independent and distinct. J Biol Chem. 2007;282(14):10311–24. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- [16].Lasagna-Reeves CA, Glabe CG, Kayed R. Amyloid-beta annular protofibrils evade fibrillar fate in Alzheimer disease brain. J Biol Chem. 2011;286(25):22122–30. doi: 10.1074/jbc.M111.236257. Epub 2011/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tomic JL, Pensalfini A, Head E, Glabe CG. Soluble fibrillar oligomer levels are elevated in Alzheimer’s disease brain and correlate with cognitive dysfunction. Neurobiol Dis. 2009;35(3):352–8. doi: 10.1016/j.nbd.2009.05.024. Epub 2009/06/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kayed R, Glabe CG. Conformation-dependent anti-amyloid oligomer antibodies. Methods Enzymol. 2006;413:326–44. doi: 10.1016/S0076-6879(06)13017-7. [DOI] [PubMed] [Google Scholar]
- [19].Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nature reviews Neuroscience. 2010;11(3):155–9. doi: 10.1038/nrn2786. Epub 2009/12/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Brundin P, Melki R, Kopito R. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol. 2010;11(4):301–7. doi: 10.1038/nrm2873. Epub 2010/03/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lotz GP, Legleiter J, Aron R, Mitchell EJ, Huang SY, Ng C, et al. Hsp70 and Hsp40 functionally interact with soluble mutant huntingtin oligomers in a classic ATP-dependent reaction cycle. J Biol Chem. 2010;285(49):38183–93. doi: 10.1074/jbc.M110.160218. Epub 2010/09/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat Struct Mol Biol. 2004;11(12):1215–22. doi: 10.1038/nsmb860. Epub 2004/11/16. [DOI] [PubMed] [Google Scholar]
- [23].Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci U S A. 2000;97(14):7841–6. doi: 10.1073/pnas.140202897. Epub 2000/06/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Weiss A, Klein C, Woodman B, Sathasivam K, Bibel M, Regulier E, et al. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington’s disease. J Neurochem. 2008;104(3):846–58. doi: 10.1111/j.1471-4159.2007.05032.x. [DOI] [PubMed] [Google Scholar]
- [25].Legleiter J, Mitchell E, Lotz GP, Sapp E, Ng C, DiFiglia M, et al. Mutant huntingtin fragments form oligomers in a polyglutamine length-dependent manner in vitro and in vivo. J Biol Chem. 2010;285(19):14777–90. doi: 10.1074/jbc.M109.093708. Epub 2010/03/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Miller J, Arrasate M, Brooks E, Libeu CP, Legleiter J, Hatters D, et al. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nature chemical biology. 2011;7(12):925–34. doi: 10.1038/nchembio.694. Epub 2011/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Apostol BL, Kazantsev A, Raffioni S, Illes K, Pallos J, Bodai L, et al. A cell-based assay for aggregation inhibitors as therapeutics of polyglutamine-repeat disease and validation in Drosophila. Proc Natl Acad Sci U S A. 2003;100(10):5950–5. doi: 10.1073/pnas.2628045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87(3):493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- [29].Agrawal N, Pallos J, Slepko N, Apostol BL, Bodai L, Chang LW, et al. Identification of combinatorial drug regimens for treatment of Huntington’s disease using Drosophila. Proc Natl Acad Sci U S A. 2005;102(10):3777–81. doi: 10.1073/pnas.0500055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Apostol BL, Illes K, Pallos J, Bodai L, Wu J, Strand A, et al. Mutant huntingtin alters MAPK signaling pathways in PC12 and striatal cells: ERK1/2 protects against mutant huntingtin-associated toxicity. Hum Mol Genet. 2006;15(2):273–85. doi: 10.1093/hmg/ddi443. [DOI] [PubMed] [Google Scholar]
- [31].Zhang X, Smith DL, Meriin AB, Engemann S, Russel DE, Roark M, et al. A potent small molecule inhibits polyglutamine aggregation in Huntington’s disease neurons and suppresses neurodegeneration in vivo. Proc Natl Acad Sci U S A. 2005;102(3):892–7. doi: 10.1073/pnas.0408936102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ehrnhoefer DE, Duennwald M, Markovic P, Wacker JL, Engemann S, Roark M, et al. Green tea (-)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington’s disease models 10.1093/hmg/ddl210. Hum Mol Genet. 2006:ddl210. doi: 10.1093/hmg/ddl210. [DOI] [PubMed] [Google Scholar]
- [33].Maroney AC, Glicksman MA, Basma AN, Walton KM, Knight E, Jr., Murphy CA, et al. Motoneuron apoptosis is blocked by CEP-1347 (KT 7515), a novel inhibitor of the JNK signaling pathway. J Neurosci. 1998;18(1):104–11. doi: 10.1523/JNEUROSCI.18-01-00104.1998. Epub 1998/01/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Roux PP, Dorval G, Boudreau M, Angers-Loustau A, Morris SJ, Makkerh J, et al. K252a and CEP1347 are neuroprotective compounds that inhibit mixed-lineage kinase-3 and induce activation of Akt and ERK. J Biol Chem. 2002;277(51):49473–80. doi: 10.1074/jbc.M203428200. Epub 2002/10/22. [DOI] [PubMed] [Google Scholar]
- [35].Xu Z, Maroney AC, Dobrzanski P, Kukekov NV, Greene LA. The MLK family mediates c-Jun N-terminal kinase activation in neuronal apoptosis. Molecular and cellular biology. 2001;21(14):4713–24. doi: 10.1128/MCB.21.14.4713-4724.2001. Epub 2001/06/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang LH, Besirli CG, Johnson EM., Jr. Mixed-lineage kinases: a target for the prevention of neurodegeneration. Annual review of pharmacology and toxicology. 2004;44:451–74. doi: 10.1146/annurev.pharmtox.44.101802.121840. Epub 2004/01/28. [DOI] [PubMed] [Google Scholar]
- [37].Apostol BL, Simmons DA, Zuccato C, Illes K, Pallos J, Casale M, et al. CEP-1347 reduces mutant huntingtin-associated neurotoxicity and restores BDNF levels in R6/2 mice. Mol Cell Neurosci. 2008 doi: 10.1016/j.mcn.2008.04.007. [DOI] [PubMed] [Google Scholar]
- [38].Chopra V, Fox JH, Lieberman G, Dorsey K, Matson W, Waldmeier P, et al. A small-molecule therapeutic lead for Huntington’s disease: preclinical pharmacology and efficacy of C2-8 in the R6/2 transgenic mouse. Proc Natl Acad Sci U S A. 2007;104(42):16685–9. doi: 10.1073/pnas.0707842104. Epub 2007/10/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, Lurz R, et al. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat Struct Mol Biol. 2008;15(6):558–66. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- [40].Rich T, Assier E, Skepper J, Segard HB, Allen RL, Charron D, et al. Disassembly of nuclear inclusions in the dividing cell--a novel insight into neurodegeneration. Hum Mol Genet. 1999;8(13):2451–9. doi: 10.1093/hmg/8.13.2451. Epub 1999/11/11. [DOI] [PubMed] [Google Scholar]
- [41].Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends in cell biology. 2000;10(12):524–30. doi: 10.1016/s0962-8924(00)01852-3. Epub 2000/12/21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure 1. Representative data for EGCG treatment. A-E, Representative images used to count visible inclusions in 14A2.6 cells treated with EGCG.
Supplemental figure 3. Representative data for CEP-1347 treatment. A-E, Representative images used to count visible inclusions in 14A2.6 cells treated with CEP-1347.
Supplemental figure 4. Representative video illustrating the lag phase of inclusion body formation in 14A2.6 cells treated with CEP-1347. Treatment with CEP-1347 does not appear to affect the rate of inclusion body formation.
Supplemental figue 2. Representative video illustrating the lag phase of inclusion body formation in 14A2.6 cells treated with EGCG. Treatment with EGCG does not appear to affect the rate of inclusion body formation.
Supplemental figure 5. Cystamine decreases oligomers and SDS-insoluble Htt in a stably expressing truncated Httex1p model of HD. A, Cystamine appears to decrease the levels of soluble fibrillar oligomers in 14A2.6 cells by SDS-AGE analysis. B, Cystamine decreases the amount of SDS-insoluble Htt in 14A2.6 cells by filter-retardation assay.